SpSrtA-Catalyzed Isopeptide Ligation on Lysine Residues
Abstract
:1. Introduction
2. Materials and Methods
2.1. Reagents and Materials
2.2. Recombinant Protein Expression and Purification
2.3. Substrate LPAT-isoK Prediction and Molecular Docking
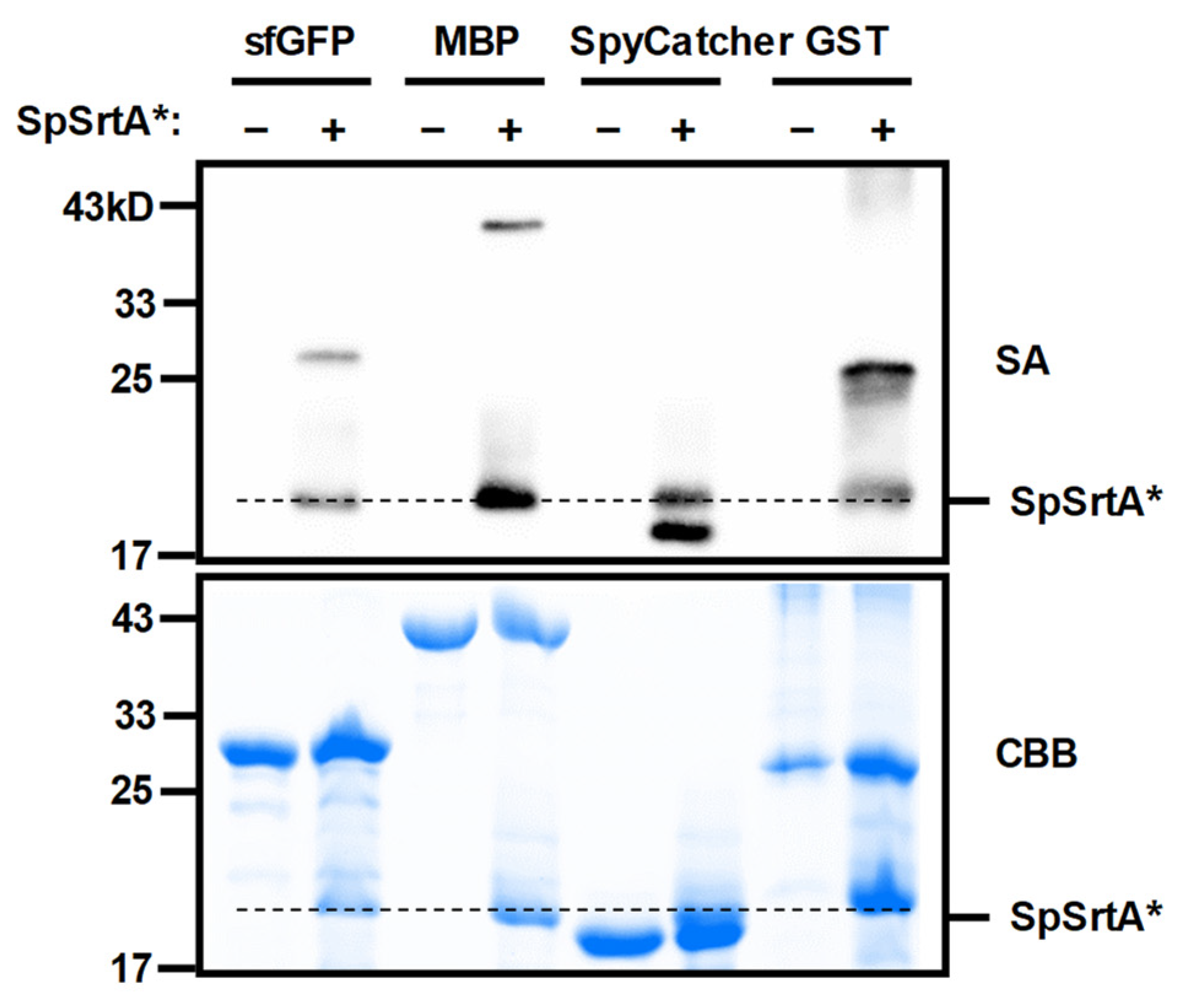
2.4. Western Blot Analysis of Protein Ligation Products
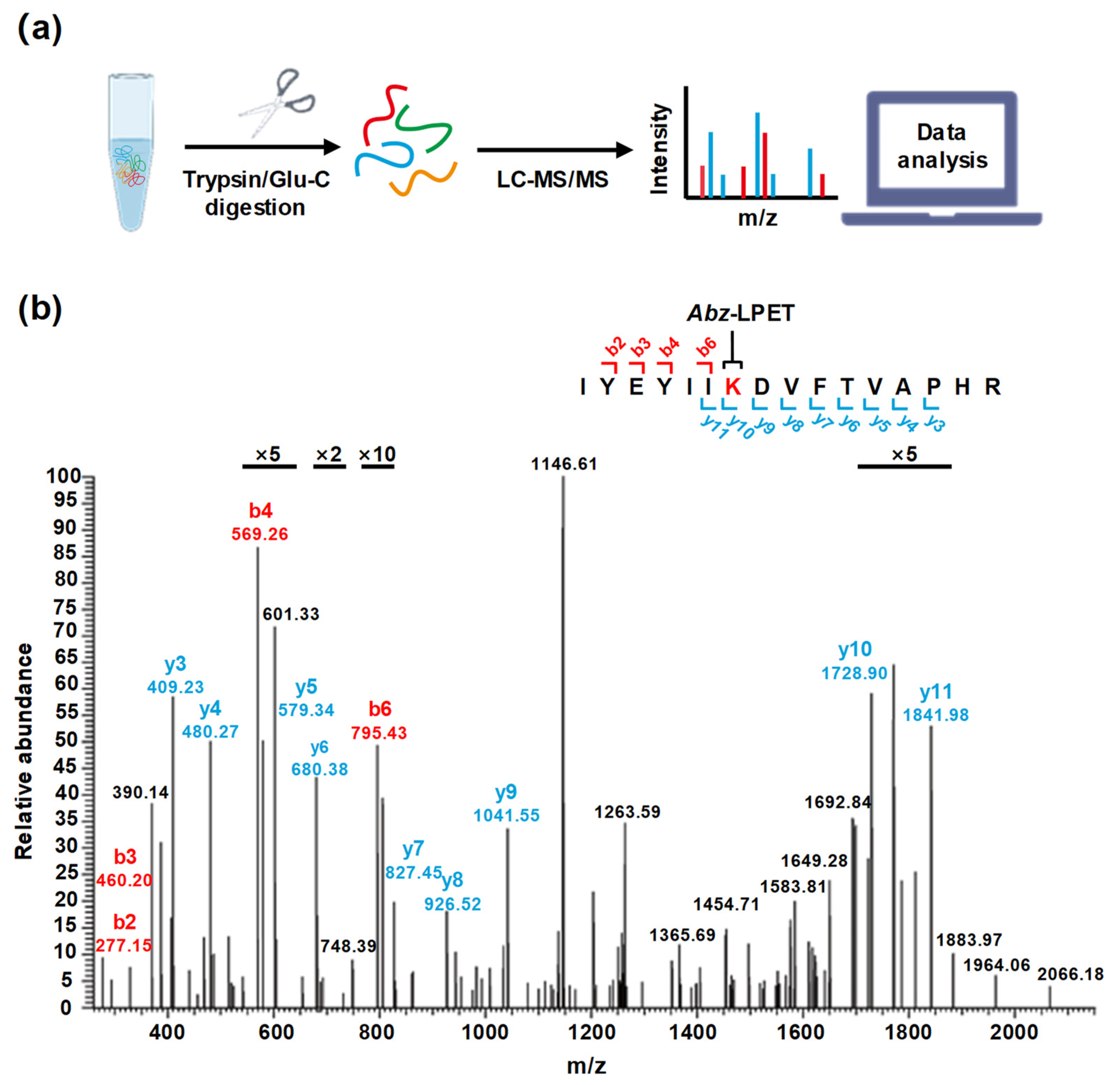
2.5. LC-MS/MS and Data Analysis for Identification of Modification Sites
2.6. Evaluation of the Efficiency of Lysine-Containing Peptide Tag
2.7. Surface Modification of Living Microorganisms
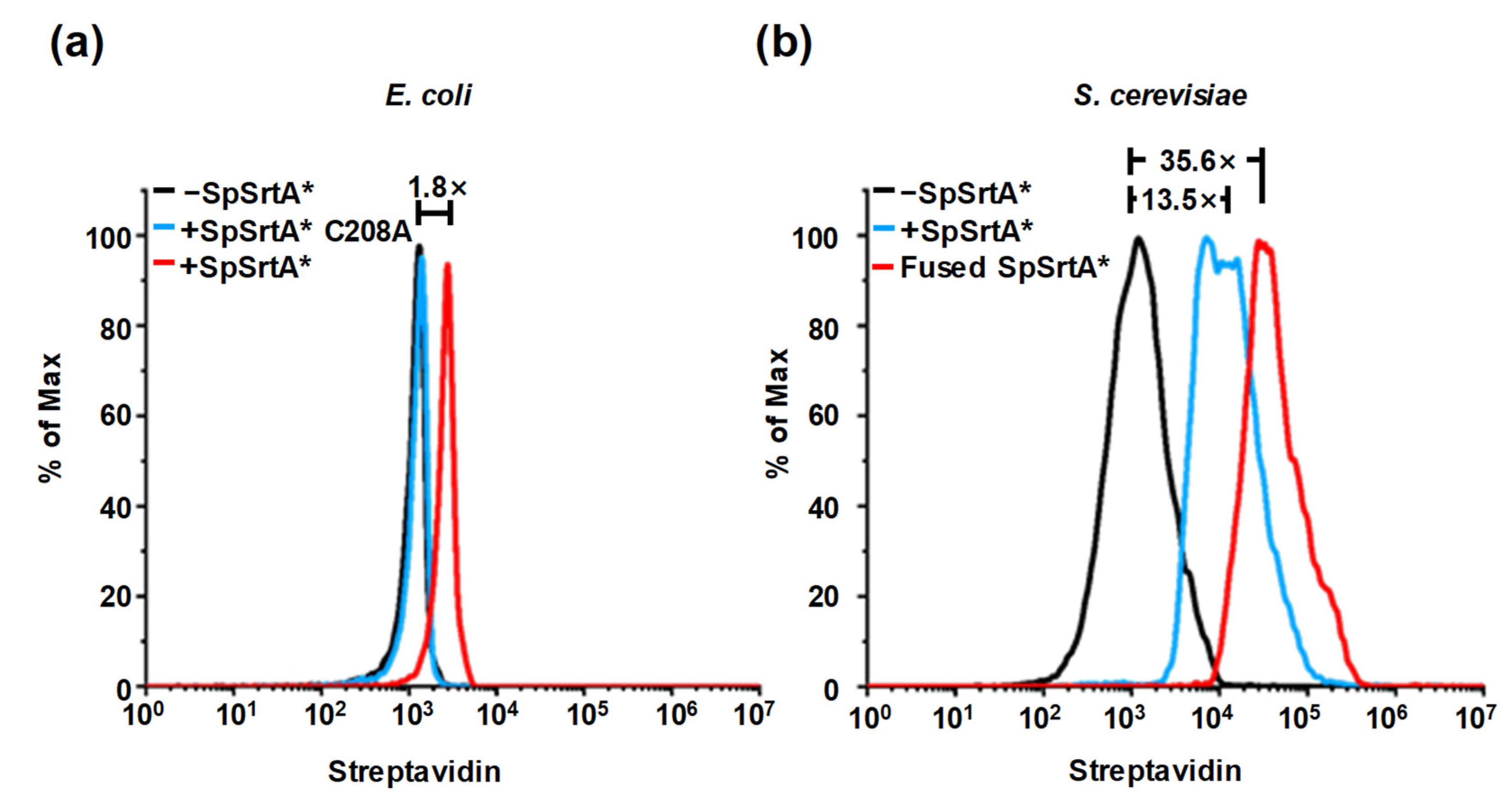
2.8. Flow Cytometry Analysis of Yeast Surface Display Proteins
2.9. Generation of Random Mutant Libraries
2.10. Streptavidin Blot Characterization of SpSrtA* Labeling on Yeast Surface
3. Results
3.1. Lysine Residue as a Nucleophilic Receptor for SpSrtA*
3.2. Identification of Lysine Residue Modification Sites in SpSrtA*
3.3. Isopeptide Ligation with Diverse Proteins Lacking N-Terminal Glycine
3.4. Screening for Optimal Lysine-Containing Peptide Tag
3.5. Application of SML for Labeling of Living Microorganism Surfaces
3.6. Directed Evolution of SpSrtA* for Enhanced Surface Protein Labeling
4. Discussion
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hoyt, E.A.; Cal, P.M.S.D.; Oliveira, B.L.; Bernardes, G.J.L. Contemporary approaches to site-selective protein modification. Nat. Rev. Chem. 2019, 3, 147–171. [Google Scholar] [CrossRef]
- Zou, Z.; Nöth, M.; Jakob, F.; Schwaneberg, U. Designed Streptococcus pyogenes Sortase A Accepts Branched Amines as Nucleophiles in Sortagging. Bioconjug. Chem. 2020, 31, 2476–2481. [Google Scholar] [CrossRef] [PubMed]
- Krall, N.; da Cruz, F.P.; Boutureira, O.; Bernardes, G.J. Site-selective protein-modification chemistry for basic biology and drug development. Nat. Chem. 2016, 8, 103–113. [Google Scholar] [CrossRef]
