Improvement of Intestinal Immune Cell Function by Lactic Acid Bacteria for Dairy Products
Abstract
:1. Introduction
2. Materials and Methods
2.1. Mice
2.2. Lactic Acid Bacteria
2.3. Cell Isolation from Peyer’s Patches (PPs)
2.4. Isolation of CD11c+ Cells
2.5. Isolation of Naïve T Cells
2.6. Generation of Bone Marrow-Derived Dendritic Cells (BMDCs)
2.7. CD11c+ Cells and Naïve T Cells Co-Culture
2.8. Measurement of Cytokine
2.9. RNA Extraction, cDNA Synthesis, and qRT-PCR
2.10. Statistical Analysis
3. Results
3.1. CD4+ Cells Activation in Peyer’s Patches by LAB
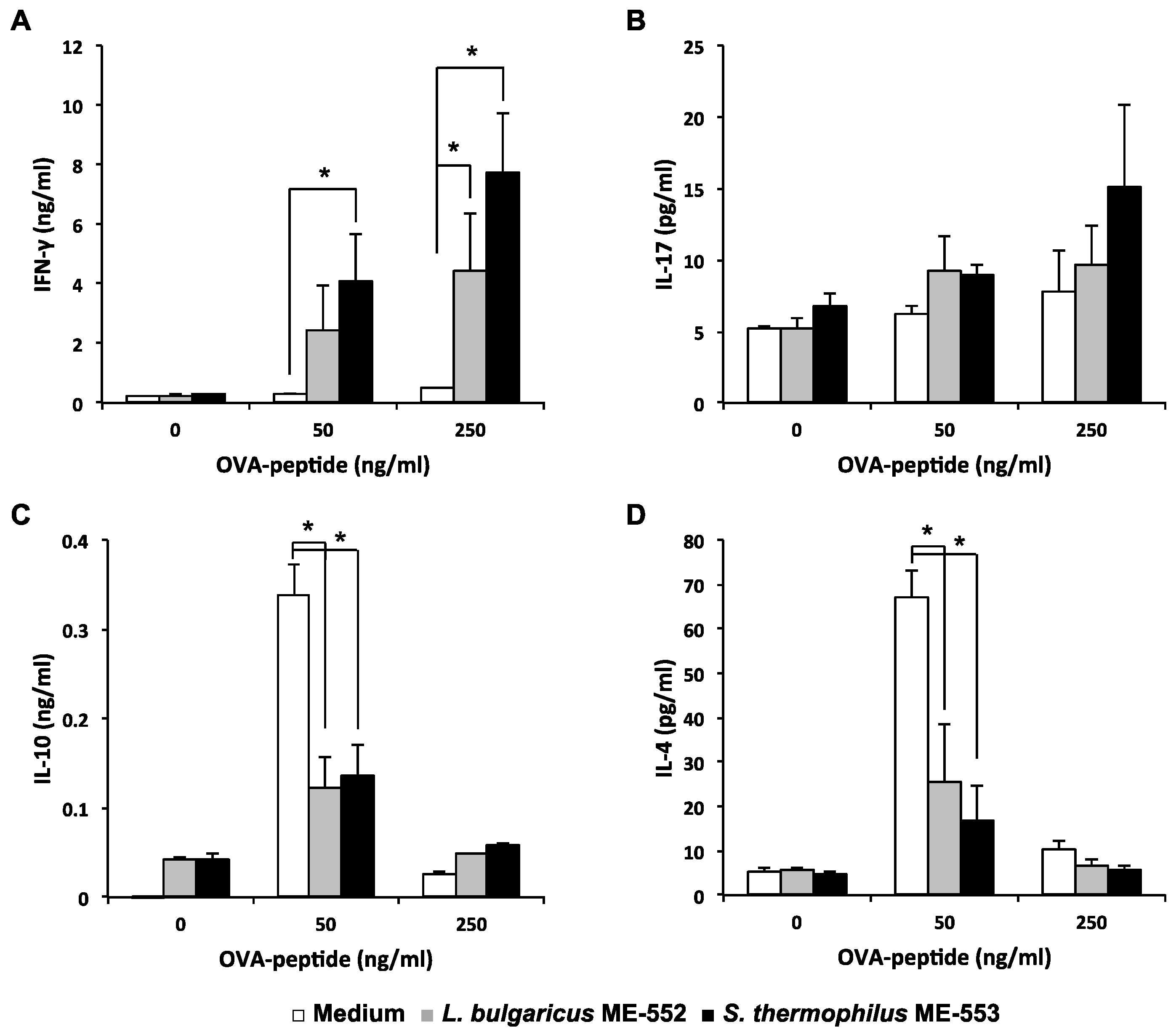
3.2. Antigen-Specific T Helper Cell Differentiation by LAB
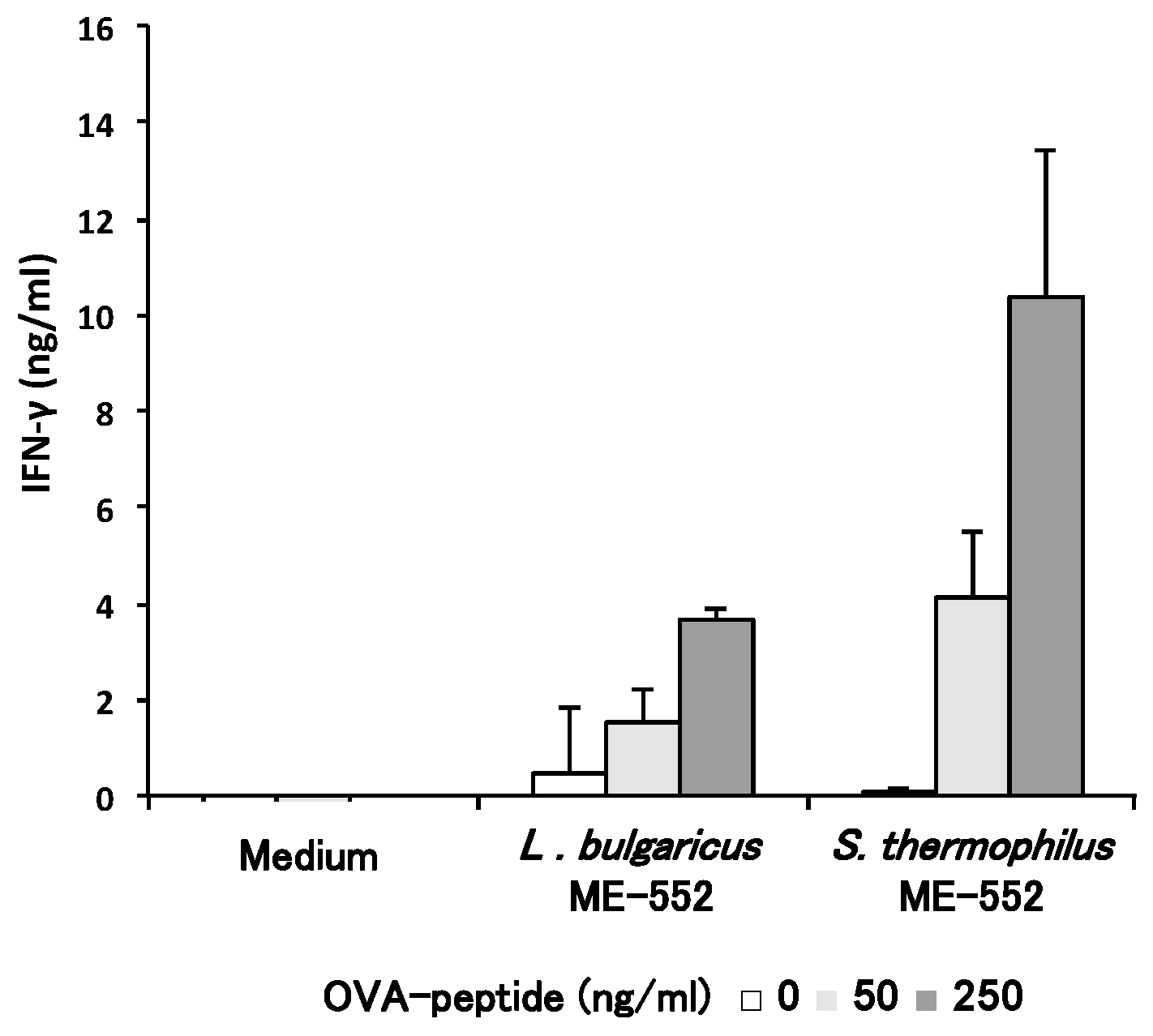
3.3. Th1 Cell Differentiation with CD11c+ Cell from Peyer’s Patches by LAB
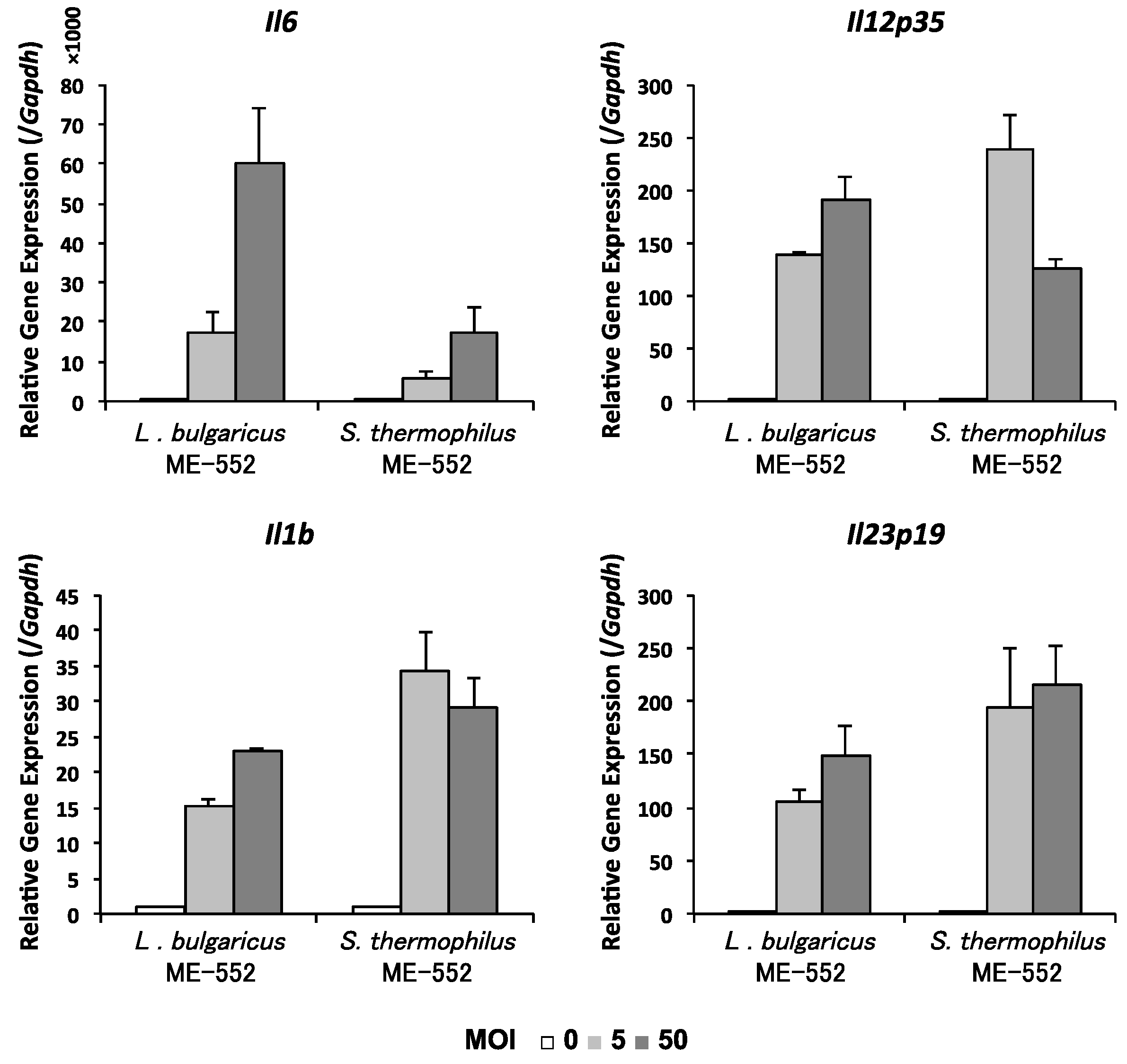
3.4. Induction of Cytokine in Dendritic Cell by LAB
4. Discussion
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Quinto, E.J.; Jiménez, P.; Caro, I.; Tejero, J.; Mateo, J.; Girbés, T. Probiotic Lactic Acid Bacteria: A Review. Food Nutr. Sci. 2014, 5, 1765–1775. [Google Scholar] [CrossRef]
- Heyman, M.; Ménard, S. Probiotic microorganisms: How they affect intestinal pathophysiology. Cell. Mol. Life Sci. 2002, 59, 1151–1165. [Google Scholar] [CrossRef] [PubMed]
- Selhub, E.M.; Logan, A.C.; Bested, A.C. Fermented foods, microbiota, and mental health: Ancient practice meets nutritional psychiatry. J. Physiol. Anthropol. 2014, 33, 1. [Google Scholar] [CrossRef] [PubMed]
- Yoshimatsu, Y.; Yamada, A.; Furukawa, R.; Sono, K.; Osamura, A.; Nakamura, K.; Aoki, H.; Tsuda, Y.; Hosoe, N.; Takada, N.; Suzuki, Y. Effectiveness of probiotic therapy for the prevention of relapse in patients with inactive ulcerative colitis. World J. Gastroenterol. 2015, 21, 5985–5994. [Google Scholar] [PubMed]
