LPS-Dephosphorylating Cobetia amphilecti Alkaline Phosphatase of PhoA Family Divergent from the Multiple Homologues of Cobetia spp.
Abstract
:1. Introduction
2. Materials and Methods
2.1. Recombinant Production of the C. amphilecti KMM 296 Alkaline Phosphatase CmAP
2.2. Alkaline Phosphatase Activity Assay
2.3. Dephosphorylation Activity Assay towards E. coli LPS
2.4. Phylogenetic and Biosynthetic Gene Cluster Analyses
3. Results and Discussion
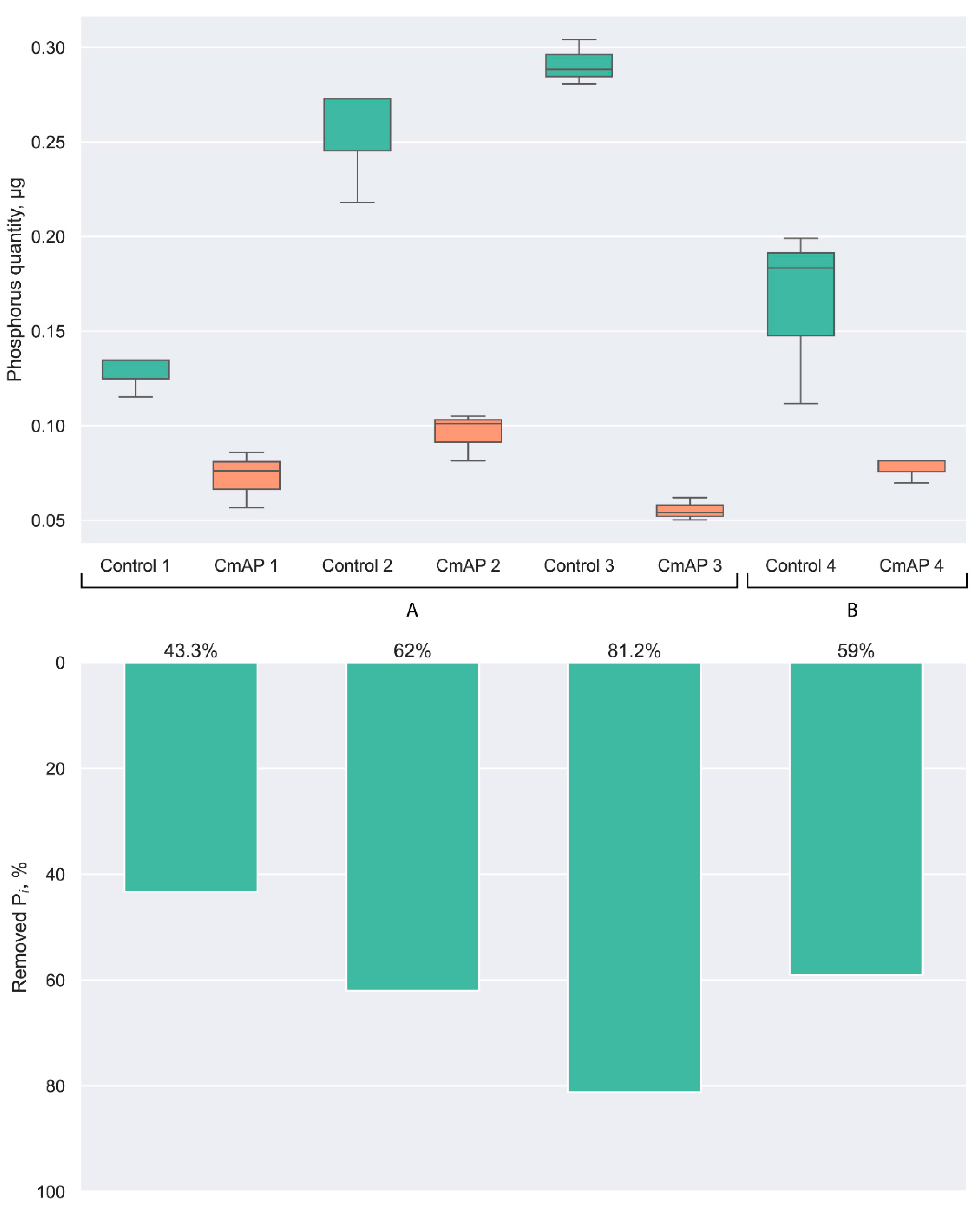
3.1. Dephosphorylation Activity of the Recombinant Alkaline Phosphatase CmAP towards E. coli LPS
3.2. Multiplicity of Alkaline Phosphatase Phylotypes in the Species Cobetia amphilecti
3.2.1. Structural Classification of the Bacterial Alkaline Phosphatases
3.2.2. Searching for Homologues of the C. amphilecti KMM 296 Alkaline Phosphatase CmAP
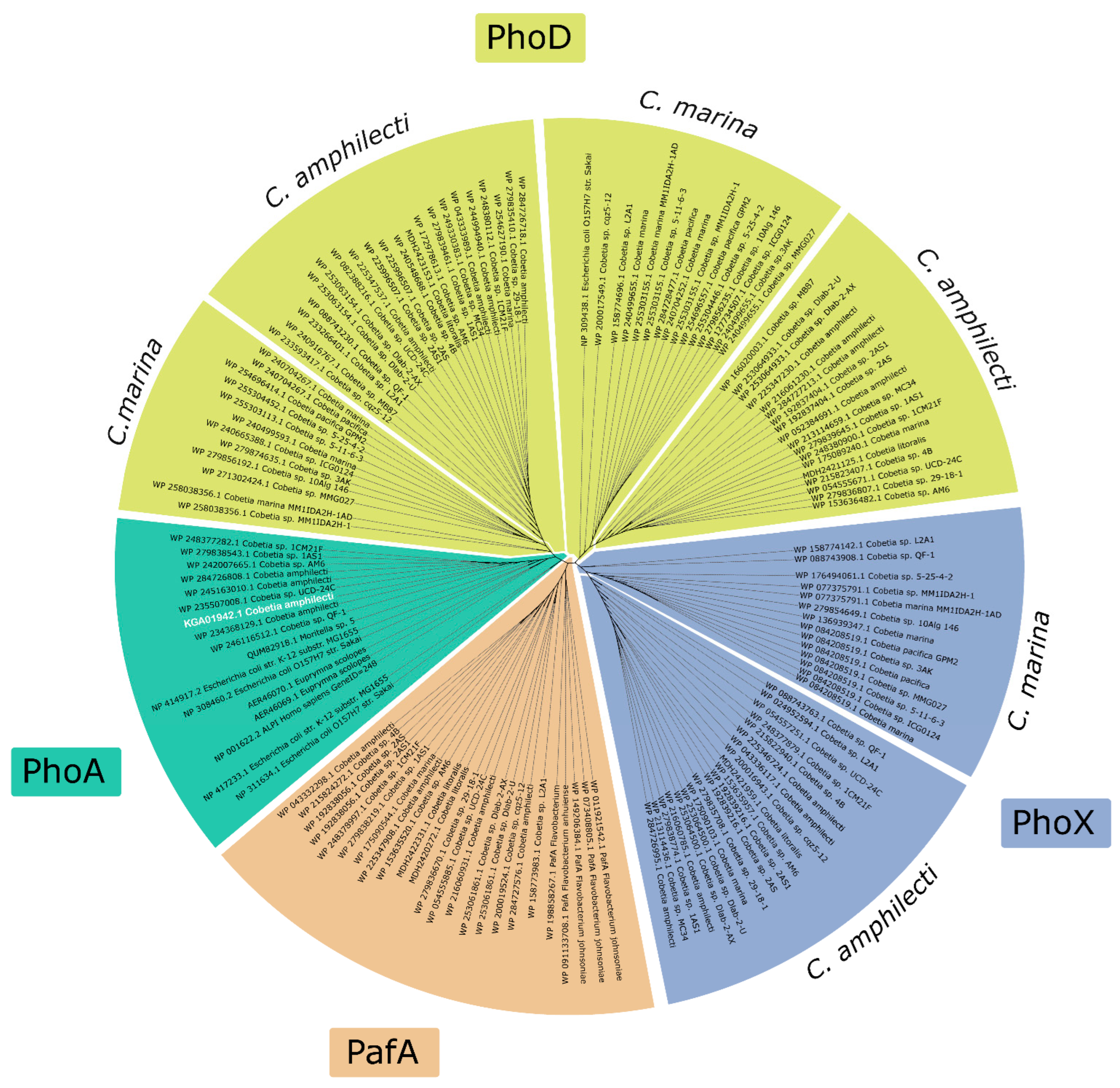
3.2.3. Phylogenetic Analysis of Alkaline Phosphatases in the Genus Cobetia
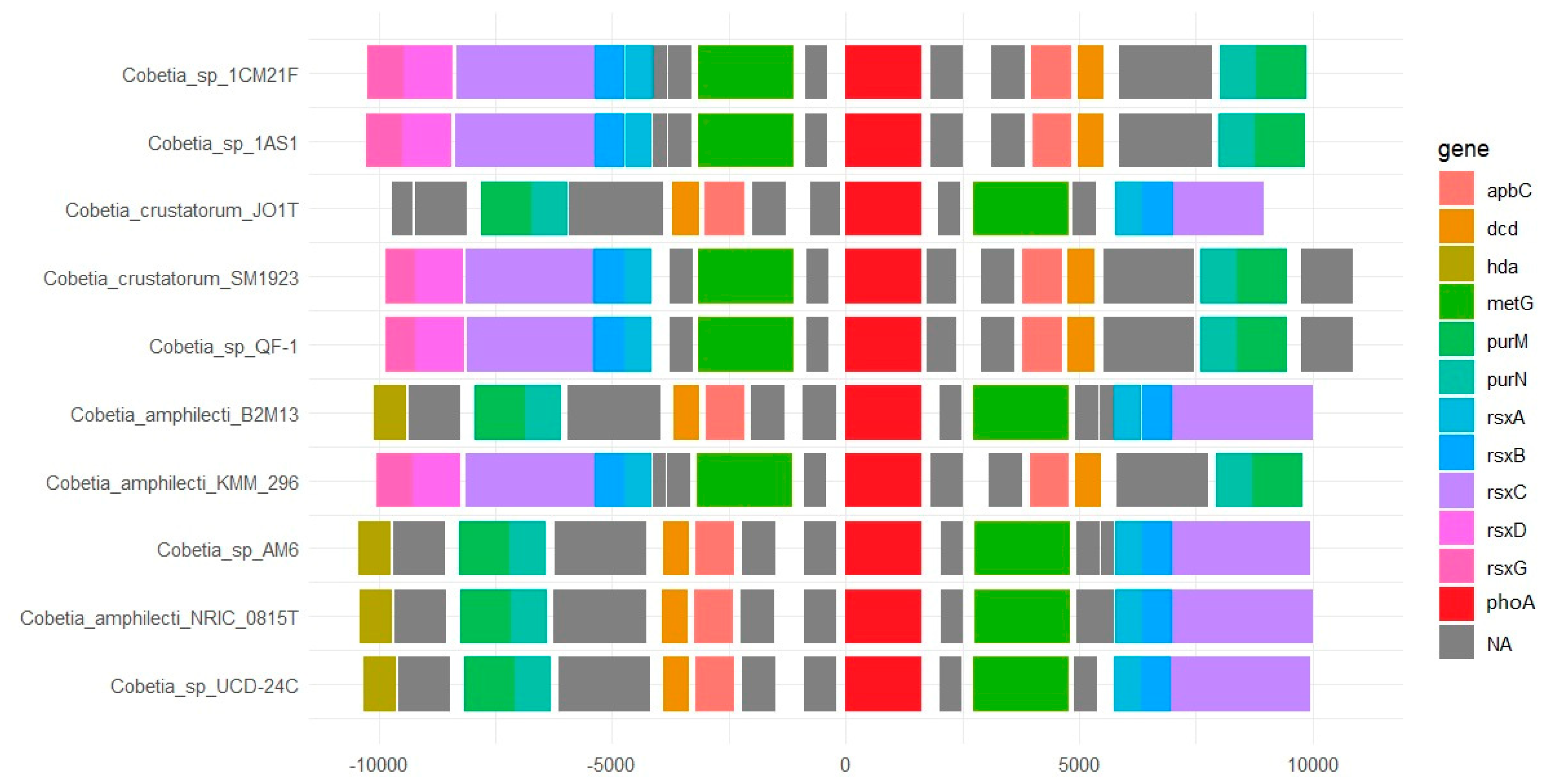
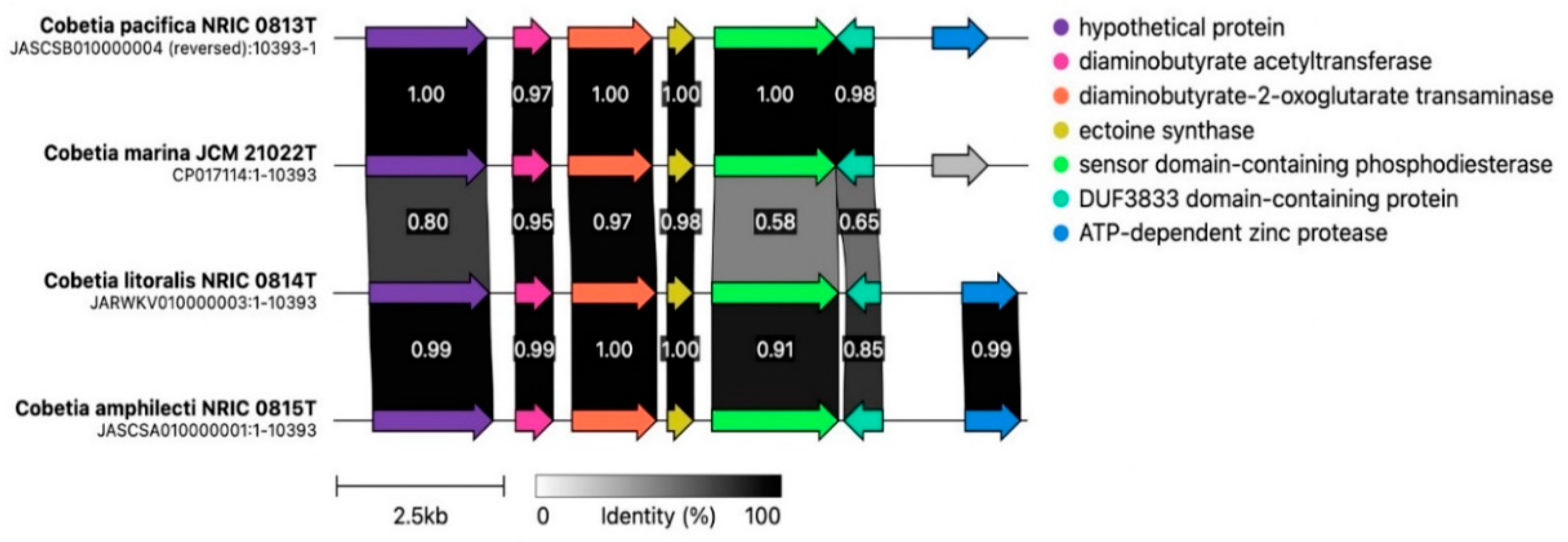
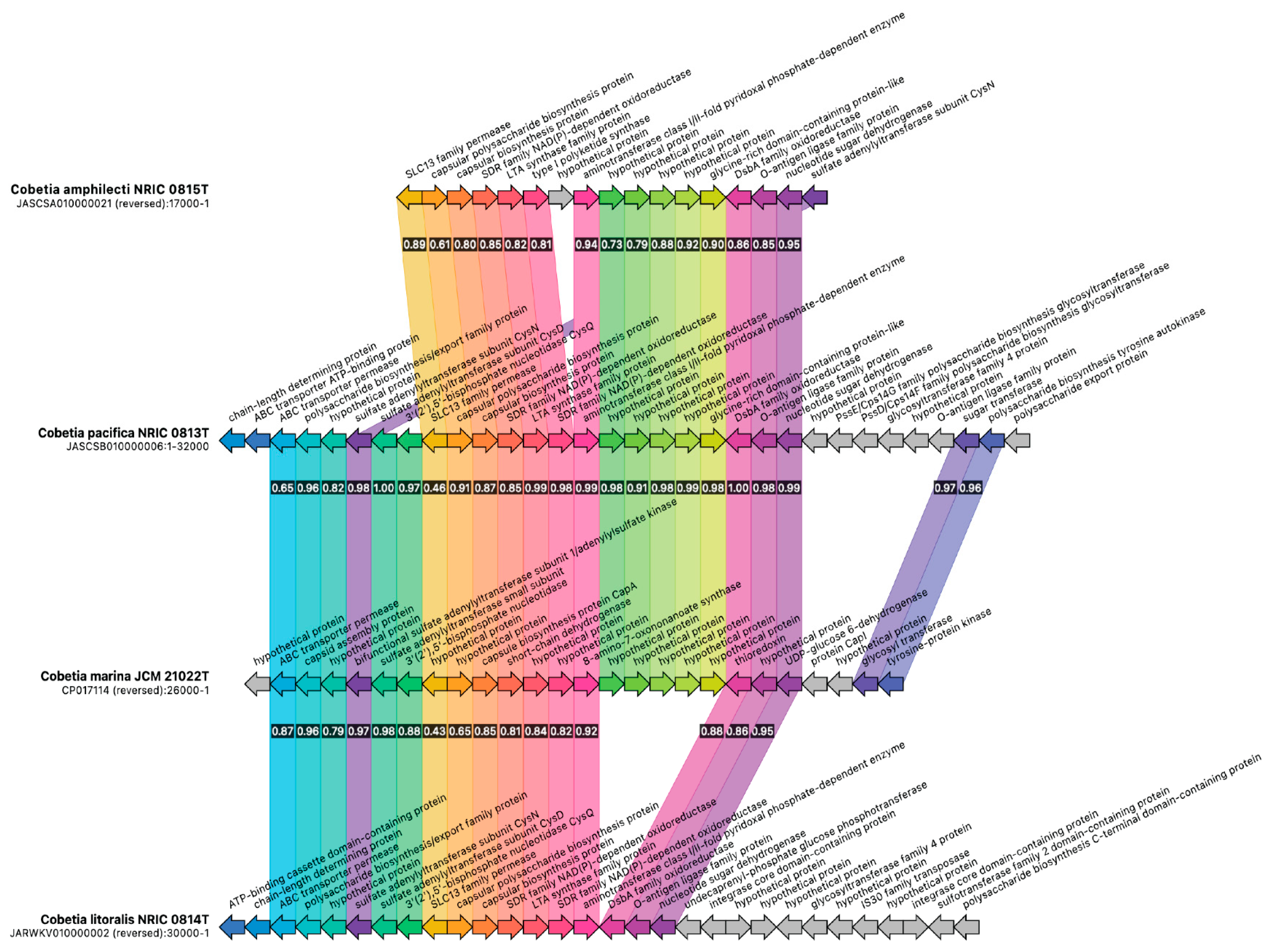
3.3. Biosynthetic Gene Cluster Analysis in the Genus Cobetia
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Millán, J.L. Alkaline Phosphatases: Structure, Substrate Specificity and Functional Relatedness to Other Members of a Large Superfamily of Enzymes. Purinergic Signal. 2006, 2, 335–341. [Google Scholar] [CrossRef]
- Chen, S.-L.; Liao, R.-Z. Phosphate Monoester Hydrolysis by Trinuclear Alkaline Phosphatase; DFT Study of Transition States and Reaction Mechanism. ChemPhysChem 2014, 15, 2321–2330. [Google Scholar] [CrossRef]
- Harroun, S.G.; Vallée-Bélisle, A. Methods to Characterise Enzyme Kinetics with Biological and Medicinal Substrates: The Case of Alkaline Phosphatase. Chem. Methods 2023, 3, e202200067. [Google Scholar] [CrossRef]
- Levitt, M.D.; Hapak, S.M.; Levitt, D.G. Alkaline Phosphatase Pathophysiology with Emphasis on the Seldom-Discussed Role of Defective Elimination in Unexplained Elevations of Serum ALP—A Case Report and Literature Review. Clin. Exp. Gastroenterol. 2022, 15, 41–49. [Google Scholar] [CrossRef]
- Zaher, D.M.; El-Gamal, M.I.; Omar, H.A.; Aljareh, S.N.; Al-Shamma, S.A.; Ali, A.J.; Zaib, S.; Iqbal, J. Recent Advances with Alkaline Phosphatase Isoenzymes and Their Inhibitors. Arch. Pharm. 2020, 353, e2000011. [Google Scholar] [CrossRef]
- Malo, M.S.; Nasrin Alam, S.; Mostafa, G.; Zeller, S.; Johnson, P.V.; Mohammad, N.N.; Chen, K.T.; Moss, A.K.; Ramasamy, S.; Faruqui, A.A.; et al. Intestinal Alkaline Phosphatase Preserves the Normal Homeostasis of Gut Microbiota. Gut 2010, 59, 1476–1484. [Google Scholar] [CrossRef]
- Kühn, F.; Adiliaghdam, F.; Cavallaro, P.M.; Hamarneh, S.R.; Tsurumi, A.; Hoda, R.S.; Munoz, A.R.; Dhole, Y.; Ramirez, J.M.; Liu, E.; et al. Intestinal Alkaline Phosphatase Targets the Gut Barrier to Prevent Aging. JCI Insight 2020, 5, e134049. [Google Scholar] [CrossRef]