- Spicer, C.D.; Davis, B.G. Selective chemical protein modification. Nat. Commun. 2014, 5, 4740. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Boker, A.; Glebe, U. Broadening the scope of sortagging. RSC Adv. 2019, 9, 4700–4721. [Google Scholar] [CrossRef]
- Tsukiji, S.; Nagamune, T. Sortase-Mediated Ligation: A Gift from Gram-Positive Bacteria to Protein Engineering. Chembiochem 2009, 10, 787–798. [Google Scholar] [CrossRef] [PubMed]
- Pishesha, N.; Ingram, J.R.; Ploegh, H.L. Sortase A: A Model for Transpeptidation and Its Biological Applications. Annu. Rev. Cell Dev. Biol. 2018, 34, 163–188. [Google Scholar] [CrossRef]
- Spirig, T.; Weiner, E.M.; Clubb, R.T. Sortase enzymes in Gram-positive bacteria. Mol. Microbiol. 2011, 82, 1044–1059. [Google Scholar] [CrossRef]
- Chen, I.; Dorr, B.M.; Liu, D.R. A general strategy for the evolution of bond-forming enzymes using yeast display. Proc. Natl. Acad. Sci. USA 2011, 108, 11399–11404. [Google Scholar] [CrossRef]
- Pasqual, G.; Chudnovskiy, A.; Tas, J.M.J.; Agudelo, M.; Schweitzer, L.D.; Cui, A.; Hacohen, N.; Victora, G.D. Monitoring T cell-dendritic cell interactions in vivo by intercellular enzymatic labelling. Nature 2018, 553, 496–500. [Google Scholar] [CrossRef]
- Koniev, O.; Wagner, A. Developments and recent advancements in the field of endogenous amino acid selective bond forming reactions for bioconjugation. Chem. Soc. Rev. 2015, 44, 5495–5551. [Google Scholar] [CrossRef] [PubMed]
- Hacker, S.M.; Backus, K.M.; Lazear, M.R.; Forli, S.; Correia, B.E.; Cravatt, B.F. Global profiling of lysine reactivity and ligandability in the human proteome. Nat. Chem. 2017, 9, 1181–1190. [Google Scholar] [CrossRef]
- Fierer, J.O.; Veggiani, G.; Howarth, M. SpyLigase peptide-peptide ligation polymerizes affibodies to enhance magnetic cancer cell capture. Proc. Natl. Acad. Sci. USA 2014, 111, E1176–E1181. [Google Scholar] [CrossRef]
