The Fish Pathogen Vibrio ordalii Under Iron Deprivation Produces the Siderophore Piscibactin
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Growth Conditions
2.2. RNA Extraction and RT-PCR
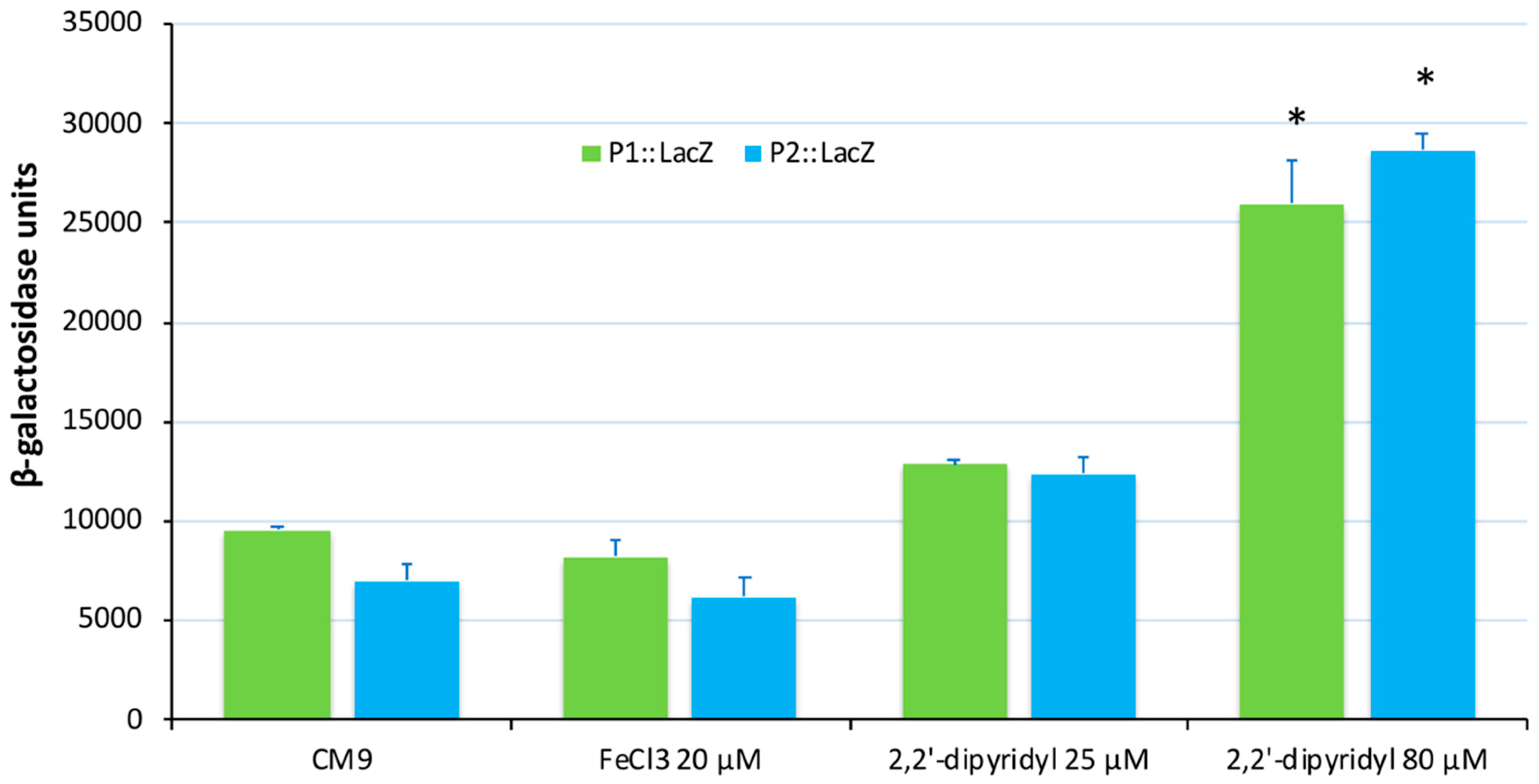
2.3. Construction of lacZ Transcriptional Fusions and β-Galactosidase Assays
2.4. Analysis of Outer Membrane Proteins (OMP) Profile of V. ordalii
2.5. Protein Identification by Peptide Mass Fingerprinting (PMF)
2.6. Bioinformatics Tools
2.7. Detection of Siderophore Piscibactin
2.8. Statistical Analysis
3. Results
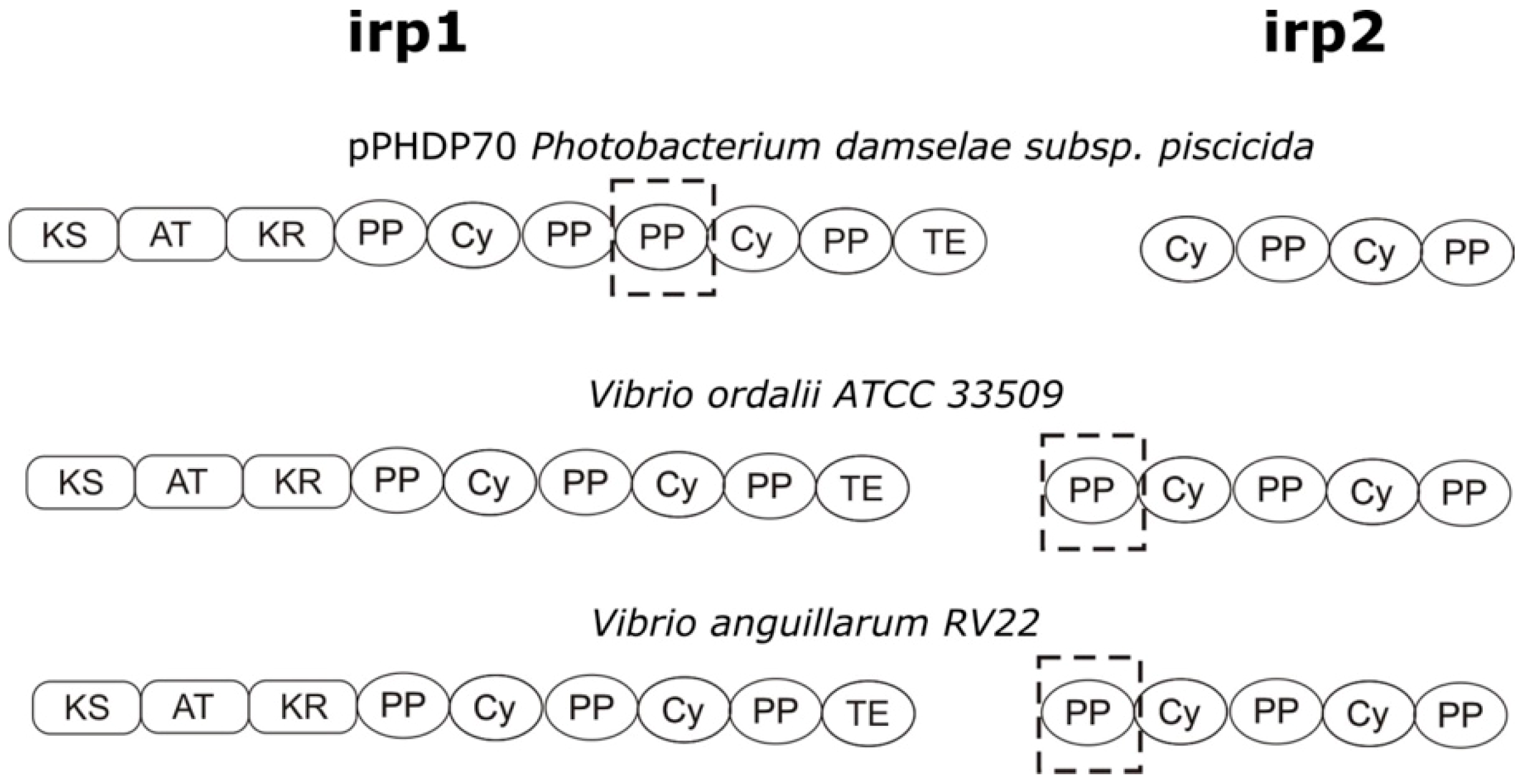
3.1. Characterization of the V. ordalii Gene Cluster Encoding a Piscibactin-Like Siderophore
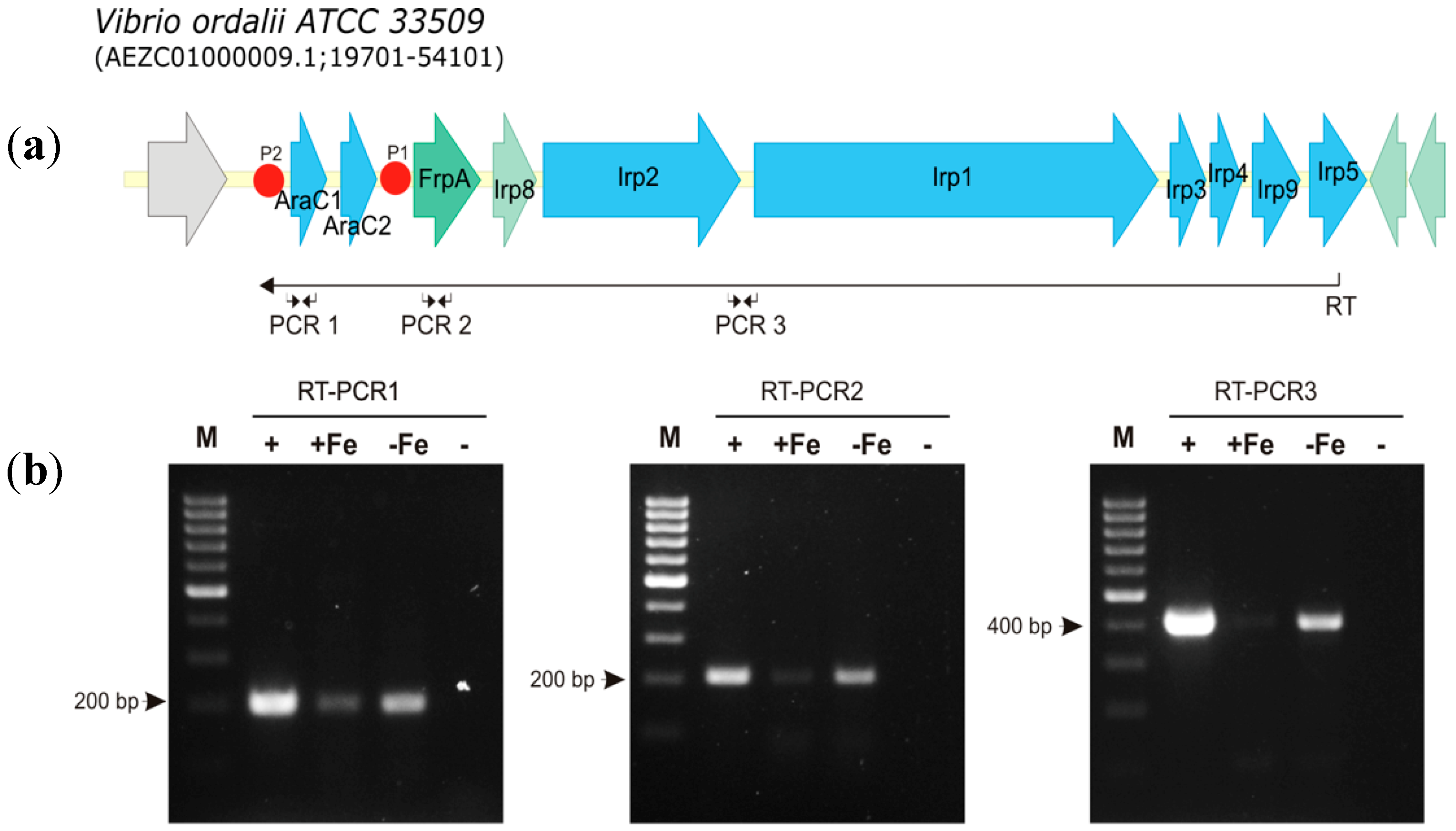
3.2. Transcriptional Analysis and Iron Regulation of the Irp Gene Cluster of V. ordalii
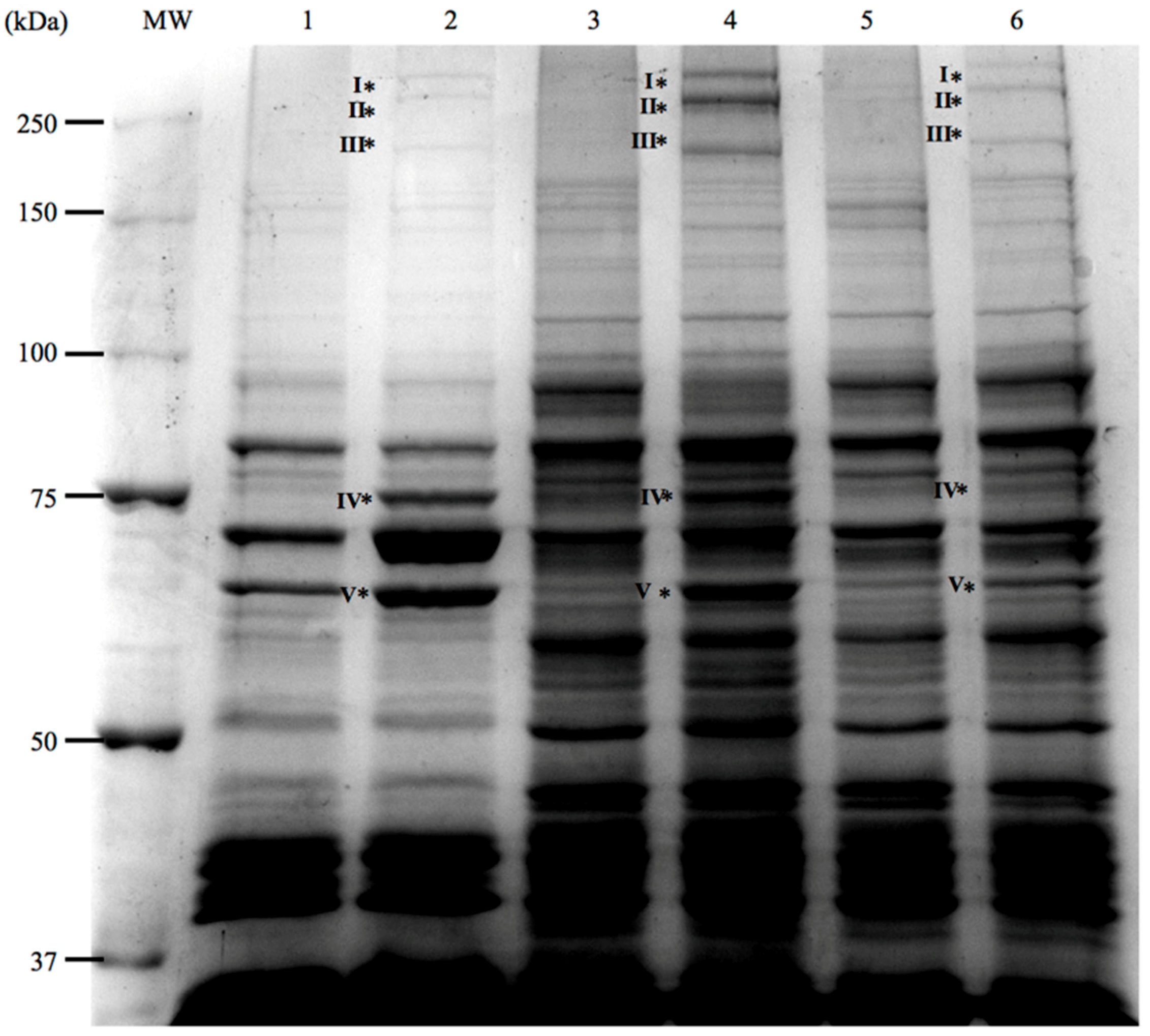
3.3. Analysis of Iron-Regulated Outer Membrane Proteins
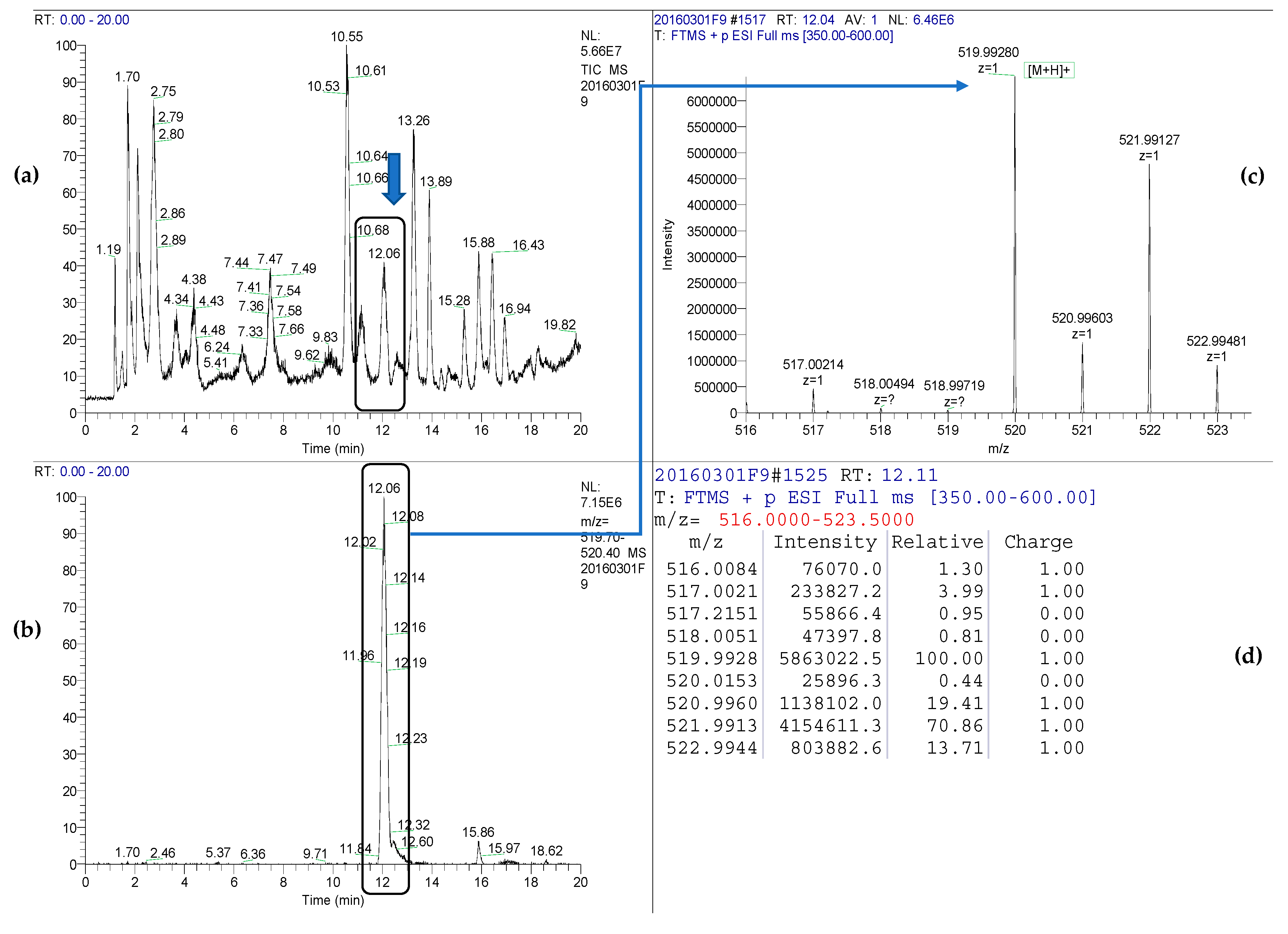
3.4. Identification of Siderophores in Cultures of V. ordalii
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Toranzo, A.E.; Magariños, B.; Avendaño-Herrera, R. Vibriosis: Vibrio anguillarum, V. ordalii and Aliivibrio salmonicida. In Fish Viruses and Bacteria: Pathobiology and Protection; Woo, P.T.K., Cipriano, R.C., Eds.; CABI: London, UK, 2017; pp. 314–333. [Google Scholar]
