A Novel Strain of Bacillus cereus with a Strong Antagonistic Effect Specific to Sclerotinia and Its Genomic and Transcriptomic Analysis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.1.1. Soil Samples
2.1.2. Fungus Strains and Oilseed Rape Variety
2.1.3. Culture Medium
2.2. Isolation and Screening of Bacteria Antagonizing Fungal Pathogens
2.3. Characterization of the Selected Bacteria
2.3.1. Physiological and Biochemical Characteristics of Strain HF10
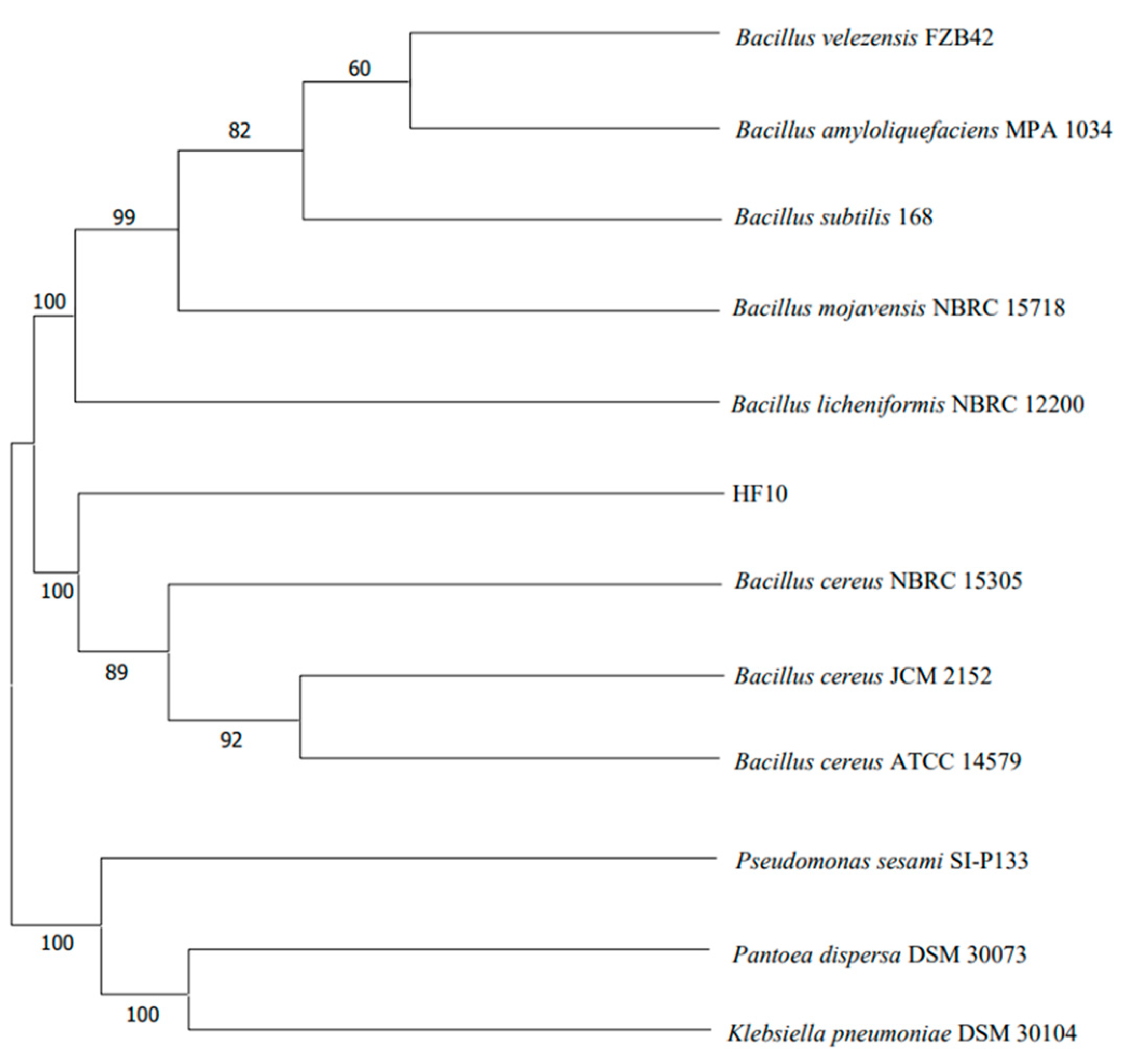
2.3.2. 16S rDNA Sequencing and Phylogenetic Tree Construction of the HF10 Strain
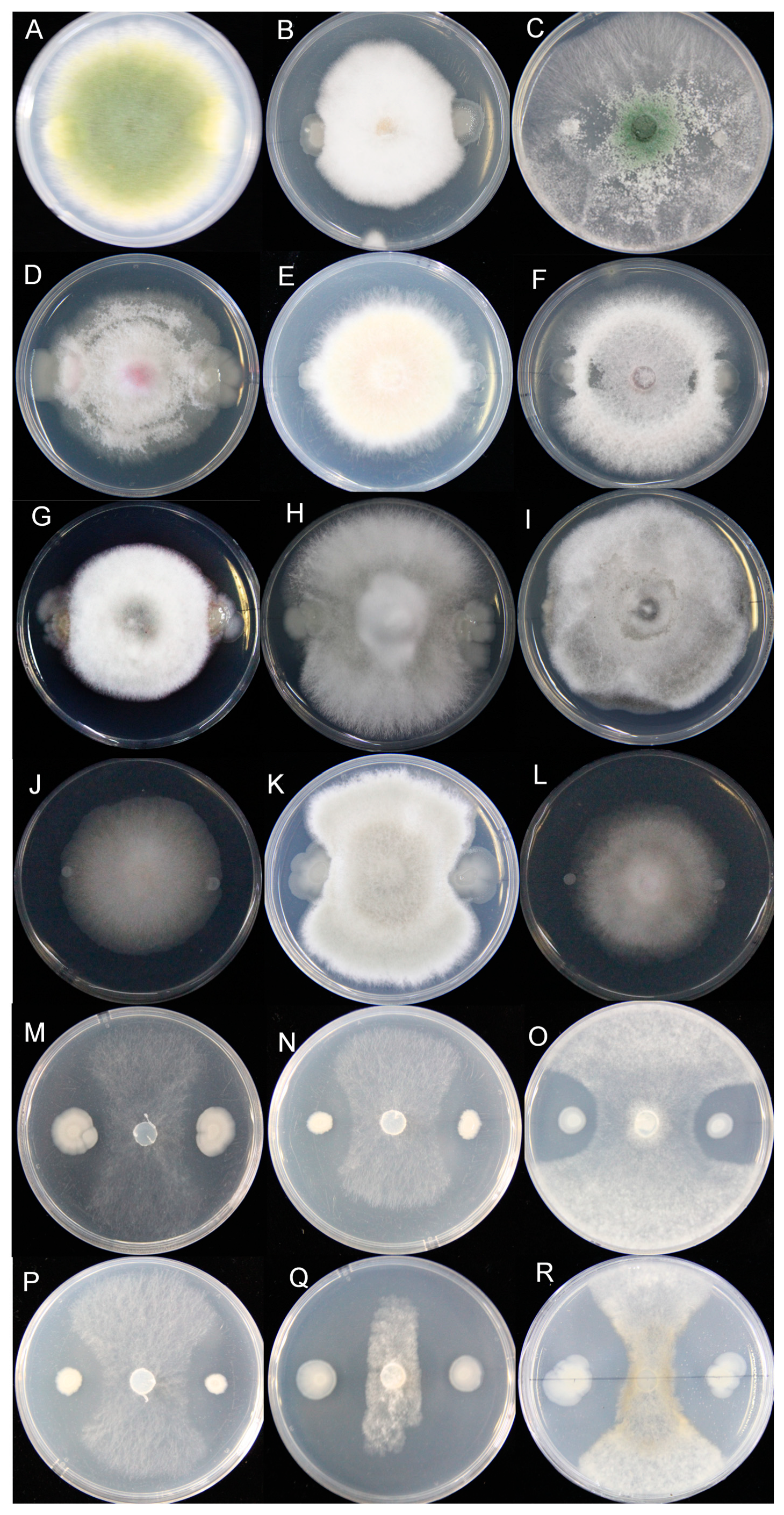
2.4. Determination of the Antifungal Activity Spectrum of the HF10 Strain
2.5. Test of the Effect of the HF10 Strain on the S. sclerotiorum Infection of Oilseed Rape In Vitro
2.6. Effect of the HF10 Strain on the S. sclerotiorum Infection of Oilseed Rape Leaves In Vivo
2.7. Colonization Ability of the HF10 Strain on Oilseed Rape Leaves
2.8. Investigation of the Antifungal Substances of the HF10 Strain
2.8.1. Isolation and Detection of Antimicrobial Lipopeptides
2.8.2. Detection of Siderophores
2.8.3. β-1,3 Glucanase Assay
2.9. Multiomics Analysis of the HF10 Strain and Its Loss of the Antifunctional Function Mutant Y11
2.9.1. Whole-Genome Sequence Analysis of HF10
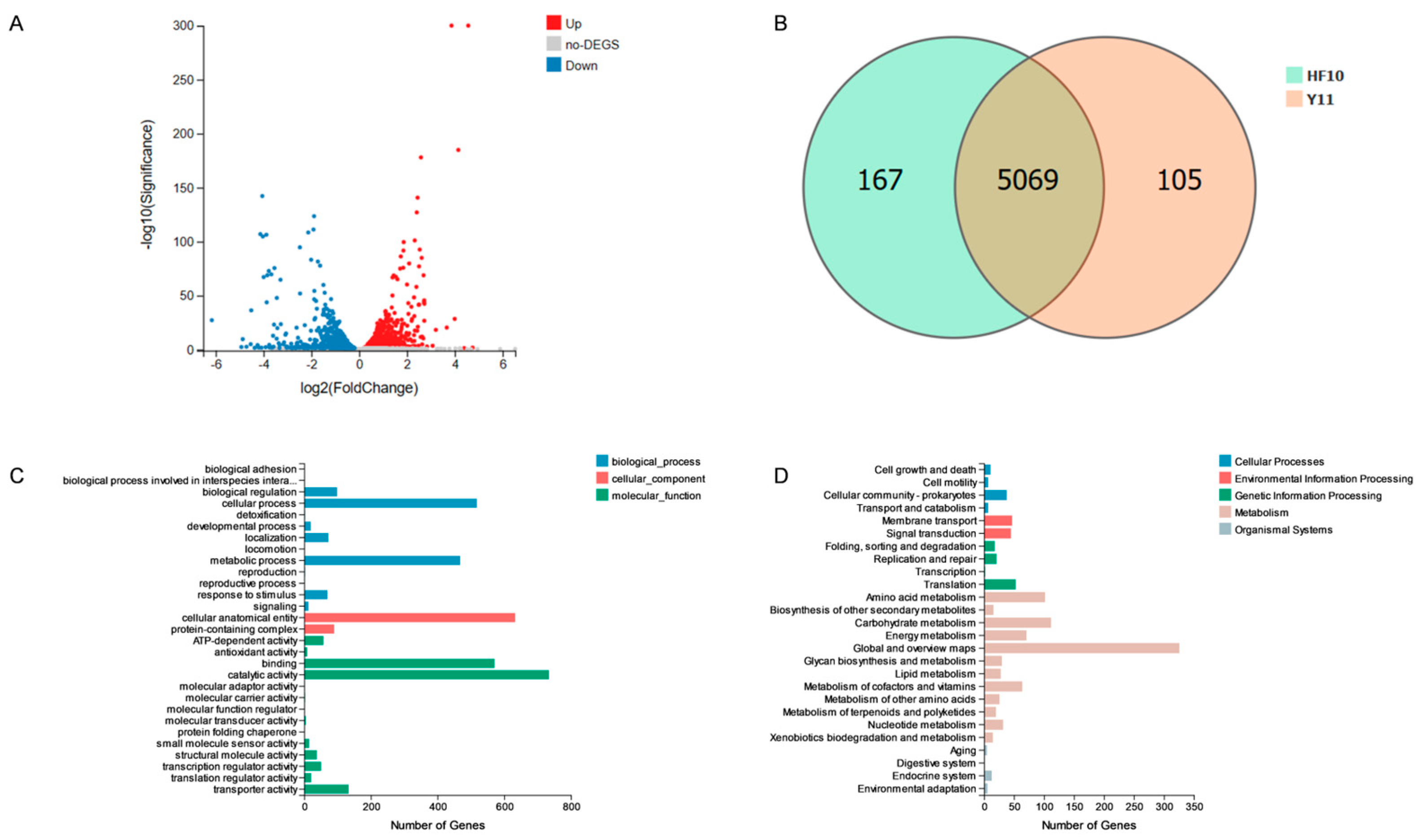
2.9.2. Transcriptome Analysis of HF10 and Y11
2.10. Statistical Analysis
3. Results
3.1. Isolation and Screening of the Antagonistic Bacterium Bacillus cereus HF10
3.2. Identification of Antagonistic Strains
3.2.1. Physiological and Biochemical Characteristics
3.2.2. Phylogenetic Tree
3.3. Determination of the Antifungal Activity Spectrum of the HF10 Strain
3.4. The HF10 Strain Strongly Inhibited S. sclerotiorum Infection in Oilseed Rape
3.5. The Colonization of Oilseed Rape by the HF10 Strain
3.6. Investigation of the Antibacterial Substances of the HF10 Strain
3.7. Genome Assembly of HF10 De Novo
3.7.1. Genomic Features of the HF10 Strain
3.7.2. Transcriptome Results for the HF10 and Y11 Strains