- Buldun, C.M.; Jean, J.X.; Bedford, M.R.; Howarth, M. SnoopLigase Catalyzes Peptide-Peptide Locking and Enables Solid-Phase Conjugate Isolation. J. Am. Chem. Soc. 2018, 140, 3008–3018. [Google Scholar] [CrossRef] [PubMed]
- Savoca, M.P.; Tonoli, E.; Atobatele, A.G.; Verderio, E.A. Biocatalysis by Transglutaminases: A Review of Biotechnological Applications. Micromachines 2018, 9, 562. [Google Scholar] [CrossRef] [PubMed]
- Roux, K.J.; Kim, D.I.; Raida, M.; Burke, B. A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J. Cell Biol. 2012, 196, 801–810. [Google Scholar] [CrossRef]
- Dasgupta, S.; Samantaray, S.; Sahal, D.; Roy, R.P. Isopeptide ligation catalyzed by quintessential sortase A: Mechanistic cues from cyclic and branched oligomers of indolicidin. J. Biol. Chem. 2011, 286, 23996–24006. [Google Scholar] [CrossRef]
- Bellucci, J.J.; Bhattacharyya, J.; Chilkoti, A. A noncanonical function of sortase enables site-specific conjugation of small molecules to lysine residues in proteins. Angew. Chem. Int. Ed. 2015, 54, 441–445. [Google Scholar] [CrossRef]
- Kruger, R.G.; Otvos, B.; Frankel, B.A.; Bentley, M.; Dostal, P.; McCafferty, D.G. Analysis of the substrate specificity of the Staphylococcus aureus sortase transpeptidase SrtA. Biochemistry 2004, 43, 1541–1551. [Google Scholar] [CrossRef]
- Schmohl, L.; Bierlmeier, J.; von Kügelgen, N.; Kurz, L.; Reis, P.; Barthels, F.; Mach, P.; Schutkowski, M.; Freund, C.; Schwarzer, D. Identification of sortase substrates by specificity profiling. Bioorg. Med. Chem. 2017, 25, 5002–5007. [Google Scholar] [CrossRef]
- Nikghalb, K.D.; Horvath, N.M.; Prelesnik, J.L.; Banks, O.G.B.; Filipov, P.A.; Row, R.D.; Roark, T.J.; Antos, J.A.-O. Expanding the Scope of Sortase-Mediated Ligations by Using Sortase Homologues. Chembiochem 2018, 19, 185–195. [Google Scholar] [CrossRef]
