Ironing out Persisters? Revisiting the Iron Chelation Strategy to Target Planktonic Bacterial Persisters Harboured in Carbapenem-Resistant Escherichia coli
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacteria
2.2. Flow Cytometry
2.2.1. Fluorochromes Used for Flow Cytometry
2.2.2. Flow Cytometric Data Acquisition and Data Analysis
2.3. Time-Kill Studies (TKS)
2.3.1. Time-Kill Assessments via Viable Plating
2.3.2. Time-Kill Assessments Using Flow Cytometry
2.4. Determining Minimum Inhibitory Concentrations (MIC) and Minimum Bactericidal Concentrations (MBC)
3. Results
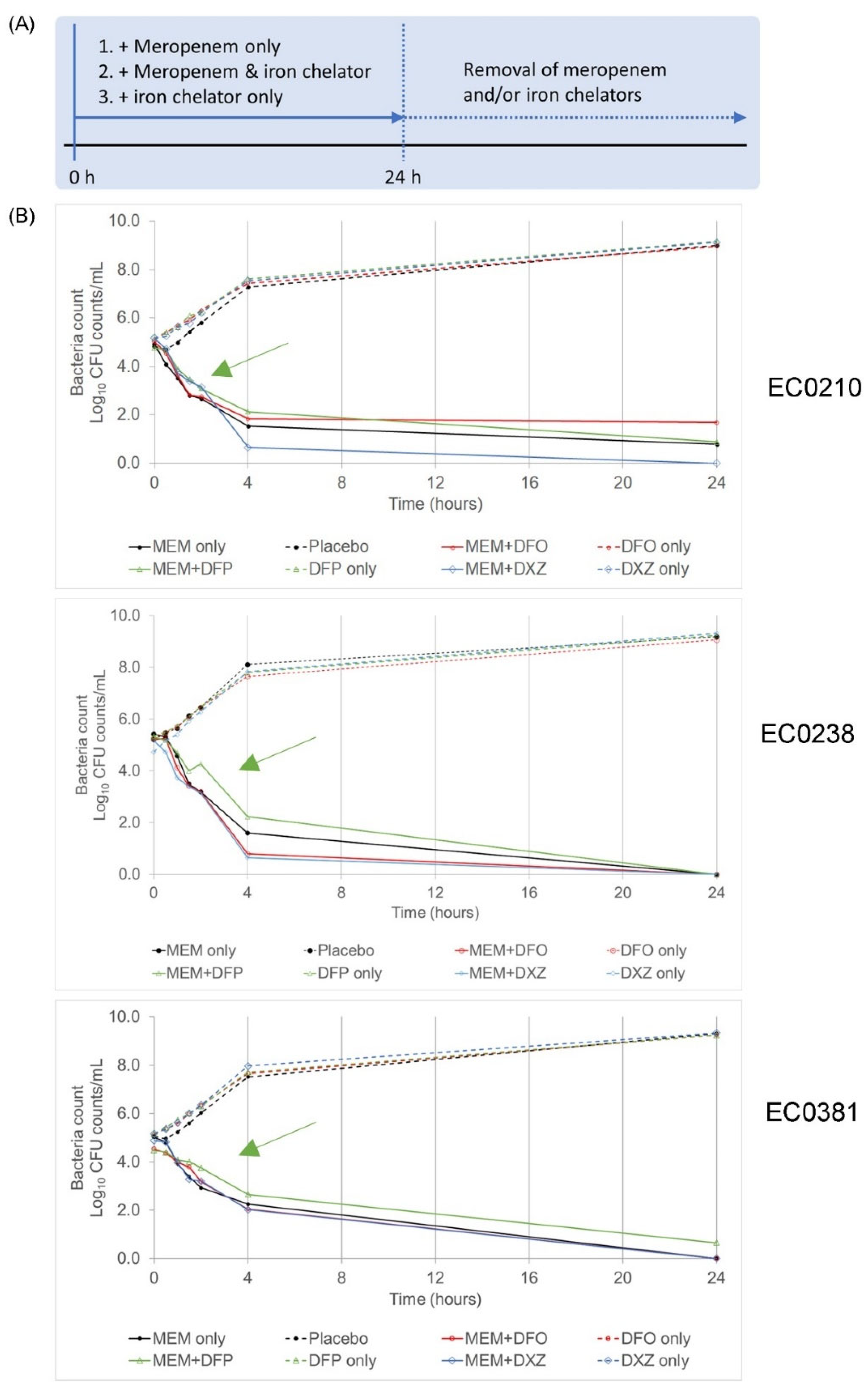
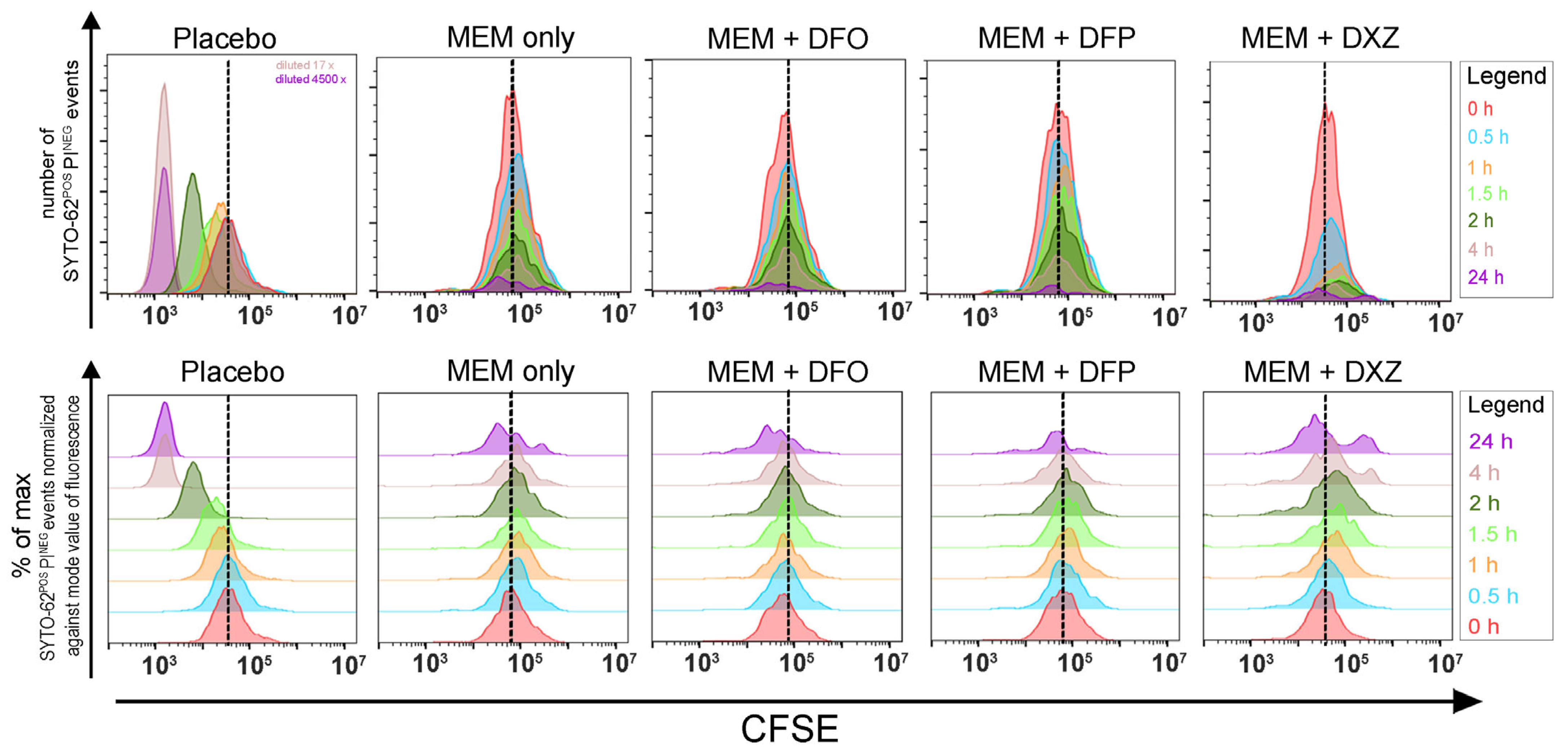
3.1. A Combination of Clinically Approved Iron Chelators and Meropenem Did Not Eradicate Bacteria Persisters
3.2. Iron Chelators Are Inconsistent in Suppressing Persister Resuscitation in All Strains
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.L.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y.; et al. Discovery, research, and development of new antibiotics: The WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet. Infect. Dis. 2018, 18, 318–327. [Google Scholar] [CrossRef] [PubMed]
- Teo, J.Q.M.; Cai, Y.; Lim, T.-P.P.; Tan, T.T.; Kwa, A.L.H. Carbapenem resistance in gram-negative bacteria: The not-so-little problem in the little red dot. Microorganisms 2016, 4, 13. [Google Scholar] [CrossRef] [PubMed]
- Marimuthu, K.; Venkatachalam, I.; Khong, W.X.; Koh, T.H.; Cherng, B.P.Z.; Van La, M.; Pratim De, P.; Krishnan, P.U.; Yen Tan, T.; Choon, R.F.K.; et al. Clinical and molecular epidemiology of carbapenem-resistant enterobacteriaceae among adult inpatients in Singapore. Clin. Infect. Dis. 2017, 64 (Suppl. S2), S68–S75. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.; Venkatachalam, I.; Tee, N.W.; Yen Tan, T.; Kurup, A.; Wong, S.Y.; Low, C.Y.; Wang, Y.; Lee, W.; Liew, Y.X.; et al. Prevalence of healthcare-associated infections and antimicrobial use among adult inpatients in Singapore acute-care hospitals: Results from the first national point prevalence survey. Clin. Infect. Dis. 2017, 64 (Suppl. S2), S61–S67. [Google Scholar] [CrossRef] [PubMed]