- Hamarneh, S.R.; Mahgoub, M.; Economopoulos, K.P.; Morrison, S.A.; Phupitakphol, T.; Tantillo, T.J.; Gul, S.; Gharedaghi, M.H.; Tao, Q.; Kaliannan, K.; et al. A Novel Approach to Maintain Gut Mucosal Integrity Using an Oral Enzyme Supplement. Ann. Surg. 2014, 260, 706–715. [Google Scholar] [CrossRef]
- Duan, R. The Role of Intestinal Alkaline Phosphatase and Bacterial Lipopolysaccharides in Patients Undergoing Pancreaticoduodenectomy; Ludwig-Maximilians-Universität: München, Germany, 2022. [Google Scholar]
- Yang, Y.; Wandler, A.M.; Postlethwait, J.H.; Guillemin, K. Dynamic Evolution of the LPS-Detoxifying Enzyme Intestinal Alkaline Phosphatase in Zebrafish and Other Vertebrates. Front. Immunol. 2012, 3, 35467. [Google Scholar] [CrossRef]
- Rader, B.A.; Kremer, N.; Apicella, M.A.; Goldman, W.E.; McFall-Ngai, M.J. Modulation of Symbiont Lipid a Signaling by Host Alkaline Phosphatases in the Squid-Vibrio Symbiosis. mBio 2012, 3, e00093-12. [Google Scholar] [CrossRef]
- Ghyoot, C.; Gypens, N.; Flynn, K.K.J.; Lancelot, C. Modelling Alkaline Phosphatase Activity in Microalgae under Orthophosphate Limitation: The Case ofPhaeocystis Globosa. J. Plankton Res. 2015, 37, 869–885. [Google Scholar] [CrossRef]
- Srivastava, A.; Saavedra, D.E.M.; Thomson, B.; García, J.A.L.; Zhao, Z.; Patrick, W.M.; Herndl, G.J.; Baltar, F. Enzyme Promiscuity in Natural Environments: Alkaline Phosphatase in the Ocean. ISME J. 2021, 15, 3375–3383. [Google Scholar] [CrossRef] [PubMed]
- Lidbury, I.; Scanlan, D.J.; Murphy, A.; Christie-Oleza, J.A.; Aguilo-Ferretjans, M.M.; Hitchcock, A.; Daniell, T.J. A Widely Distributed Phosphate-Insensitive Phosphatase Presents a Route for Rapid Organophosphorus Remineralization in the Biosphere. Proc. Natl. Acad. Sci. USA 2022, 119, e2118122119. [Google Scholar] [CrossRef]
- Plisova, Y.E.; Balabanova, L.A.; Ivanova, E.P.; Kozhemyako, V.B.; Mikhailov, V.V.; Agafonova, E.V.; Rasskazov, V.A. A Highly Active Alkaline Phosphatase from the Marine Bacterium Cobetia. Mar. Biotechnol. 2005, 7, 173–178. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhang, T.; Jin, S.; Chen, S.; Zhang, Y. Effects of Escherichia Coli Alkaline Phosphatase PhoA on the Mineralization of Dissolved Organic Phosphorus. Water 2021, 13, 3315. [Google Scholar] [CrossRef]
- Westermann, L.M.; Lidbury, I.; Li, C.-Y.; Wang, N.; Murphy, A.; Del Mar Aguilo Ferretjans, M.; Quareshy, M.; Shanmugam, M.; Torcello-Requena, A.; Silvano, E.; et al. Bacterial Catabolism of Membrane Phospholipids Links Marine Biogeochemical Cycles. Sci. Adv. 2023, 9, eadf5122. [Google Scholar] [CrossRef] [PubMed]
- Barrozo, A.; Duarte, F.; Bauer, P.; Carvalho, A.T.P.; Kamerlin, S.C.L. Cooperative Electrostatic Interactions Drive Functional Evolution in the Alkaline Phosphatase Superfamily. J. Am. Chem. Soc. 2015, 137, 9061–9076. [Google Scholar] [CrossRef]