- Colquhoun, D.J.; Aase, I.L.; Wallace, C.; Baklien, A.; Gravningen, K. First description of Vibrio ordalii from Chile. Bull. Eur. Assoc. Fish. Pathol. 2004, 24, 185–188. [Google Scholar]
- Ruiz, P.; Poblete, M.; Yañez, A.J.; Irgang, R.; Toranzo, A.E.; Avendaño-Herrera, R. Cell-surface properties of Vibrio ordalii strains isolated from Atlantic salmon Salmo salar in Chilean farms. Dis. Aquat. Organ. 2015, 113, 9–23. [Google Scholar] [CrossRef] [PubMed]
- Naka, H.; Dias, G.M.; Thompson, C.C.; Dubay, C.; Thompson, F.L.; Crosa, J.H. Complete genome sequence of the marine fish pathogen Vibrio anguillarum harboring the pJM1 virulence plasmid and genomic comparison with other virulent strains of V. anguillarum and V. ordalii. Infect. Immun. 2011, 79, 2889–2900. [Google Scholar] [CrossRef] [PubMed]
- Steinum, T.M.; Karatas, S.; Martinussen, N.T.; Meirelles, P.M.; Thompson, F.L.; Colquhoun, D.J. Multilocus Sequence Analysis of Close Relatives Vibrio anguillarum and Vibrio ordalii. Appl. Environ. Microbiol. 2016, 82, 5496–5504. [Google Scholar] [CrossRef] [PubMed]
- Ruiz, P.; Poblete-Morales, M.; Irgang, R.; Toranzo, A.E.; Avendaño-Herrera, R. Survival behaviour and virulence of the fish pathogen Vibrio ordalii in seawater microcosms. Dis. Aquat. Organ. 2016, 120, 27–38. [Google Scholar] [CrossRef] [PubMed]