- Baez, A.; Sharma, A.K.; Bryukhanov, A.; Anderson, E.D.; Rudack, L.; Olivares-Hernández, R.; Quan, D.; Shiloach, J. Iron availability enhances the cellular energetics of aerobic Escherichia coli cultures while upregulating anaerobic respiratory chains. N. Biotechnol. 2022, 71, 11–20. [Google Scholar] [CrossRef] [PubMed]
- McHugh, J.P.; Rodríguez-Quiñones, F.; Abdul-Tehrani, H.; Svistunenko, D.A.; Poole, R.K.; Cooper, C.E.; Andrews, S.C. Global iron-dependent gene regulation in Escherichia coli: A new mechanism for iron homeostasis. J. Biol. Chem. 2003, 278, 29478–29486. [Google Scholar] [CrossRef]
- Ratha, R.; Altaei, T. Therapeutic Drug Monitoring of Chelating Agent Deferoxamine for β-Thalassemia Major Patients. Int. J. Clin. Med. 2013, 04, 331–342. [Google Scholar] [CrossRef]
- Bellanti, F.; Danhof, M.; Pasqua, O. Della Population pharmacokinetics of deferiprone in healthy subjects. Br. J. Clin. Pharmacol. 2014, 78, 1397–1406. [Google Scholar] [CrossRef] [PubMed]
- Hasinoff, B.B. Dexrazoxane (ICRF-187) Protects Cardiac Myocytes Against Hypoxia-Reoxygenation Damage. Cardiovasc. Toxicol. 2002, 2, 111–118. [Google Scholar] [CrossRef]
- Kwok, J.C.; Richardson, D.R. The cardioprotective effect of the iron chelator dexrazoxane (ICRF-187) on anthracycline-mediated cardiotoxicity. Redox Rep. 2000, 5, 317–324. [Google Scholar] [CrossRef]
- Coraça-Huber, D.C.; Dichtl, S.; Steixner, S.; Nogler, M.; Weiss, G. Iron chelation destabilizes bacterial biofilms and potentiates the antimicrobial activity of antibiotics against coagulase-negative Staphylococci. Pathog. Dis. 2018, 76, fty052. [Google Scholar] [CrossRef] [PubMed]
- Mettrick, K.; Hassan, K.; Lamont, I.; Reid, D. The Iron-chelator, N,N’-bis (2-hydroxybenzyl) Ethylenediamine-N,N’-diacetic acid is an Effective Colistin Adjunct against Clinical Strains of Biofilm-Dwelling Pseudomonas aeruginosa. Antibiotics 2020, 9, 144. [Google Scholar] [CrossRef] [PubMed]
- Moreau-Marquis, S.; O’Toole, G.A.; Stanton, B.A. Tobramycin and FDA-approved iron chelators eliminate Pseudomonas aeruginosa biofilms on cystic fibrosis cells. Am. J. Respir. Cell Mol. Biol. 2009, 41, 305–313. [Google Scholar] [CrossRef] [PubMed]
- Chan, G.C.F.; Chan, S.; Ho, P.L.; Ha, S.Y. Effects of chelators (Deferoxamine, deferiprone and deferasirox) on the growth of klebsiella pneumoniae and aeromonas hydrophila isolated from transfusion-dependent thalassemia patients. Hemoglobin 2009, 33, 352–360. [Google Scholar] [CrossRef] [PubMed]