- Sunden, F.; AlSadhan, I.; Lyubimov, A.Y.; Ressl, S.; Wiersma-Koch, H.; Borland, J.; Brown, C.L.; Johnson, T.A.; Singh, Z.; Herschlag, D. Mechanistic and Evolutionary Insights from Comparative Enzymology of Phosphomonoesterases and Phosphodiesterases across the Alkaline Phosphatase Superfamily. J. Am. Chem. Soc. 2016, 138, 14273–14287. [Google Scholar] [CrossRef]
- Fuhrmann, J.; Mierzwa, B.; Trentini, D.B.; Spiess, S.; Lehner, A.; Charpentier, E.; Clausen, T. Structural Basis for Recognizing Phosphoarginine and Evolving Residue-Specific Protein Phosphatases in Gram-Positive Bacteria. Cell Rep. 2013, 3, 1832–1839. [Google Scholar] [CrossRef]
- Du, M.; Li, X.; Li, Z.; Shen, Q.; Wang, Y.; Li, G.; Zhang, D. Phosphorylation Regulated by Protein Kinase a and Alkaline Phosphatase Play Positive Roles in μ-Calpain Activity. Food Chem. 2018, 252, 33–39. [Google Scholar] [CrossRef]
- Sebastian, M.; Ammerman, J.W. The Alkaline Phosphatase PhoX Is More Widely Distributed in Marine Bacteria than the Classical PhoA. ISME J. 2009, 3, 563–572. [Google Scholar] [CrossRef]
- Ragot, S.A.; Kertesz, M.A.; Mészáros, É.; Frossard, E.; Bünemann, E.K. Soil PhoD and PhoX Alkaline Phosphatase Gene Diversity Responds to Multiple Environmental Factors. FEMS Microbiol. Ecol. 2016, 93, fiw212. [Google Scholar] [CrossRef]
- Skouri-Panet, F.; Benzerara, K.; Cosmidis, J.; Férard, C.; Caumes, G.; De Luca, G.; Heulin, T.; Duprat, E. In Vitro and in Silico Evidence of Phosphatase Diversity in the Biomineralizing Bacterium Ramlibacter Tataouinensis. Front. Microbiol. 2018, 8, 2592. [Google Scholar] [CrossRef]
- Wan, B.; Huang, R.; Diaz, J.M.; Tang, Y. Rethinking the Biotic and Abiotic Remineralization of Complex Phosphate Molecules in Soils and Sediments. Sci. Total Environ. 2022, 833, 155187. [Google Scholar] [CrossRef]
- Zeng, J.; Tu, Q.; Yu, X.; Lu, Q.; Wang, C.; Shu, L.; Liu, F.; Liu, S.; Huang, Z.; He, J.; et al. PCycDB: A Comprehensive and Accurate Database for Fast Analysis of Phosphorus Cycling Genes. Microbiome 2022, 10, 101. [Google Scholar] [CrossRef] [PubMed]
- Doing, G.; Koeppen, K.; Occipinti, P.; Harty, C.E.; Hogan, D.A. Conditional Antagonism in Co-Cultures of Pseudomonas Aeruginosa and Candida Albicans: An Intersection of Ethanol and Phosphate Signaling Distilled from Dual-Seq Transcriptomics. PLoS Genet. 2020, 16, e1008783. [Google Scholar] [CrossRef] [PubMed]
- Balabanova, L.; Podvolotskaya, A.; Slepchenko, L.; Eliseikina, M.; Noskova, Y.; Nedashkovskaya, O.; Son, O.; Tekutyeva, L.; Rasskazov, V. Nucleolytic Enzymes from the Marine Bacterium Cobetia Amphilecti KMM 296 with Antibiofilm Activity and Biopreservative Effect on Meat Products. Food Control 2017, 78, 270–278. [Google Scholar] [CrossRef]
- Zorzetto, L.; Scoppola, E.; Raguin, É.; Blank, K.; Fratzl, P.; Bidan, C.M. Induced Mineralization of Hydroxyapatite in Escherichia Coli Biofilms and the Potential Role of Bacterial Alkaline Phosphatase. Chem. Mater. 2023, 35, 2762–2772. [Google Scholar] [CrossRef]