- Johnson, D.A.; Piper, I.M.; Vogel, B.A.; Jackson, S.N.; Svendsen, J.E.; Kodama, H.M.; Lee, D.E.; Lindblom, K.M.; McCarty, J.; Antos, J.M.; et al. Structures of Streptococcus pyogenes class A sortase in complex with substrate and product mimics provide key details of target recognition. J. Biol. Chem. 2022, 298, 102446. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Eberhardt, J.; Santos-Martins, D.; Tillack, A.F.; Forli, S. AutoDock Vina 1.2.0: New Docking Methods, Expanded Force Field, and Python Bindings. J. Chem. Inf. Model. 2021, 61, 3891–3898. [Google Scholar] [CrossRef]
- Gibson, D.G.; Young, L.; Chuang, R.-Y.; Venter, J.C.; Hutchison, C.A.; Smith, H.O. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 2009, 6, 343–345. [Google Scholar] [CrossRef] [PubMed]
- Lam, S.S.; Martell, J.D.; Kamer, K.J.; Deerinck, T.J.; Ellisman, M.H.; Mootha, V.K.; Ting, A.Y. Directed evolution of APEX2 for electron microscopy and proximity labeling. Nat. Methods 2015, 12, 51–54. [Google Scholar] [CrossRef] [PubMed]
- Martell, J.D.; Yamagata, M.; Deerinck, T.J.; Phan, S.; Kwa, C.G.; Ellisman, M.H.; Sanes, J.R.; Ting, A.Y. A split horseradish peroxidase for the detection of intercellular protein–protein interactions and sensitive visualization of synapses. Nat. Biotechnol. 2016, 34, 774–780. [Google Scholar] [CrossRef]
- Li, Y.; Liu, K.; Zhou, Y.; Yang, J.; Zou, P. Protocol for Proximity-Dependent Proteomic Profiling in Yeast Cells by APEX and Alk-Ph Probe. STAR Protoc. 2020, 1, 100137. [Google Scholar] [CrossRef]
- Olsen, J.V.; Ong, S.-E.; Mann, M. Trypsin Cleaves Exclusively C-terminal to Arginine and Lysine Residues*. Mol. Cell Proteom. 2004, 3, 608–614. [Google Scholar] [CrossRef]
- Macias, L.A.; Santos, I.C.; Brodbelt, J.S. Ion Activation Methods for Peptides and Proteins. Anal. Chem. 2020, 92, 227–251. [Google Scholar] [CrossRef]
- Morgan, H.E.; Turnbull, W.B.; Webb, M.E. Challenges in the use of sortase and other peptide ligases for site-specific protein modification. Chem. Soc. Rev. 2022, 51, 4121–4145. [Google Scholar] [CrossRef]