- van Asbeck, B.S.; Marcelis, J.H.; van Kats, J.H.; Jaarsma, E.Y.; Verhoef, J. Synergy between the iron chelator deferoxamine and the antimicrobial agents gentamicin, chloramphenicol, cefalothin, cefotiam and cefsulodin. Eur. J. Clin. Microbiol. 1983, 2, 432–438. [Google Scholar] [CrossRef] [PubMed]
- Fekri, K.; Khoshdel, A.; Rasoulynezhad, M.; Kheiri, S.; Malekpour, A.; Zamanzad, B. In vitro effect of iron chelators on the growth of Escherichia coli, Staphylococcus epidermidis, Staphylococcus aureus, Yersinia enterocolitica, and Pseudomonas aeruginosa Strains. J. Shahrekord Univ. Med. Sci. 2019, 21, 244–249. [Google Scholar] [CrossRef]
- Gentile, V.; Frangipani, E.; Bonchi, C.; Minandri, F.; Runci, F.; Visca, P. Iron and Acinetobacter baumannii Biofilm formation. Pathogens 2014, 3, 704–719. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, C.; Islam, S.; Jarosch, S.; Zhou, J.; Hoskin, D.; Greenshields, A.; Al-Banna, N.; Sharawy, N.; Sczcesniak, A.; Kelly, M.; et al. The utility of iron chelators in the management of inflammatory disorders. Mediators Inflamm. 2015, 2015, 516740. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.J.; Wei, H.; Frei, B. The iron chelator, desferrioxamine, reduces inflammation and atherosclerotic lesion development in experimental mice. Exp. Biol. Med. 2010, 235, 633–641. [Google Scholar] [CrossRef]
- Fokam, D.; Dickson, K.; Kamali, K.; Holbein, B.; Colp, P.; Stueck, A.; Zhou, J.; Lehmann, C. Iron chelation in murine models of systemic inflammation induced by gram-positive and gram-negative toxins. Antibiotics 2020, 9, 283. [Google Scholar] [CrossRef]
- Kester, J.C.; Fortune, S.M. Persisters and beyond: Mechanisms of phenotypic drug resistance and drug tolerance in bacteria. Crit. Rev. Biochem. Mol. Biol. 2014, 49, 91–101. [Google Scholar] [CrossRef]
- Banks, H.T.; Sutton, K.L.; Thompson, W.C.; Bocharov, G.; Roose, D.; Schenkel, T.; Meyerhans, A. Estimation of cell proliferation dynamics using CFSE data. Bull. Math. Biol. 2011, 73, 116–150. [Google Scholar] [CrossRef]