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Niedbala, G.; Piekutowska, M.; Weres, J.; Korzeniewicz, R.; Witaszek, K.; Adamski, M.; Pilarski, K.; Czechowska-Kosacka, A.; Krysztofiak-Kaniewska, A. Application of artificial neural networks for yield modeling of winter rapeseed based on combined quantitative and qualitative data. Agronomy 2019, 9, 781. [Google Scholar] [CrossRef]
- Adamczyk-Chauvat, K.; Delaunay, S.; Vannier, A.; François, C.; Thomas, G.; Eber, F.; Lodé, M.; Gilet, M.; Huteau, V.; Morice, J.; et al. Gene introgression in weeds depends on initial gene location in the crop: Brassica napus-Raphanus raphanistrum Model. Genetics 2017, 206, 1361–1372. [Google Scholar] [CrossRef]
- Zhao, D.H.; Cao, Z.W.; Chen, L.J.; Zhang, G.X.; Zhu, Y.Z.; Han, J. Optimising the effect of nitrogen on winter oilseed rape grain yield in China: A meta-analysis. Eur. J. Agron. 2023, 144, 10. [Google Scholar] [CrossRef]
- Hu, X.J.; Roberts, D.P.; Xie, L.H.; Yu, C.B.; Li, Y.S.; Qi, L.; Hu, L.; Zhang, Y.B.; Liao, X. Biological control of Sclerotinia disease by Aspergillus sp. on oilseed rape in the field. Biocontrol Sci. Technol. 2016, 26, 1526–1537. [Google Scholar] [CrossRef]
- Boland, G.J. Epidemiology of Sclerotinia stem rot of soybean in ontario. Phytopathology 1988, 78, 1241–1245. [Google Scholar] [CrossRef]
- Kohn, L.M. A monographic revision of the genus Sclerotinia. Mycotaxon 1979, 9, 365–444. [Google Scholar]
- Boland, G.J.; Hall, R. Index of plant hosts of Sclerotinia sclerotiorum. Can. J. Plant Pathol. 1994, 16, 93–108. [Google Scholar] [CrossRef]
- Hu, X.J.; Roberts, D.P.; Xie, L.H.; Maul, J.E.; Yu, C.B.; Li, Y.S.; Zhang, S.J.; Liao, X. Bacillus megaterium A6 suppresses Sclerotinia sclerotiorum on oilseed rape in the field and promotes oilseed rape growth. Crop Prot. 2013, 52, 151–158. [Google Scholar] [CrossRef]
- Barro, J.P.; Meyer, M.C.; Godoy, C.V.; Dias, A.R.; Utiamada, C.M.; Jaccoud, D.D.; Wruck, D.S.M.; Borges, E.P.; Siqueri, F.; Juliatti, F.C.; et al. Performance and profitability of fungicides for managing soybean white mold: A 10-year summary of cooperative trials. Plant Dis. 2019, 103, 2212–2220. [Google Scholar] [CrossRef]