- Amagase, H. Current Marketplace for Probiotics: A Japanese Perspective. Clin. Infect. Dis. 2008, 46, S73–S75. [Google Scholar] [CrossRef] [PubMed]
- Behnsen, J.; Jellbauer, S.; Wong, C.P.; Edwards, R.A.; George, M.D.; Ouyang, W.; Raffatellu, M. The Cytokine IL-22 Promotes Pathogen Colonization by Suppressing Related Commensal Bacteria. Immunity 2014, 40, 262–273. [Google Scholar] [CrossRef] [PubMed]
- Macpherson, A.J.; Harris, N.L. Opinion: Interactions between commensal intestinal bacteria and the immune system. Nat. Rev. Immunol. 2004, 4, 478–485. [Google Scholar] [CrossRef] [PubMed]
- Cogen, A.L.; Nizet, V.; Gallo, R.L. Skin microbiota: A source of disease or defence? Br. J. Dermatol. 2008, 158, 442–455. [Google Scholar] [CrossRef] [PubMed]
- Naik, S.; Bouladoux, N.; Wilhelm, C.; Molloy, M.J.; Salcedo, R.; Kastenmuller, W.; Deming, C.; Quinones, M.; Koo, L.; Conlan, S.; et al. Compartmentalized Control of Skin Immunity by Resident Commensals. Science 2012, 337, 1115–1119. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, I.I.; Atarashi, K.; Manel, N.; Brodie, E.L.; Shima, T.; Karaoz, U.; Wei, D.; Goldfarb, K.C.; Santee, C.A.; Lynch, S.V.; et al. Induction of Intestinal Th17 Cells by Segmented Filamentous Bacteria. Cell 2009, 139, 485–498. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.-J.; Ivanov, I.I.; Darce, J.; Hattori, K.; Shima, T.; Umesaki, Y.; Littman, D.R.; Benoist, C.; Mathis, D. Gut-Residing Segmented Filamentous Bacteria Drive Autoimmune Arthritis via T Helper 17 Cells. Immunity 2010, 32, 815–827. [Google Scholar] [CrossRef] [PubMed]