- Wong, F.H.-S.; Cai, Y.; Leck, H.; Lim, T.-P.; Teo, J.Q.-M.; Lee, W.; Koh, T.H.; Tan, T.T.; Tan, K.W.; Kwa, A.L.-H. Determining the Development of Persisters in Extensively Drug-Resistant Acinetobacter baumannii upon Exposure to Polymyxin B-Based Antibiotic Combinations Using Flow Cytometry. Antimicrob. Agents Chemother. 2020, 64, e01712-19. [Google Scholar] [CrossRef]
- Ueckert, J.E.; Nebe Von-Caron, G.; Bos, A.P.; Ter Steeg, P.F. Flow cytometric analysis of Lactobacillus plantarum to monitor lag times, cell division and injury. Lett. Appl. Microbiol. 1997, 25, 295–299. [Google Scholar] [CrossRef]
- Tod, M.; Lortholary, O.; Seytre, D.; Semaoun, R.; Uzzan, B.; Guillevin, L.; Casassus, P.; Petitjean, O. Population pharmacokinetic study of amikacin administered once or twice daily to febrile, severely neutropenic adults. Antimicrob. Agents Chemother. 1998, 42, 849–856. [Google Scholar] [CrossRef]
- Stein, G.E.E.; Smith, C.L.L.; Scharmen, A.; Kidd, J.M.M.; Cooper, C.; Kuti, J.; Mitra, S.; Nicolau, D.P.P.; Havlichek, D.H.H. Pharmacokinetic and pharmacodynamic analysis of ceftazidime/avibactam in critically ill patients. Surg. Infect. 2019, 20, 55–61. [Google Scholar] [CrossRef]
- Jirkovský, E.; Jirkovská, A.; Bures, J.; Chládek, J.; Lencová, O.; Stariat, J.; Pokorná, Z.; Karabanovich, G.; Roh, J.; Brázdová, P.; et al. Pharmacokinetics of the cardioprotective drug dexrazoxane and its active metabolite adr-925 with focus on cardiomyocytes and the hearts. J. Pharmacol. Exp. Ther. 2018, 364, 433–446. [Google Scholar] [CrossRef]
- Rebuck, J.A.; Fish, D.N.; Abraham, E. Pharmacokinetics of intravenous and oral levofloxacin in critically ill adults in a medical intensive care unit. Pharmacotherapy 2002, 22, 1216–1225. [Google Scholar] [CrossRef]
- Tam, V.H.; Schilling, A.N.; Nikolaou, M. Modelling time-kill studies to discern the pharmacodynamics of meropenem. J. Antimicrob. Chemother. 2005, 55, 699–706. [Google Scholar] [CrossRef]
- Kwa, A.L.H.; Lim, T.P.; Low, J.G.H.; Hou, J.G.; Kurup, A.; Prince, R.A.; Tam, V.H. Pharmacokinetics of polymyxin B1 in patients with multidrug-resistant Gram-negative bacterial infections. Diagn. Microbiol. Infect. Dis. 2008, 60, 163–167. [Google Scholar] [CrossRef]