- Miethke, M. Molecular strategies of microbial iron assimilation: From high-affinity complexes to cofactor assembly systems. Metallomics 2013, 5, 15–28. [Google Scholar] [CrossRef] [PubMed]
- Skaar, E.P.; Raffatellu, M. Metals in infectious diseases and nutritional immunity. Metallomics 2015, 7, 926–928. [Google Scholar] [CrossRef] [PubMed]
- Wilson, B.R.; Bogdan, A.R.; Miyazawa, M.; Hashimoto, K.; Tsuji, Y. Siderophores in Iron Metabolism: From Mechanism to Therapy Potential. Trends Mol. Med. 2016, 22, 1077–1090. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hood, M.I.; Skaar, E.P. Nutritional immunity: Transition metals at the pathogen-host interface. Nat. Rev. Microbiol. 2012, 10, 525–537. [Google Scholar] [CrossRef]
- Fillat, M.F. The FUR (ferric uptake regulator) superfamily: Diversity and versatility of key transcriptional regulators. Arch. Biochem. Biophys. 2014, 546, 41–52. [Google Scholar] [CrossRef]
- Richard, K.L.; Kelley, B.R.; Johnson, J.G. Heme Uptake and Utilization by Gram-Negative Bacterial Pathogens. Front. Cell. Infect. Microbiol. 2019, 9, 81. [Google Scholar] [CrossRef] [PubMed]
- Holden, V.I.; Bachman, M.A. Diverging roles of bacterial siderophores during infection. Metallomics 2015, 7, 986–995. [Google Scholar] [CrossRef] [PubMed]
- Ellermann, M.; Arthur, J.C. Siderophore-mediated iron acquisition and modulation of host-bacterial interactions. Free Radic. Biol. Med. 2017, 105, 68–78. [Google Scholar] [CrossRef] [PubMed]