- Atarashi, K.; Tanoue, T.; Shima, T.; Imaoka, A.; Kuwahara, T.; Momose, Y.; Cheng, G.; Yamasaki, S.; Saito, T.; Ohba, Y.; et al. Induction of Colonic Regulatory T Cells by Indigenous Clostridium Species. Science 2011, 331, 337–341. [Google Scholar] [CrossRef] [PubMed]
- Kawashima, T.; Kosaka, A.; Yan, H.; Guo, Z.; Uchiyama, R.; Fukui, R.; Kaneko, D.; Kumagai, Y.; You, D.-J.; Carreras, J.; et al. Double-Stranded RNA of Intestinal Commensal but Not Pathogenic Bacteria Triggers Production of Protective Interferon-β. Immunity 2013, 38, 1187–1197. [Google Scholar] [CrossRef] [PubMed]
- Mohamadzadeh, M.; Pfeiler, E.A.; Brown, J.B.; Zadeh, M.; Gramarossa, M.; Managlia, E.; Bere, P.; Sarraj, B.; Khan, M.W.; Pakanati, K.C.; et al. Regulation of induced colonic inflammation by Lactobacillus acidophilus deficient in lipoteichoic acid. Proc. Natl. Acad. Sci. 2011, 108, 4623–4630. [Google Scholar] [CrossRef] [PubMed]
- Herve-Jimenez, L.; Guillouard, I.; Guedon, E.; Boudebbouze, S.; Hols, P.; Monnet, V.; Maguin, E.; Rul, F. Postgenomic Analysis of Streptococcus thermophilus Cocultivated in Milk with Lactobacillus delbrueckii subsp. bulgaricus: Involvement of Nitrogen, Purine, and Iron Metabolism. Appl. Environ. Microbiol. 2009, 75, 2062–2073. [Google Scholar] [CrossRef] [PubMed]
- del Carmen, S.; de Moreno de LeBlanc, A.; Martin, R.; Chain, F.; Langella, P.; Bermudez-Humaran, L.G.; LeBlanc, J.G. Genetically Engineered Immunomodulatory Streptococcus thermophilus Strains Producing Antioxidant Enzymes Exhibit Enhanced Anti-Inflammatory Activities. Appl. Environ. Microbiol. 2014, 80, 869–877. [Google Scholar] [CrossRef] [PubMed]
- del Carmen, S.; de Moreno de LeBlanc, A.; LeBlanc, J.G. Development of a potential probiotic yoghurt using selected anti-inflammatory lactic acid bacteria for prevention of colitis and carcinogenesis in mice. J. Appl. Microbiol. 2016, 121, 821–830. [Google Scholar] [CrossRef] [PubMed]