- Ruiz, S.A.-O.; van’t Klooster, J.S.; Bianchi, F.; Poolman, B. Growth Inhibition by Amino Acids in Saccharomyces cerevisiae. Microorganisms 2021, 9, 7. [Google Scholar] [CrossRef] [PubMed]
- Fukuchi, S.; Nishikawa, K. Protein surface amino acid compositions distinctively differ between thermophilic and mesophilic bacteria11Edited by F. E. Cohen. J. Mol. Biol. 2001, 309, 835–843. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Moysey, R.; Molloy, P.E.; Vuidepot, A.-L.; Mahon, T.; Baston, E.; Dunn, S.; Liddy, N.; Jacob, J.; Jakobsen, B.K.; et al. Directed evolution of human T-cell receptors with picomolar affinities by phage display. Nat. Biotechnol. 2005, 23, 349–354. [Google Scholar] [CrossRef] [PubMed]
- Löfblom, J. Bacterial display in combinatorial protein engineering. Biotechnol. J. 2011, 6, 1115–1129. [Google Scholar] [CrossRef]
- Podracky, C.J.; An, C.; DeSousa, A.; Dorr, B.M.; Walsh, D.M.; Liu, D.R. Laboratory evolution of a sortase enzyme that modifies amyloid-beta protein. Nat. Chem. Biol. 2021, 17, 317–325. [Google Scholar] [CrossRef]
- Pluchinsky, A.J.; Wackelin, D.J.; Huang, X.; Arnold, F.H.; Mrksich, M. High Throughput Screening with SAMDI Mass Spectrometry for Directed Evolution. J. Am. Chem. Soc. 2020, 142, 19804–19808. [Google Scholar] [CrossRef] [PubMed]
- Mao, H.; Hart, S.A.; Schink, A.; Pollok, B.A. Sortase-Mediated Protein Ligation: A New Method for Protein Engineering. J. Am. Chem. Soc. 2004, 126, 2670–2671. [Google Scholar] [CrossRef]
- Antos, J.M.; Miller, G.M.; Grotenbreg, G.M.; Ploegh, H.L. Lipid Modification of Proteins through Sortase-Catalyzed Transpeptidation. J. Am. Chem. Soc. 2008, 130, 16338–16343. [Google Scholar] [CrossRef]