- Michel, L.V.; Snyder, J.; Schmidt, R.; Milillo, J.; Grimaldi, K.; Kalmeta, B.; Khan, M.N.; Sharma, S.; Wright, L.K.; Pichichero, M.E. Dual orientation of the outer membrane lipoprotein P6 of nontypeable Haemophilus influenzae. J. Bacteriol. 2013, 195, 3252–3259. [Google Scholar] [CrossRef]
- Girardello, R.; Bispo, P.J.M.; Yamanaka, T.M.; Gales, A.C. Cation concentration variability of four distinct Mueller-Hinton agar brands influences polymyxin B susceptibility results. J. Clin. Microbiol. 2012, 50, 2414–2418. [Google Scholar] [CrossRef]
- Thompson, M.G.; Corey, B.W.; Si, Y.; Craft, D.W.; Zurawski, D.V. Antibacterial activities of iron chelators against common nosocomial pathogens. Antimicrob. Agents Chemother. 2012, 56, 5419–5421. [Google Scholar] [CrossRef]
- Lewis, K. Persister cells and the riddle of biofilm survival. Biochem. 2005, 70, 267–274. [Google Scholar] [CrossRef]
- Roberts, M.E.; Stewart, P.S. Modelling protection from antimicrobial agents in biofilms through the formation of persister cells. Microbiology 2005, 151, 75–80. [Google Scholar] [CrossRef]
- Deng, S.; Yan, T.; Jendrny, C.; Nemecek, A.; Vincetic, M.; Gödtel-Armbrust, U.; Wojnowski, L. Dexrazoxane may prevent doxorubicin-induced DNA damage via depleting both Topoisomerase II isoforms. BMC Cancer 2014, 14, 842. [Google Scholar] [CrossRef] [PubMed]
- Hasinoff, B.B.; Kuschak, T.I.; Yalowich, J.C.; Creighton, A.M. A QSAR study comparing the cytotoxicity and DNA topoisomerase II inhibitory effects of bisdioxopiperazine analogs of ICRF-187 (dexrazoxane). Biochem. Pharmacol. 1995, 50, 953–958. [Google Scholar] [CrossRef]
- Van Acker, H.; Gielis, J.; Acke, M.; Cools, F.; Cos, P.; Coenye, T. The role of reactive oxygen species in antibiotic-induced cell death in Burkholderia cepacia complex bacteria. PLoS ONE 2016, 11, e0159837. [Google Scholar] [CrossRef]
- McBee, M.E.; Chionh, Y.H.; Sharaf, M.L.; Ho, P.; Cai, M.W.L.; Dedon, P.C. Production of superoxide in bacteria is stress- and cell state-dependent: A gating-optimized flow cytometry method that minimizes ROS measurement artifacts with fluorescent dyes. Front. Microbiol. 2017, 8, 459. [Google Scholar] [CrossRef]
- Kohanski, M.A.; Dwyer, D.J.; Hayete, B.; Lawrence, C.A.; Collins, J.J. A Common Mechanism of Cellular Death Induced by Bactericidal Antibiotics. Cell 2007, 130, 797–810. [Google Scholar] [CrossRef]