- Stork, M.; Di Lorenzo, M.; Mouriño, S.; Osorio, C.R.; Lemos, M.L.; Crosa, J.H. Two tonB systems function in iron transport in Vibrio anguillarum, but only one is essential for virulence. Infect. Immun. 2004, 72, 7326–7329. [Google Scholar] [CrossRef] [PubMed]
- Kuehl, C.J.; Crosa, J.H. The TonB energy transduction systems in Vibrio species. Future Microbiol. 2010, 5, 1403–1412. [Google Scholar] [CrossRef] [PubMed]
- Krewulak, K.D.; Vogel, H.J. TonB or not TonB: Is that the question? Biochem. Cell Biol. 2011, 89, 87–97. [Google Scholar] [CrossRef] [PubMed]
- Avendaño-Herrera, R.; Toranzo, A.E.; Romalde, J.L.; Lemos, M.L.; Magariños, B. Iron uptake mechanisms in the fish pathogen Tenacibaculum maritimum. Appl. Environ. Microbiol. 2005, 71, 6947–6953. [Google Scholar] [CrossRef] [PubMed]
- Soengas, R.G.; Anta, C.; Espada, A.; Paz, V.; Ares, I.R.; Balado, M.; Rodríguez, J.; Lemos, M.L.; Jiménez, C. Structural characterization of vanchrobactin, a new catechol siderophore produced by the fish pathogen Vibrio anguillarum serotype O2. Tetrahedron Lett. 2006, 47, 7113–7116. [Google Scholar] [CrossRef]
- Lemos, M.L.; Osorio, C.R. Heme, an iron supply for vibrios pathogenic for fish. Biometals 2007, 20, 615–626. [Google Scholar] [CrossRef]