- Samantaray, S.; Marathe, U.; Dasgupta, S.; Nandicoori, V.K.; Roy, R.P. Peptide−Sugar Ligation Catalyzed by Transpeptidase Sortase: A Facile Approach to Neoglycoconjugate Synthesis. J. Am. Chem. Soc. 2008, 130, 2132–2133. [Google Scholar] [CrossRef]
- Parthasarathy, R.; Subramanian, S.; Boder, E.T. Sortase A as a Novel Molecular “Stapler” for Sequence-Specific Protein Conjugation. Bioconjug. Chem. 2007, 18, 469–476. [Google Scholar] [CrossRef]
- Chan, L.; Cross, H.F.; She, J.K.; Cavalli, G.; Martins, H.F.; Neylon, C. Covalent attachment of proteins to solid supports and surfaces via Sortase-mediated ligation. PLoS ONE 2007, 2, e1164. [Google Scholar] [CrossRef] [PubMed]
- Pritz, S.; Wolf, Y.; Kraetke, O.; Klose, J.; Bienert, M.; Beyermann, M. Synthesis of Biologically Active Peptide Nucleic Acid−Peptide Conjugates by Sortase-Mediated Ligation. J. Org. Chem. 2007, 72, 3909–3912. [Google Scholar] [CrossRef] [PubMed]
- Dennler, P.; Chiotellis, A.; Fischer, E.; Brégeon, D.; Belmant, C.; Gauthier, L.; Lhospice, F.; Romagne, F.; Schibli, R. Transglutaminase-Based Chemo-Enzymatic Conjugation Approach Yields Homogeneous Antibody–Drug Conjugates. Bioconjug. Chem. 2014, 25, 569–578. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Stefano, J.E.; Manning, C.; Kyazike, J.; Chen, B.; Gianolio, D.A.; Park, A.; Busch, M.; Bird, J.; Zheng, X.; et al. Site-Specific Antibody–Drug Conjugation through Glycoengineering. Bioconjug. Chem. 2014, 25, 510–520. [Google Scholar] [CrossRef] [PubMed]
- Drake, P.M.; Albers, A.E.; Baker, J.; Banas, S.; Barfield, R.M.; Bhat, A.S.; de Hart, G.W.; Garofalo, A.W.; Holder, P.; Jones, L.C.; et al. Aldehyde Tag Coupled with HIPS Chemistry Enables the Production of ADCs Conjugated Site-Specifically to Different Antibody Regions with Distinct in Vivo Efficacy and PK Outcomes. Bioconjug. Chem. 2014, 25, 1331–1341. [Google Scholar] [CrossRef] [PubMed]
- Ge, Y.; Chen, L.; Liu, S.; Zhao, J.; Zhang, H.; Chen, P.R. Enzyme-Mediated Intercellular Proximity Labeling for Detecting Cell-Cell Interactions. J. Am. Chem. Soc. 2019, 141, 1833–1837. [Google Scholar] [CrossRef]