- Cao, S.; Jiang, B.X.; Yang, G.G.; Pan, G.X.; Pan, Y.M.; Chen, F.X.; Gao, Z.M.; Dai, Y.L. Isolation and evaluation of Bacillus subtilis RSS-1 as a potential biocontrol agent against Sclerotinia sclerotiorum on oilseed rape. Eur. J. Plant Pathol. 2023, 166, 9–25. [Google Scholar] [CrossRef]
- O’Sullivan, C.A.A.; Belt, K.; Thatcher, L.F.F. Tackling control of a cosmopolitan phytopathogen: Sclerotinia. Front. Plant Sci. 2021, 12, 18. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Pan, Y.; Zhang, H.; Li, X.; Dai, Y.; Cao, S.; Gao, Z. Detection and characterization of carbendazim resistance in Sclerotinia sclerotiorum isolates from oilseed rape in Anhui Province of China. Genet. Mol. Res. 2015, 14, 16627–16638. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.R.; ElKomy, M.H.; Ibrahim, Y.E.; Hamad, Y.K.; Molan, Y.Y.; Saleh, A.A. Organic management of tomato fusarium wilt using a native Bacillus subtilis strain and compost combination in saudi arabia. Int. J. Agric. Biol. 2020, 23, 1003–1012. [Google Scholar]
- Delfosse, E.S. Risk and ethics in biological control. Biol. Control 2005, 35, 319–329. [Google Scholar] [CrossRef]
- Ye, G.Y.; Xiao, Q.; Chen, M.; Chen, X.X.; Yuan, Z.J.; Stanley, D.W.; Hu, C. Tea: Biological control of insect and mite pests in China. Biol. Control 2014, 68, 73–91. [Google Scholar] [CrossRef]
- Pearson, D.E.; Callaway, R.M. Indirect nontarget effects of host-specific biological control agents: Implications for biological control. Biol. Control 2005, 35, 288–298. [Google Scholar] [CrossRef]
- Fokkema, N.J. Biological control of fungal plant diseases. Entomophaga 1996, 41, 333–342. [Google Scholar] [CrossRef]
- Bonaterra, A.; Badosa, E.; Daranas, N.; Francés, J.; Roselló, G.; Montesinos, E. Bacteria as biological control agents of plant diseases. Microorganisms 2022, 10, 1759. [Google Scholar] [CrossRef]
- Hu, X.J.; Roberts, D.P.; Xie, L.H.; Maul, J.E.; Yu, C.B.; Li, Y.S.; Jiang, M.L.; Liao, X.S.; Che, Z.; Liao, X. Formulations of Bacillus subtilis BY-2 suppress Sclerotinia sclerotiorum on oilseed rape in the field. Biol. Control 2014, 70, 54–64. [Google Scholar] [CrossRef]