- Lemos, M.L.; Balado, M.; Osorio, C.R. Anguibactin- versus vanchrobactin-mediated iron uptake in Vibrio anguillarum: Evolution and ecology of a fish pathogen. Environ. Microbiol. Rep. 2010, 2, 19–26. [Google Scholar] [CrossRef]
- Retamales, J.; González-Contreras, A.; Salazar, S.; Toranzo, A.E.; Avendaño-Herrera, R. Iron utilization and siderophore production by Streptococcus phocae isolated from diseased Atlantic salmon (Salmo salar). Aquaculture 2012, 364–365, 305–311. [Google Scholar] [CrossRef]
- Souto, A.; Montaos, M.A.; Rivas, A.J.; Balado, M.; Osorio, C.R.; Rodríguez, J.; Lemos, M.L.; Jiménez, C. Structure and Biosynthetic Assembly of Piscibactin, a Siderophore from Photobacterium damselae subsp. piscicida, Predicted from Genome Analysis. Eur. J. Org. Chem. 2012, 2012, 5693–5700. [Google Scholar]
- Balado, M.; Souto, A.; Vences, A.; Careaga, V.P.; Valderrama, K.; Segade, Y.; Rodríguez, J.; Osorio, C.R.; Jiménez, C.; Lemos, M.L. Two Catechol Siderophores, Acinetobactin and Amonabactin, Are Simultaneously Produced by Aeromonas salmonicida subsp. salmonicida Sharing Part of the Biosynthetic Pathway. ACS Chem. Biol. 2015, 10, 2850–2860. [Google Scholar] [PubMed]
- Bethke, J.; Poblete-Morales, M.; Irgang, R.; Yañez, A.; Avendaño-Herrera, R. Iron acquisition and siderophore production in the fish pathogen Renibacterium salmoninarum. J. Fish. Dis. 2016, 39, 1275–1283. [Google Scholar] [CrossRef] [PubMed]