- Perdigón, G.; Maldonado Galdeano, C.; Valdez, J.C.; Medici, M. Interaction of lactic acid bacteria with the gut immune system. Eur. J. Clin. Nutr. 2002, 56, S21–S26. [Google Scholar] [CrossRef] [PubMed]
- Makino, S.; Ikegami, S.; Kano, H.; Sashihara, T.; Sugano, H.; Horiuchi, H.; Saito, T.; Oda, M. Immunomodulatory Effects of Polysaccharides Produced by Lactobacillus delbrueckii ssp. bulgaricus OLL1073R-1. J. Dairy Sci. 2006, 89, 2873–2881. [Google Scholar] [CrossRef]
- Sashihara, T.; Sueki, N.; Furuichi, K.; Ikegami, S. Effect of growth conditions of Lactobacillus gasseri OLL2809 on the immunostimulatory activity for production of interleukin-12 (p70) by murine splenocytes. Int. J. Food Microbiol. 2007, 120, 274–281. [Google Scholar] [CrossRef] [PubMed]
- Mitsuoka, T. Development of Functional Foods. Biosci. Microbiota Food Heal. 2014, 33, 117–128. [Google Scholar] [CrossRef] [PubMed]
- Mitsuoka, T. Establishment of Intestinal Bacteriology. Biosci. Microbiota Food Heal. 2014, 33, 99–116. [Google Scholar] [CrossRef] [PubMed]
- Shimazu, T.; Villena, J.; Tohno, M.; Fujie, H.; Hosoya, S.; Shimosato, T.; Aso, H.; Suda, Y.; Kawai, Y.; Saito, T.; et al. Immunobiotic Lactobacillus jensenii Elicits Anti-Inflammatory Activity in Porcine Intestinal Epithelial Cells by Modulating Negative Regulators of the Toll-Like Receptor Signaling Pathway. Infect. Immun. 2012, 80, 276–288. [Google Scholar] [CrossRef] [PubMed]
- Tsuji, N.M.; Kosaka, A. Oral tolerance: Intestinal homeostasis and antigen-specific regulatory T cells. Trends Immunol. 2008, 29, 532–540. [Google Scholar] [CrossRef] [PubMed]