- Dwyer, D.J.; Belenky, P.A.; Yang, J.H.; MacDonald, I.C.; Martell, J.D.; Takahashi, N.; Chan, C.T.Y.; Lobritz, M.A.; Braff, D.; Schwarz, E.G.; et al. Antibiotics induce redox-related physiological alterations as part of their lethality. Proc. Natl. Acad. Sci. USA 2014, 111, E2100–E2109. [Google Scholar] [CrossRef]
- Zager, R.A.A.; Johnson, A.C.M.C.M.; Hanson, S.Y.S.Y. Parenteral iron therapy exacerbates experimental sepsis Rapid Communication. Kidney Int. 2004, 65, 2108–2112. [Google Scholar] [CrossRef]
- Tomaras, A.P.; Dorsey, C.W.; Edelmann, R.E.; Actis, L.A. Attachment to and biofilm formation on abiotic surfaces by Acinetobacter baumannii: Involvement of a novel chaperone-usher pili assembly system. Microbiology 2003, 149, 3473–3484. [Google Scholar] [CrossRef]
- Nairz, M.; Weiss, G. Infections Associated with Iron Administration. Met. Ions Life Sci. 2019, 19, 123–156. [Google Scholar]
- Rando, E.; Segala, F.V.; Vargas, J.; Seguiti, C.; De Pascale, G.; Murri, R.; Fantoni, M. Cefiderocol for severe carbapenem-resistant A. baumannii pneumonia: Towards the comprehension of its place in therapy. Antibiotics 2022, 11, 3. [Google Scholar] [CrossRef]
- Wang, Q.; Jin, L.; Sun, S.; Yin, Y.; Wang, R.; Chen, F.; Wang, X.; Zhang, Y.Y.; Hou, J.; Zhang, Y.Y.; et al. Occurrence of High Levels of Cefiderocol Resistance in Carbapenem-Resistant Escherichia coli before Its Approval in China: A Report from China CRE-Network. Microbiol. Spectr. 2022, 10, e0267021. [Google Scholar] [CrossRef] [PubMed]
- Falcone, M.; Tiseo, G.; Leonildi, A.; Sala, L.D.; Vecchione, A.; Barnini, S.; Farcomeni, A.; Menichetti, F. Cefiderocol-Compared to Colistin-Based Regimens for the Treatment of Severe Infections Caused by Carbapenem-Resistant Acinetobacter baumannii. Antimicrob. Agents Chemother. 2022, 66, e0214221. [Google Scholar] [CrossRef]
- Klein, S.; Boutin, S.; Kocer, K.; Fiedler, M.O.; Störzinger, D.; Weigand, M.A.; Tan, B.; Richter, D.; Rupp, C.; Mieth, M.; et al. Rapid Development of Cefiderocol Resistance in Carbapenem-resistant Enterobacter cloacae During Therapy Is Associated with Heterogeneous Mutations in the Catecholate Siderophore Receptor cirA. Clin. Infect. Dis. 2022, 74, 905–908. [Google Scholar] [CrossRef] [PubMed]