- Ruiz, P.; Balado, M.; Toranzo, A.E.; Poblete-Morales, M.; Lemos, M.L.; Avendaño-Herrera, R. Iron assimilation and siderophore production by Vibrio ordalii strains isolated from diseased Atlantic salmon Salmo salar in Chile. Dis. Aquat. Organ. 2016, 118, 217–226. [Google Scholar] [CrossRef]
- Osorio, C.R.; Rivas, A.J.; Balado, M.; Fuentes-Monteverde, J.C.; Rodríguez, J.; Jiménez, C.; Lemos, M.L.; Waldor, M.K. A Transmissible Plasmid-Borne Pathogenicity Island Confers Piscibactin Biosynthesis in the Fish Pathogen Photobacterium damselae subsp. piscicida. Appl. Environ. Microbiol. 2015, 81, 5867–5879. [Google Scholar] [CrossRef] [PubMed]
- Thode, S.K.; Rojek, E.; Kozlowski, M.; Ahmad, R.; Haugen, P. Distribution of siderophore gene systems on a Vibrionaceae phylogeny: Database searches, phylogenetic analyses and evolutionary perspectives. PLoS ONE 2018, 13, e0191860. [Google Scholar] [CrossRef]
- Balado, M.; Lages, M.A.; Fuentes-Monteverde, J.C.; Martínez-Matamoros, D.; Rodríguez, J.; Jiménez, C.; Lemos, M.L. The Siderophore Piscibactin Is a Relevant Virulence Factor for Vibrio anguillarum Favored at Low Temperatures. Front. Microbiol. 2018, 9, 1766. [Google Scholar] [CrossRef] [PubMed]