- Dong, H.; Huang, L.; Zhao, L.; Zeng, Q.; Liu, X.; Sheng, Y.; Shi, L.; Wu, G.; Jiang, H.; Li, F.; et al. A Critical Review of Mineral–Microbe Interaction and Co-Evolution: Mechanisms and Applications. Natl. Sci. Rev. 2022, 9, nwac128. [Google Scholar] [CrossRef] [PubMed]
- Noskova, Y.; Likhatskaya, G.; Terentieva, N.; Son, O.; Tekutyeva, L.; Balabanova, L. A Novel Alkaline Phosphatase/Phosphodiesterase, CamPhoD, from Marine Bacterium Cobetia amphilecti KMM 296. Mar. Drugs 2019, 17, 657. [Google Scholar] [CrossRef] [PubMed]
- Kamennaya, N.A.; Geraki, K.; Scanlan, D.J.; Zubkov, M.V. Accumulation of Ambient Phosphate into the Periplasm of Marine Bacteria Is Proton Motive Force Dependent. Nat. Commun. 2020, 11, 2642. [Google Scholar] [CrossRef]
- Golotin, V.; Balabanova, L.; Likhatskaya, G.; Rasskazov, V. Recombinant Production and Characterization of a Highly Active Alkaline Phosphatase from Marine Bacterium Cobetia Marina. Mar. Biotechnol. 2014, 17, 130–143. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of Structural Proteins during the Assembly of the Head of Bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef]
- Vaskovsky, V.E.; Kostetsky, E.Y.; Vasendin, I.M. A Universal Reagent for Phospholipid Analysis. J. Chromatogr. A 1975, 114, 129–141. [Google Scholar] [CrossRef]
- Jones, D.T.; Taylor, W.R.; Thornton, J.M. The Rapid Generation of Mutation Data Matrices from Protein Sequences. Bioinformatics 1992, 8, 275–282. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Zimmermann, J.; Kaleta, C.; Waschina, S. Gapseq: Informed Prediction of Bacterial Metabolic Pathways and Reconstruction of Accurate Metabolic Models. Genome Biol. 2021, 22, 81. [Google Scholar] [CrossRef] [PubMed]
- Gilchrist, C.L.M.; Chooi, Y.-H. Clinker & Clustermap.js: Automatic Generation of Gene Cluster Comparison Figures. Bioinformatics 2021, 37, 2473–2475. [Google Scholar] [CrossRef] [PubMed]
- Blin, K.; Shaw, S.; Augustijn, H.E.; Reitz, Z.L.; Biermann, F.; Alanjary, M.; Fetter, A.; Terlouw, B.R.; Metcalf, W.W.; Helfrich, E.J.N.; et al. AntiSMASH 7.0: New and Improved Predictions for Detection, Regulation, Chemical Structures and Visualisation. Nucleic Acids Res. 2023, 51, W46–W50. [Google Scholar] [CrossRef] [PubMed]
- Hashem, K.A.; Authman, S.H.; Mahdi, L.H. In Vivo Antibacterial Activity of Alkaline Phosphatase Isolates from Escherichia Coli Isolated from Diarrhea Patients against Pseudomonas aeruginosa. Pharma Innov. J. 2016, 5, 32–36. [Google Scholar]
- Avila-Calderón, E.D.; Ruiz-Palma, M.D.S.; Aguilera-Arreola, M.G.; Velázquez-Guadarrama, N.; Ruiz, E.A.; Gomez-Lunar, Z.; Witonsky, S.; Contreras-Rodríguez, A. Outer Membrane Vesicles of Gram-Negative Bacteria: An Outlook on Biogenesis. Front. Microbiol. 2021, 12, 557902. [Google Scholar] [CrossRef]