- Smolinska, U.; Kowalska, B. Biological control of the soil-borne fungal pathogen Sclerotinia sclerotiorum—A review. J. Plant Pathol. 2018, 100, 1–12. [Google Scholar] [CrossRef]
- Siddiqui, S.; Siddiqui, Z.A.; Ahmad, I. Evaluation of fluorescent Pseudomonads and Bacillus isolates for the biocontrol of a wilt disease complex of pigeonpea. World J. Microbiol. Biotechnol. 2005, 21, 729–732. [Google Scholar] [CrossRef]
- Zhao, S.; Chen, X.; Deng, S.P.; Dong, X.N.; Song, A.P.; Yao, J.J.; Fang, W.M.; Chen, F.D. The effects of fungicide, soil fumigant, bio-organic fertilizer and their combined application on chrysanthemum Fusarium wilt controlling, soil enzyme activities and microbial properties. Molecules 2016, 21, 15. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.S.; Qin, L.; Roberts, D.P.; Hu, X.J.; Xie, L.H.; Gu, C.M.; Liao, X.S.; Han, P.P.; Liao, X. Biological fertilizer containing Bacillus subtilis BY-2 for control of Sclerotinia sclerotiorum on oilseed rape. Crop Prot. 2020, 138, 3. [Google Scholar] [CrossRef]
- Perry, E.K.; Meirelles, L.A.; Newman, D.K. From the soil to the clinic: The impact of microbial secondary metabolites on antibiotic tolerance and resistance. Nat. Rev. Microbiol. 2022, 20, 129–142. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Cao, X.; Chen, R.; Wu, H.; Wang, J.; Ma, G.; Bao, Z.; Peng, H. Screening and identification of a marine bacteria with high efficiency against Neurosporacrasa sp. and Trichoderma viride. J. Agric. Sci. 2019, 50, 1519–1526. [Google Scholar]
- Li, S.; Ma, J.; Li, S.; Chen, F.; Song, C.; Zhang, H.; Jiang, M.; Shen, N. Comparative transcriptome analysis unravels the response mechanisms of fusarium oxysporum f.sp. cubense to a biocontrol agent, Pseudomonas aeruginosa Gxun-2. Int. J. Mol. Sci. 2022, 23, 15432. [Google Scholar] [CrossRef]
- Ma, M.; Feng, J.; Li, R.; Chen, S.W.; Xu, H. Synthesis and antifungal activity of ethers, alcohols, and iodohydrin derivatives of sclareol against phytopathogenic fungi in vitro. Bioorg. Med. Chem. Lett. 2015, 25, 2773–2777. [Google Scholar] [CrossRef]
- Ruan, J. Bergey’s manual of systematic bacteriology (second edition) volume 5 and the study of actinomycetes systematic in China. Acta Microbiol. Sin. 2013, 53, 521–530. [Google Scholar]