- Lan, P.; Lu, Y.; Chen, Z.; Wu, X.; Hua, X.; Jiang, Y.; Zhou, J.; Yu, Y. Emergence of High-Level Cefiderocol Resistance in Carbapenem-Resistant Klebsiella pneumoniae from Bloodstream Infections in Patients with Hematologic Malignancies in China. Microbiol. Spectr. 2022, 10, e0008422. [Google Scholar] [CrossRef]
- Ou, F.; McGoverin, C.; Swift, S.; Vanholsbeeck, F. Absolute bacterial cell enumeration using flow cytometry. J. Appl. Microbiol. 2017, 123, 464–477. [Google Scholar] [CrossRef]
- Clarke, R.G.; Pinder, A.C. Improved detection of bacteria by flow cytometry using a combination of antibody and viability markers. J. Appl. Microbiol. 1998, 84, 577–584. [Google Scholar] [CrossRef] [PubMed]
- Ou, F.; McGoverin, C.; Swift, S.; Vanholsbeeck, F. Near real-time enumeration of live and dead bacteria using a fibre-based spectroscopic device. Sci. Rep. 2019, 9, 4807. [Google Scholar] [CrossRef] [PubMed]
- Kerstens, M.; Boulet, G.; Tritsmans, C.; Horemans, T.; Hellings, M.; Delputte, P.; Maes, L.; Cos, P. Flow Cytometric Enumeration of Bacteria Using TO-PRO®-3 Iodide as a Single-Stain Viability Dye. J. Lab. Autom. 2014, 19, 555–561. [Google Scholar] [CrossRef] [PubMed]
- Gunasekera, T.S.; Attfield, P.V.; Veal, D.A. A flow cytometry method for rapid detection and enumeration of total bacteria in milk. Appl. Environ. Microbiol. 2000, 66, 1228–1232. [Google Scholar] [CrossRef] [PubMed]
- Balaban, N.Q.; Helaine, S.; Lewis, K.; Ackermann, M.; Aldridge, B.; Andersson, D.I.; Brynildsen, M.P.; Bumann, D.; Camilli, A.; Collins, J.J.; et al. Definitions and guidelines for research on antibiotic persistence. Nat. Rev. Microbiol. 2019, 17, 441–448. [Google Scholar] [CrossRef]
- Bigger, J.W. Treatment of Staphylococcal Infections With Penicillin By Intermittent Sterilisation. Lancet 1944, 244, 497–500. [Google Scholar] [CrossRef]

| Antibiotics (Abbreviations) | Company | Final Concentration (mg/L) | Reference | |
|---|---|---|---|---|
| Amikacin | TRC 1 | 65 | Tod et al., 1998 [25] | |
| Ceftazidime–avibactam (CZA) | Ceftazidime | TRC 1 | 21 | Stein et al., 2019 [26] |
| Avibactam | Pfizer | 5.25 | ||
| Deferiprone (DFP) 2 | ChemScene | 26.49 | Bellanti et al., 2014 [8] | |