- Richter, W.; Vogel, V.; Howe, J.; Steiniger, F.; Brauser, A.; Koch, M.H.J.; Roessle, M.; Gutsmann, T.; Garidel, P.; Mäntele, W.; et al. Morphology, size distribution, and aggregate structure of lipopolysaccharide and lipid A dispersions from enterobacterial origin. Innate Immun. 2011, 17, 427–438. [Google Scholar] [CrossRef]
- Paracini, N.; Schneck, E.; Imberty, A.; Micciulla, S. Lipopolysaccharides at Solid and Liquid Interfaces: Models for Biophysical Studies of the Gram-Negative Bacterial Outer Membrane. Adv. Colloid Interface Sci. 2022, 301, 102603. [Google Scholar] [CrossRef]
- Rodriguez, F.; Lillington, J.; Johnson, S.G.; Timmel, C.R.; Lea, S.M.; Berks, B.C. Crystal Structure of the Bacillus Subtilis Phosphodiesterase PhoD Reveals an Iron and Calcium-Containing Active Site. J. Biol. Chem. 2014, 289, 30889–30899. [Google Scholar] [CrossRef] [PubMed]
- Noskova, Y.; Seitkalieva, A.; Nedashkovskaya, O.; Shevchenko, L.; Tekutyeva, L.; Son, O.; Balabanova, L. Are the Closely Related Cobetia Strains of Different Species? Molecules 2021, 26, 690. [Google Scholar] [CrossRef]
- Nedashkovskaya, O.; Balabanova, L.; Otstavnykh, N.; Zhukova, N.; Detkova, E.; Seitkalieva, A.; Bystritskaya, E.; Noskova, Y.; Tekutyeva, L.; Isaeva, M. In-Depth Genome Characterization and Pan-Genome Analysis of Strain KMM 296, a Producer of Highly Active Alkaline Phosphatase; Proposal for the Reclassification of Cobetia litoralis and Cobetia pacifica as the Later Heterotypic Synonyms of Cobetia amphilecti and Cobetia marina, and Emended Description of the Species Cobetia amphilecti and Cobetia marina. Biomolecules 2024, 14, 196. [Google Scholar]
- Burschel, S.; Kreuzer Decovic, D.; Nuber, F.; Stiller, M.; Hofmann, M.; Zupok, A.; Siemiatkowska, B.; Gorka, M.; Leimkühler, S.; Friedrich, T. Iron-sulfur Cluster Carrier Proteins Involved in the Assembly of Escherichia coli NADH:ubiquinone Oxidoreductase (Complex I). Mol. Microbiol. 2019, 111, 31–45. [Google Scholar] [CrossRef]
- Vermassen, A.; Leroy, S.; Talon, R.; Provot, C.; Popowska, M.; Desvaux, M. Cell Wall Hydrolases in Bacteria: Insight on the Diversity of Cell Wall Amidases, Glycosidases and Peptidases Toward Peptidoglycan. Front. Microbiol. 2019, 10, 331. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Li, J.; Zhou, Z.; Huang, R.; Lin, S. Roles of Alkaline Phosphatase PhoA in Algal Metabolic Regulation under Phosphorus-replete Conditions. J. Phycol. 2021, 57, 703–707. [Google Scholar] [CrossRef]
- Xiao, W.; Wang, R.-S.; Handy, D.E.; Loscalzo, J. NAD(H) and NADP(H) Redox Couples and Cellular Energy Metabolism. Antioxid. Redox Signal. 2018, 28, 251–272. [Google Scholar] [CrossRef]
- Zhang, Q.; Padayatti, P.S.; Leung, J.H. Proton-Translocating Nicotinamide Nucleotide Transhydrogenase: A Structural Perspective. Front. Physiol. 2017, 8, 293323. [Google Scholar] [CrossRef] [PubMed]
- Kämäräinen, J.; Huokko, T.; Kreula, S.; Jones, P.R.; Aro, E.-M.; Kallio, P. Pyridine Nucleotide Transhydrogenase PntAB Is Essential for Optimal Growth and Photosynthetic Integrity under Low-Light Mixotrophic Conditions in Synechocystis Sp. PCC 6803. New Phytol. 2016, 214, 194–204. [Google Scholar] [CrossRef] [PubMed]