- Wang, B.; Wan, C.X.; Zeng, H. Colonization on cotton plants with a GFP labeled strain of Bacillus axarquiensis. Curr. Microbiol. 2020, 77, 3085–3094. [Google Scholar] [CrossRef]
- Yang, H.; Li, X.; Li, X.; Yu, H.; Shen, Z. Identification of lipopeptide isoforms by MALDI-TOF-MS/MS based on the simultaneous purification of iturin, fengycin, and surfactin by RP-HPLC. Anal. Bioanal. Chem. 2015, 407, 2529–2542. [Google Scholar] [CrossRef]
- Schwyn, B.; Neilands, J.B. Universal chemical-assay for the detection and determination of siderophores. Anal. Biochem. 1987, 160, 47–56. [Google Scholar] [CrossRef]
- Pérez-Miranda, S.; Cabirol, N.; George-Téllez, R.; Zamudio-Rivera, L.S.; Fernández, F.J. O-CAS, a fast and universal method for siderophore detection. J. Microbiol. Methods 2007, 70, 127–131. [Google Scholar] [CrossRef] [PubMed]
- Baakza, A.; Vala, A.K.; Dave, B.P.; Dube, H.C. A comparative study of siderophore production by fungi from marine and terrestrial habitats. J. Exp. Mar. Biol. Ecol. 2004, 311, 1–9. [Google Scholar] [CrossRef]
- Zeng, C.; Zhao, R.B.; Ma, M.M.; Zeng, Z.L.; Gong, D.M. Mutagenesis and characterization of a Bacillus amyloliquefaciens strain for Cinnamomum camphora seed kernel oil extraction by aqueous enzymatic method. AMB Express 2017, 7, 10. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.D.; Watanabe, C.K. GMAP: A genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics 2005, 21, 1859–1875. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef] [PubMed]
- Johnson, L.S.; Eddy, S.R.; Portugaly, E. Hidden Markov model speed heuristic and iterative HMM search procedure. BMC Bioinform. 2010, 11, 431. [Google Scholar] [CrossRef]
- Finn, R.D.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Mistry, J.; Mitchell, A.L.; Potter, S.C.; Punta, M.; Qureshi, M.; Sangrador-Vegas, A.; et al. The Pfam protein families database: Towards a more sustainable future. Nucleic Acids Res. 2016, 44, D279–D285. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef] [PubMed]