| Deferoxamine mesylate (DFO) 2 | ChemScene | 43.7 | Ratha et al., 2013 [7] | |
| Dexrazoxane (DXZ) 2 | ApexBio | 36.5 | Jirkovský et al., 2018 [27] | |
| Levofloxacin (LVX) | Daiichi | 8 | Rebuck et al., 2002 [28] | |
| Meropenem (MEM) | TRC 1 | 20 | Tam et al., 2005 [29] | |
| Polymyxin B (PMB) | TRC 1 | 2 | Kwa et al., 2008 [30] |
| Conditions | EC0210 | EC0238 | EC0381 | |
|---|---|---|---|---|
| No antibiotic | Log10 CFU/mL at 24 h | 9.03 | 9.24 | 9.36 |
| Meropenem (MEM) only | Log10 CFU/mL at 24 h | 1.69 | 0 | 0 |
| Resuscitate after removal of MEM? | Yes | Yes | Yes | |
| MEM + Deferoxamine mesylate (DFO) | Log10 CFU/mL at 24 h | 1.69 | 0 | 0 |
| Resuscitate after removal of MEM + DFO? | Yes | Yes | Yes | |
| MEM + Deferiprone (DFP) | Log10 CFU/mL at 24 h | 0.89 | 0.95 | 0.65 |
| Resuscitate after removal of MEM + DFP? | Yes | Yes | Yes | |
| MEM + Dexrazoxane (DXZ) | Log10 CFU/mL at 24 h | 0 | 0 | 0 |
| Resuscitate after removal of MEM + DXZ? | Yes | Yes | Yes |
| Resuscitate at 48 h in the Presence of Iron Chelators? | |||
|---|---|---|---|
| EC0210 | EC0238 | EC0381 | |
| Deferoxamine mesylate (DFO) | Yes | No | Yes |
| Deferiprone (DFP) | Yes | No | Yes |
| Dexrazoxane (DXZ) | No | Yes | No |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yeo, J.H.; Begam, N.; Leow, W.T.; Goh, J.X.; Zhong, Y.; Cai, Y.; Kwa, A.L.-H. Ironing out Persisters? Revisiting the Iron Chelation Strategy to Target Planktonic Bacterial Persisters Harboured in Carbapenem-Resistant Escherichia coli. Microorganisms 2024, 12, 972. https://doi.org/10.3390/microorganisms12050972
Yeo JH, Begam N, Leow WT, Goh JX, Zhong Y, Cai Y, Kwa AL-H. Ironing out Persisters? Revisiting the Iron Chelation Strategy to Target Planktonic Bacterial Persisters Harboured in Carbapenem-Resistant Escherichia coli. Microorganisms. 2024; 12(5):972. https://doi.org/10.3390/microorganisms12050972
Chicago/Turabian StyleYeo, Jia Hao, Nasren Begam, Wan Ting Leow, Jia Xuan Goh, Yang Zhong, Yiying Cai, and Andrea Lay-Hoon Kwa. 2024. "Ironing out Persisters? Revisiting the Iron Chelation Strategy to Target Planktonic Bacterial Persisters Harboured in Carbapenem-Resistant Escherichia coli" Microorganisms 12, no. 5: 972. https://doi.org/10.3390/microorganisms12050972