- Avendaño-Herrera, R.; Maldonado, J.P.; Tapia-Cammas, D.; Feijoo, C.G.; Calleja, F.; Toranzo, A.E. PCR protocol for detection of Vibrio ordalii by amplification of the vohB (hemolysin) gene. Dis. Aquat. Organ. 2014, 107, 223–234. [Google Scholar] [CrossRef]
- Lemos, M.L.; Salinas, P.; Toranzo, A.E.; Barja, J.L.; Crosa, J.H. Chromosome-mediated iron uptake system in pathogenic strains of Vibrio anguillarum. J. Bacteriol. 1988, 170, 1920–1925. [Google Scholar] [CrossRef]
- Solovyev, V.; Salamov, A. Automatic Annotation of Microbial Genomes and Metagenomic Sequences. In Metagenomics and Its Applications in Agriculture, Biomedicine and Environmental Studies; Li, R.W., Ed.; Nova Science Publishers: New York, NY, USA, 2011; pp. 61–78. [Google Scholar]
- Escolar, L.; Perez-Martin, J.; de Lorenzo, V. Opening the iron box: Transcriptional metalloregulation by the Fur protein. J. Bacteriol. 1999, 181, 6223–6229. [Google Scholar] [PubMed]
- Parales, R.E.; Harwood, C.S. Construction and use of a new broad-host-range lacZ transcriptional fusion vector, pHRP309, for Gram- bacteria. Gene 1993, 133, 23–30. [Google Scholar] [CrossRef]
- Miller, J.H. Experiments in Molecular Genetics; Cold Spring Harbor Laboratory: Cold Spring Harbor, NY, USA, 1972. [Google Scholar]
- Crosa, J.H.; Hodges, L.L. Outer membrane proteins induced under conditions of iron limitation in the marine fish pathogen Vibrio anguillarum 775. Infect. Immun. 1981, 31, 223–227. [Google Scholar] [PubMed]