- Koonin, E.V.; Fedorova, N.D.; Jackson, J.D.; Jacobs, A.R.; Krylov, D.M.; Makarova, K.S.; Mazumder, R.; Mekhedov, S.L.; Nikolskaya, A.N.; Rao, B.S.; et al. A comprehensive evolutionary classification of proteins encoded in complete eukaryotic genomes. Genome Biol. 2004, 5, R7. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Boeckmann, B.; Bairoch, A.; Apweiler, R.; Blatter, M.C.; Estreicher, A.; Gasteiger, E.; Martin, M.J.; Michoud, K.; O’Donovan, C.; Phan, I.; et al. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res. 2003, 31, 365–370. [Google Scholar] [CrossRef]
- Conesa, A.; Götz, S.; García-Gómez, J.M.; Terol, J.; Talón, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef] [PubMed]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Swift, M.L. GraphPad Prism, Data Analysis, and Scientific Graphing. J. Chem. Inf. Comput. Sci. 1997, 37, 411–412. [Google Scholar] [CrossRef]
- Jiang, C.; Shi, J.; Liu, Y.; Zhu, C. Inhibition of Aspergillus carbonarius and fungal contamination in table grapes using Bacillus subtilis. Food Control 2014, 35, 41–48. [Google Scholar] [CrossRef]
- Cerda, R.; Avelino, J.; Gary, C.; Tixier, P.; Lechevallier, E.; Allinne, C. Primary and secondary yield losses caused by pests and diseases: Assessment and modeling in coffee. PLoS ONE 2017, 12, 17. [Google Scholar] [CrossRef] [PubMed]
- Ding, L.N.; Li, T.; Guo, X.J.; Li, M.; Liu, X.Y.; Cao, J.; Tan, X.L. Sclerotinia stem rot resistance in rapeseed: Recent progress and future prospects. J. Agric. Food Chem. 2021, 69, 2965–2978. [Google Scholar] [CrossRef] [PubMed]
- McQuilken, M.P.; Mitchell, S.J.; Archer, S.A. Origin of early attacks of Sclerotinia stem rot on winter oilseet rape (Brassica-napus sup sp oleifera var biensis) in the UK. J. Phytopathol. 1994, 140, 179–186. [Google Scholar] [CrossRef]
- Atallah, Z.K.; Johnson, D.A. Development of Sclerotinia stem rot in potato fields in south-central Washington. Plant Dis. 2004, 88, 419–423. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Li, M.; Wang, D.; Wang, F.; Shen, H.; Sun, G.; Feng, C.; Wang, X.; Chen, D.; Sun, X. Biocontrol efficacy of Bacillus siamensis LZ88 against brown spot disease of tobacco caused by Alternaria alternata. Biol. Control 2021, 154, 104508. [Google Scholar] [CrossRef]