- Hill, C.; Guarner, F.; Reid, G.; Gibson, G.R.; Merenstein, D.J.; Pot, B.; Morelli, L.; Canani, R.B.; Flint, H.J.; Salminen, S.; et al. Expert consensus document: The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 506–514. [Google Scholar] [CrossRef] [PubMed]
- Jang, M.H.; Kweon, M.-N.; Iwatani, K.; Yamamoto, M.; Terahara, K.; Sasakawa, C.; Suzuki, T.; Nochi, T.; Yokota, Y.; Rennert, P.D.; et al. Intestinal villous M cells: An antigen entry site in the mucosal epithelium. Proc. Natl. Acad. Sci. 2004, 101, 6110–6115. [Google Scholar] [CrossRef] [PubMed]
- Park, M.-J.; Min, S.-Y.; Park, K.-S.; Cho, Y.-G.; Cho, M.-L.; Jung, Y.-O.; Park, H.-S.; Chang, S.-H.; Cho, S.; Min, J.-K.; et al. Indoleamine 2,3-dioxygenase-expressing dendritic cells are involved in the generation of CD4+CD25+ regulatory T cells in Peyer’s patches in an orally tolerized, collagen-induced arthritis mouse model. Arthritis Res. Ther. 2008, 10, R11. [Google Scholar] [CrossRef] [PubMed]
- Tsuji, N.M. Antigen-specific, CD4+CD25+ regulatory T cell clones induced in Peyer’s patches. Int. Immunol. 2003, 15, 525–534. [Google Scholar] [CrossRef] [PubMed]
- Jung, C.; Hugot, J.-P.; Barreau, F. Peyer’s Patches: The Immune Sensors of the Intestine. Int. J. Inflam. 2010, 2010, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Tsuji, N.M.; Mizumachi, K.; Kurisaki, J. Interleukin-10-secreting Peyer’s patch cells are responsible for active suppression in low-dose oral tolerance. Immunology 2001, 103, 458–464. [Google Scholar] [CrossRef] [PubMed]
- Hibi, M.; Hachimura, S.; Ise, W.; Sato, A.; Yoshida, T.; Takayama, T.; Sasaki, K.; Senga, T.; Hashizume, S.; Totsuka, M.; Kaminogawa, S. Dendritic Cells from Spleen, Mesenteric Lymph Node and Peyer’s Patch Can Induce the Production of Both IL-4 and IFN- from Primary Cultures of Naive CD4+ T Cells in a Dose-Dependent Manner. Cytotechnology 2003, 43, 49–55. [Google Scholar] [CrossRef] [PubMed]