- Ju, H.-Q.; Lin, J.-F.; Tian, T.; Xie, D.; Xu, R.-H. NADPH Homeostasis in Cancer: Functions, Mechanisms and Therapeutic Implications. Signal Transduct. Target. Ther. 2020, 5, 231. [Google Scholar] [CrossRef] [PubMed]
- Mailloux, R.J.; Lemire, J.; Appanna, V.D. Metabolic Networks to Combat Oxidative Stress in Pseudomonas Fluorescens. Antonie Leeuwenhoek 2010, 99, 433–442. [Google Scholar] [CrossRef] [PubMed]
- Yamauchi, Y.; Hirasawa, T.; Nishii, M.; Furusawa, C.; Shimizu, H. Enhanced Acetic Acid and Succinic Acid Production under Microaerobic Conditions by Corynebacterium Glutamicum Harboring Escherichia Coli Transhydrogenase Gene PntAB. J. Gen. Appl. Microbiol. 2014, 60, 112–118. [Google Scholar] [CrossRef] [PubMed]
- Anantharaman, V.; Iyer, L.M.; Aravind, L. Ter-Dependent Stress Response Systems: Novel Pathways Related to Metal Sensing, Production of a Nucleoside-like Metabolite, and DNA-Processing. Mol. Biosyst. 2012, 8, 3142. [Google Scholar] [CrossRef]
- Boullet, A.; Meynial-Salles, I. An Overview of the Ferredoxin NAD+ Reductases Used for Energy Conservation in Various Anaerobic Microorganisms. Preprints 2018, 2018070473. [Google Scholar] [CrossRef]
- Zhang, Y.; Morar, M.; Ealick, S.E. Structural Biology of the Purine Biosynthetic Pathway. Cell. Mol. Life Sci. CMLS 2008, 65, 3699–3724. [Google Scholar] [CrossRef]
- Kato, J.; Katayama, T. Hda, a Novel DnaA-Related Protein, Regulates the Replication Cycle in Escherichia coli. EMBO J. 2001, 20, 4253–4262. [Google Scholar] [CrossRef]
- Deere, T. Characterization of Iron-Sulfur Cluster Biogenesis in Methanogenic Archaea. Ph.D. Thesis, University of Arkansas, Fayetteville, AR, USA, 2021. [Google Scholar]
- Ham, H.; Park, D.S. Novel Approach toward the Understanding of Genetic Diversity Based on the Two Types of Amino Acid Repeats in Erwinia Amylovora. Sci. Rep. 2023, 13, 17876. [Google Scholar] [CrossRef]
- Shoemaker, K.M.; McCliment, E.A.; Moisander, P.H. Copepod-Associated Gammaproteobacterial Alkaline Phosphatases in the North Atlantic Subtropical Gyre. Front. Microbiol. 2020, 11, 514518. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, A.K.; Kashyap, P.L.; Santoyo, G.; Newcombe, G. Editorial: Plant Microbiome: Interactions, Mechanisms of Action, and Applications. Front. Microbiol. 2021, 12, 706049. [Google Scholar] [CrossRef] [PubMed]
- Markússon, S.; Hjörleifsson, J.G.; Kursula, P.; Ásgeirsson, B. Structural Characterization of Functionally Important Chloride Binding Sites in the Marine Vibrio Alkaline Phosphatase. Biochemistry 2022, 61, 2248–2260. [Google Scholar] [CrossRef]
- Gauthier, A.E.; Chandler, C.E.; Poli, V.; Gardner, F.M.; Tekiau, A.; Smith, R.D.; Bonham, K.S.; Cordes, E.E.; Shank, T.M.; Zanoni, I.; et al. Deep-Sea Microbes as Tools to Refine the Rules of Innate Immune Pattern Recognition. Sci. Immunol. 2021, 6, eabe0531. [Google Scholar] [CrossRef]