- Hernández-León, R.; Rojas-Solís, D.; Contreras-Pérez, M.; Orozco-Mosqueda, M.D.; Macías-Rodríguez, L.I.; Reyes-de la Cruz, H.; Valencia-Cantero, E.; Santoyo, G. Characterization of the antifungal and plant growth-promoting effects of diffusible and volatile organic compounds produced by Pseudomonas fluorescens strains. Biol. Control 2015, 81, 83–92. [Google Scholar] [CrossRef]
- Mazhar, R.; Ilyas, N.; Saeed, M.; Bibi, F.; Batool, N. Biocontrol and salinity tolerance potential of Azospirillum lipoferum and its inoculation effect in wheat crop. Int. J. Agric. Biol. 2016, 18, 494–500. [Google Scholar] [CrossRef]
- Xu, D.; Côté, J.C. Phylogenetic relationships between Bacillus species and related genera inferred from comparison of 3′end 16S rDNA and 5′end 16S-23S ITS nucleotide sequences. Int. J. Syst. Evol. Microbiol. 2003, 53, 695–704. [Google Scholar] [CrossRef] [PubMed]
- da Silva, L.R.; Mello, S.C.M.d.; Valadares-Inglis, M.C.; Costa, M.M.d.C.; Saraiva, M.A.d.P.; Rego, E.C.S.; Zacaroni, A.B.; Muniz, P.H.P.C.; Pappas, M.d.C.R. Transcriptional responses and reduction in carpogenic germination of Sclerotinia sclerotiorum exposed to volatile organic compounds of Trichoderma azevedoi. Biol. Control 2022, 169, 104897. [Google Scholar] [CrossRef]
- Yang, M.; Zhang, W.; Lv, Z.; Shi, L.; Zhang, K.; Ge, B. Evaluation of the inhibitory effects of wuyiencin, a secondary metabolite of Streptomyces albulus CK-15, against Sclerotinia sclerotiorum in vitro. Plant Dis. 2022, 106, 156–164. [Google Scholar] [CrossRef]
- Zhang, H.; Xie, J.; Fu, Y.; Cheng, J.; Qu, Z.; Zhao, Z.; Cheng, S.; Chen, T.; Li, B.; Wang, Q.; et al. A 2-kb mycovirus converts a pathogenic fungus into a beneficial endophyte for brassica protection and yield enhancement. Mol. Plant 2020, 13, 1420–1433. [Google Scholar] [CrossRef]
- Ji, S.; Tian, Y.; Li, J.; Xu, G.; Zhang, Y.; Chen, S.; Chen, Y.; Tang, X. Complete genome sequence of Bacillus cereus Z4, a biocontrol agent against tobacco black shank, isolated from the Western Pacific Ocean. Mar. Genom. 2023, 72, 101071. [Google Scholar] [CrossRef]