- Mbongue, J.; Nicholas, D.; Firek, A.; Langridge, W. The Role of Dendritic Cells in Tissue-Specific Autoimmunity. J. Immunol. Res. 2014, 2014, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Wenzel, U.A.; Jonstrand, C.; Hansson, G.C.; Wick, M.J. CD103+CD11b+ Dendritic Cells Induce Th17 T Cells in Muc2-Deficient Mice with Extensively Spread Colitis. PLoS One 2015, 10, e0130750. [Google Scholar] [CrossRef] [PubMed]
- Mohammed, J.; Beura, L.K.; Bobr, A.; Astry, B.; Chicoine, B.; Kashem, S.W.; Welty, N.E.; Igyártó, B.Z.; Wijeyesinghe, S.; Thompson, E.A.; et al. Stromal cells control the epithelial residence of DCs and memory T cells by regulated activation of TGF-β. Nat. Immunol. 2016, 17, 414–421. [Google Scholar] [CrossRef] [PubMed]
- Dillon, S.; Agrawal, A.; Van Dyke, T.; Landreth, G.; McCauley, L.; Koh, A.; Maliszewski, C.; Akira, S.; Pulendran, B. A Toll-Like Receptor 2 Ligand Stimulates Th2 Responses In Vivo, via Induction of Extracellular Signal-Regulated Kinase Mitogen-Activated Protein Kinase and c-Fos in Dendritic Cells. J. Immunol. 2004, 172, 4733–4743. [Google Scholar] [CrossRef] [PubMed]
- Netea, M.G.; Van der Meer, J.W.M.; Sutmuller, R.P.; Adema, G.J.; Kullberg, B.-J. From the Th1/Th2 Paradigm towards a Toll-Like Receptor/T-Helper Bias. Antimicrob. Agents Chemother. 2005, 49, 3991–3996. [Google Scholar] [CrossRef] [PubMed]
- Kosaka, A.; Yan, H.; Ohashi, S.; Gotoh, Y.; Sato, A.; Tsutsui, H.; Kaisho, T.; Toda, T.; Tsuji, N.M. Lactococcus lactis subsp. cremoris FC triggers IFN-γ production from NK and T cells via IL-12 and IL-18. Int. Immunopharmacol. 2012, 14, 729–733. [Google Scholar] [CrossRef] [PubMed]