- Liu, Q.; Zheng, J.; Sun, W.; Huo, Y.; Zhang, L.; Hao, P.; Wang, H.; Zhuang, M. A proximity-tagging system to identify membrane protein-protein interactions. Nat. Methods 2018, 15, 715–722. [Google Scholar] [CrossRef]
- Bechtel, T.J.; Reyes-Robles, T.; Fadeyi, O.O.; Oslund, R.C. Strategies for monitoring cell-cell interactions. Nat. Chem. Biol. 2021, 17, 641–652. [Google Scholar] [CrossRef]
- Chen, D.; Guo, J.; Li, A.; Sun, C.; Lin, H.; Lin, H.; Yang, C.; Wang, W.; Gao, J. Metabolic fluorine labeling and hotspot imaging of dynamic gut microbiota in mice. Sci. Adv. 2023, 9, eabg6808. [Google Scholar] [CrossRef]



| Primer | Sequence from 5′ to 3′ |
|---|---|
| ep-F | TGGAGGAGGCTCTGGTGCTAGC |
| ep-R | TAGTCTGGAACGTCGTATGGGTAGGATCC |
| Homo-F | CAAGGTCTGCAGGCTAGTGGTGGAG-GAGGCTCTGGTGCTAGC |
| Homo-R | TGTTGTTATCAGATCTCGAGCTATTAGGCATAGTCTGGAACGTCGTATGGGTAGGATCC |
| Position | Sequence | Calculated Mass (Da) | Precursor Mass (Da) |
|---|---|---|---|
| 181 | IYEYIIKDVFTVAPHR | 2522.32 | 2522.32 |
| 220 | IIVKGELK | 1457.85 | 1457.85 |
| 224 | GELKTEYDFDKAPADVLK | 2597.29 | 2597.29 |
| 181 | YIIKDVFTVAPHRVDVIDDTAGLKE | 3372.76 | 3372.76 |
| 201 | YIIKDVFTVAPHRVDVIDDTAGLKE | 3372.76 | 3372.76 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, J.; Chu, T.; Hao, J.; Lin, L. SpSrtA-Catalyzed Isopeptide Ligation on Lysine Residues. Microorganisms 2024, 12, 179. https://doi.org/10.3390/microorganisms12010179
Wu J, Chu T, Hao J, Lin L. SpSrtA-Catalyzed Isopeptide Ligation on Lysine Residues. Microorganisms. 2024; 12(1):179. https://doi.org/10.3390/microorganisms12010179
Chicago/Turabian StyleWu, Jiajia, Tianyu Chu, Jian Hao, and Liang Lin. 2024. "SpSrtA-Catalyzed Isopeptide Ligation on Lysine Residues" Microorganisms 12, no. 1: 179. https://doi.org/10.3390/microorganisms12010179