- Vitorino, L.C.; Silva, F.O.D.; Cruvinel, B.G.; Bessa, L.A.; Rosa, M.; Souchie, E.L.; Silva, F.G. Biocontrol potential of Sclerotinia sclerotiorum and physiological changes in soybean in response to butia archeri palm rhizobacteria. Plants 2020, 9, 64. [Google Scholar] [CrossRef]
- Ramírez, V.; Martínez, J.; Bustillos-Cristales, M.D.R.; Catañeda-Antonio, D.; Munive, J.A.; Baez, A. Bacillus cereus MH778713 elicits tomato plant protection against Fusarium oxysporum. J. Appl. Microbiol. 2022, 132, 470–482. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Ren, Z.H.; Zu, X.; Yu, X.Y.; Zhu, H.J.; Li, X.J.; Zhong, J.; Liu, E.M. Efficacy of plant growth-promoting bacteria Bacillus cereus YN917 for biocontrol of rice blast. Front. Microbiol. 2021, 12, 684888. [Google Scholar] [CrossRef] [PubMed]




| Gene Cluster | Pathway | Function | Identity (%) |
|---|---|---|---|
| Petrobactin | Other | Ironophore; accumulates and takes up iron ions | 100% |
| Bacillibactin | NRP | Accumulates and takes up iron ions | 85% |
| Fengycin | NRP | Anti-fungal; signal for plant cell | 40% |
| Molybdenum cofactor | Other | Nitrogen metabolism; sulfur metabolism | 17% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, W.; Ding, J.; Jia, Q.; Li, Q.; Jiao, S.; Guo, X.; Fan, C.; Chen, Y.; Hu, Z. A Novel Strain of Bacillus cereus with a Strong Antagonistic Effect Specific to Sclerotinia and Its Genomic and Transcriptomic Analysis. Microorganisms 2024, 12, 611. https://doi.org/10.3390/microorganisms12030611
Ma W, Ding J, Jia Q, Li Q, Jiao S, Guo X, Fan C, Chen Y, Hu Z. A Novel Strain of Bacillus cereus with a Strong Antagonistic Effect Specific to Sclerotinia and Its Genomic and Transcriptomic Analysis. Microorganisms. 2024; 12(3):611. https://doi.org/10.3390/microorganisms12030611
Chicago/Turabian StyleMa, Wanfu, Jinhao Ding, Qingyun Jia, Qianru Li, Shanhai Jiao, Xupeng Guo, Chengming Fan, Yuhong Chen, and Zanmin Hu. 2024. "A Novel Strain of Bacillus cereus with a Strong Antagonistic Effect Specific to Sclerotinia and Its Genomic and Transcriptomic Analysis" Microorganisms 12, no. 3: 611. https://doi.org/10.3390/microorganisms12030611