- Saijo, S.; Ikeda, S.; Yamabe, K.; Kakuta, S.; Ishigame, H.; Akitsu, A.; Fujikado, N.; Kusaka, T.; Kubo, S.; Chung, S.; et al. Dectin-2 Recognition of α-Mannans and Induction of Th17 Cell Differentiation Is Essential for Host Defense against Candida albicans. Immunity 2010, 32, 681–691. [Google Scholar] [CrossRef] [PubMed]
- Taylor, P.R.; Roy, S.; Leal, S.M.; Sun, Y.; Howell, S.J.; Cobb, B.A.; Li, X.; Pearlman, E. Activation of neutrophils by autocrine IL-17A–IL-17RC interactions during fungal infection is regulated by IL-6, IL-23, RORγt and dectin-2. Nat. Immunol. 2013, 15, 143–151. [Google Scholar] [CrossRef] [PubMed]

| Gene | Forward (5’-, -3’) | Reverse (5’-, -3’) |
|---|---|---|
| Il1b | TCCAGGATGAGGACATGAGCAC | GAACGTCACACACCAGCAGGTTA |
| Il6 | CCACTTCACAAGTCGGAGGCTTA | CCAGTTTGGTAGCATCCATCATTTC |
| Il12p35 | GTCTTAGCCAGTCCCGAAACC | TCTTCATGATCGATGTCTTCAGCAG |
| Il23p19 | ACATGCACCAGCGGGACATA | CCTTGTGGGTCACAACCATCTTC |
© 2016 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kamiya, T.; Watanabe, Y.; Makino, S.; Kano, H.; Tsuji, N.M. Improvement of Intestinal Immune Cell Function by Lactic Acid Bacteria for Dairy Products. Microorganisms 2017, 5, 1. https://doi.org/10.3390/microorganisms5010001
Kamiya T, Watanabe Y, Makino S, Kano H, Tsuji NM. Improvement of Intestinal Immune Cell Function by Lactic Acid Bacteria for Dairy Products. Microorganisms. 2017; 5(1):1. https://doi.org/10.3390/microorganisms5010001
Chicago/Turabian StyleKamiya, Tomonori, Yohei Watanabe, Seiya Makino, Hiroshi Kano, and Noriko M Tsuji. 2017. "Improvement of Intestinal Immune Cell Function by Lactic Acid Bacteria for Dairy Products" Microorganisms 5, no. 1: 1. https://doi.org/10.3390/microorganisms5010001