- Liu, M.; Liu, H.; Shi, M.; Jiang, M.; Li, L.; Zheng, Y. Microbial Production of Ectoine and Hydroxyectoine as High-value Chemicals. Microb. Cell Fact. 2021, 20, 76. [Google Scholar] [CrossRef]
- Kokoulin, M.S.; Sigida, E.N.; Kuzmich, A.S.; Ibrahim, I.M.; Fedonenko, Y.P.; Konnova, S.A. Structure and Antiproliferative Activity of the Polysaccharide from Halomonas aquamarina Related to Cobetia pacifica. Carbohydr. Polym. 2022, 298, 120125. [Google Scholar] [CrossRef] [PubMed]
- Kokoulin, M.S.; Kuzmich, A.S.; Kalinovsky, A.I.; Tomshich, S.V.; Romanenko, L.A.; Mikhailov, V.V.; Komandrova, N.A. Structure and Anticancer Activity of Sulfated O-polysaccharide from Marine Bacterium Cobetia litoralis KMM 3880T. Carbohydr. Polym. 2016, 154, 55–61. [Google Scholar] [CrossRef] [PubMed]


| Original Strain Title | Actual Species * | Accession ID | ID Protein | ALP Family | Isolation Source |
|---|---|---|---|---|---|
| C. marina JCM 21022T | C. marina | GCF_001720485.1 | WP_240499655.1 | PhoD | Littoral water sample (USA: Woods Hole, MA, USA) |
| WP_084208519.1 | PhoX | ||||
| WP_240499593.1 | PhoD | ||||
| C. pacifica NRIC 0813T | C. marina | GCA_030010515.1 | WP_284728477.1 | PhoD | Sandy sediment (Russia: The Sea of Japan) |
| WP_240704267.1 | PhoX | ||||
| WP_084208519.1 | PhoX | ||||
| C. litoralis NRIC 0814T | C. amphilecti | GCF_029846315.1 | WP_249330383.1 | PhoD | Sandy sediment (Russia: The Sea of Japan) |
| WP_279830791.1 | PafA | ||||
| WP_279833222.1 | PhoX | ||||
| WP_279832006.1 | PhoD | ||||
| C. amphilecti NRIC 0815T ** | C. amphilecti | GCA_030010415.1 | WP_284726718.1 | PhoD | The finger sponge A. digitatus (The Sea of Okhotsk, Sakhalin Island, Piltun Bay) |
| WP_284726808.1 | PhoA ** | ||||
| WP_284726995.1 | PhoX | ||||
| WP_284727213.1 | PhoD | ||||
| WP_284727576.1 | PafA | ||||
| C. amphilecti KMM 296 ** | C. amphilecti | GCF_000754225.1 | WP_043332298.1 | PafA | Coelomic fluid of mussel C. grayanus (Russia: The Sea of Japan) |
| WP_043333989.1 | PhoD | ||||
| WP_245163010.1 | PhoA ** | ||||
| WP_043336117.1 | PhoX | ||||
| WP_052384691.1 | PhoD | ||||
| C. amphilecti B2M13 ** | C. amphilecti | GCF_018860945.1 | WP_244994940.1 | PhoD | Alginate 40–100 m particle (artificial) (USA: Canoe Beach) |
| WP_234368129.1 | PhoA ** | ||||
| WP_216060785.1 | PhoX | ||||
| WP_216060931.1 | PafA | ||||
| WP_216061230.1 | PhoD | ||||
| Cobetia sp. 1AS1 ** | C. amphilecti | GCF_029846435.1 | WP_279838219.1 | PafA | Coastal seawater (Russia: the Sea of Japan, Vostok Bay) |
| WP_279838543.1 | PhoA ** | ||||
| WP_279838774.1 | PhoX | ||||
| WP_279839461.1 | PhoD | ||||
| WP_279839645.1 | PhoD | ||||
| Cobetia sp. 1CM21F ** | C. amphilecti | GCF_023161745.1 | WP_248377282.1 | PhoA ** | Sea cave (Portugal: Algarve) |
| WP_248377879.1 | PhoX | ||||
| WP_248378997.1 | PafA | ||||
| WP_248380112.1 | PhoD | ||||
| WP_248380900.1 | PhoD | ||||
| Cobetia sp. AM6 ** | C. amphilecti | GCF_009617955.1 | WP_172978613.1 | PhoD | Exterior surface of the shell of an abalone sold in a fish market (Tokyo, Japan) |
| WP_153635520.1 | PafA | ||||
| WP_153635957.1 | PhoX | ||||
| WP_242007665.1 | PhoA ** | ||||
| WP_153636482.1 | PhoD | ||||
| Cobetia sp. UCD-24C ** | C. amphilecti | GCF_001306765.1 | WP_082388216.1 | PhoD | Seagrass Zostera sp. sediment |
| WP_054555671.1 | PhoD | ||||
| WP_054555885.1 | PafA | ||||
| WP_235507008.1 | PhoA ** | ||||
| WP_054557251.1 | PhoX | ||||
| C. crustatorum JO1T ** | C. crustatorum | GCF_000591415.1 | WP_248623642.1 | PhoA | Fermented shrimp (South Korea: Daejeon) |
| WP_282705494.1 | PhoD | ||||
| WP_282705495.1 | PhoD | ||||
| WP_024952594.1 | PhoX | ||||
| C. crustatorum SM1923 ** | C.crustatorum | GCF_007786215.1 | WP_144726746.1 | PhoD | Surface seawater (Kongsfjorden, Arctic) |
| WP_088743763.1 | PhoD | ||||
| WP_144727163.1 | PhoX | ||||
| WP_246116512.1 | PhoA ** | ||||
| WP_144728015.1 | PhoD |
| Strain_ID Genome | Mismatch |
|---|---|
| Cobetia_amphilecti_B2M13_GCF_018860945.1 | 240 F > Y, 308 F > Y |
| Cobetia_amphilecti_NRIC_0815T_GCA_030010415.1 | 240 F > Y, 308 F > Y, 372 E > G, 399 A > T |
| Cobetia_crustatorum_JO1T_GCF_000591415.1 | Down start (+45 amino acids) missing key position 21D, Tblastn_Identities 413/466 (89%) |
| Cobetia_crustatorum_SM1923_GCF_007786215.1 | Tblastn_Identities 456/511 (89%) |
| Cobetia_sp_1AS1_GCF_029846435.1 | 240 F > Y, 308 F > Y |
| Cobetia_sp_1CM21F_GCF_023161745.1 | 240 F > Y, 308 F > Y |
| Cobetia_sp_AM6_GCF_009617955.1 | 240 F > Y, 308 F > Y, 419 A > V, 422 P > L |
| Cobetia_sp_UCD-24C_GCF_001306765.1 | 240 F > Y, 308 F > Y, 390 G > D |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Balabanova, L.; Bakholdina, S.; Buinovskaya, N.; Noskova, Y.; Kolpakova, O.; Vlasova, V.; Bondarev, G.; Seitkalieva, A.; Son, O.; Tekutyeva, L. LPS-Dephosphorylating Cobetia amphilecti Alkaline Phosphatase of PhoA Family Divergent from the Multiple Homologues of Cobetia spp. Microorganisms 2024, 12, 631. https://doi.org/10.3390/microorganisms12030631
Balabanova L, Bakholdina S, Buinovskaya N, Noskova Y, Kolpakova O, Vlasova V, Bondarev G, Seitkalieva A, Son O, Tekutyeva L. LPS-Dephosphorylating Cobetia amphilecti Alkaline Phosphatase of PhoA Family Divergent from the Multiple Homologues of Cobetia spp. Microorganisms. 2024; 12(3):631. https://doi.org/10.3390/microorganisms12030631
Chicago/Turabian StyleBalabanova, Larissa, Svetlana Bakholdina, Nina Buinovskaya, Yulia Noskova, Oksana Kolpakova, Vanessa Vlasova, Georgii Bondarev, Aleksandra Seitkalieva, Oksana Son, and Liudmila Tekutyeva. 2024. "LPS-Dephosphorylating Cobetia amphilecti Alkaline Phosphatase of PhoA Family Divergent from the Multiple Homologues of Cobetia spp." Microorganisms 12, no. 3: 631. https://doi.org/10.3390/microorganisms12030631