- Toranzo, A.E.; Barja, J.L.; Potter, S.A.; Colwell, R.R.; Hetrick, F.M.; Crosa, J.H. Molecular factors associated with virulence of marine vibrios isolated from striped bass in Chesapeake Bay. Infect. Immun. 1983, 39, 1220–1227. [Google Scholar] [PubMed]
- Puentes, B.; Balado, M.; Bermudez-Crespo, J.; Osorio, C.R.; Lemos, M.L. A proteomic analysis of the iron response of Photobacterium damselae subsp. damselae reveals metabolic adaptations to iron levels changes and novel potential virulence factors. Vet. Microbiol. 2017, 201, 257–264. [Google Scholar]
- Finn, R.D.; Bateman, A.; Clements, J.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Heger, A.; Hetherington, K.; Holm, L.; Mistry, J.; et al. Pfam: The protein families database. Nucleic Acids Res. 2014, 42, D222–D230. [Google Scholar] [CrossRef] [PubMed]
- Osorio, C.R.; Juiz-Rio, S.; Lemos, M.L. A siderophore biosynthesis gene cluster from the fish pathogen Photobacterium damselae subsp. piscicida is structurally and functionally related to the Yersinia high-pathogenicity island. Microbiology 2006, 152, 3327–3341. [Google Scholar]
- Crosa, J.H.; Walsh, C.T. Genetics and assembly line enzymology of siderophore biosynthesis in bacteria. Microbiol. Mol. Biol. Rev. 2002, 66, 223–249. [Google Scholar] [CrossRef]
- Imperi, F.; Visca, P. Subcellular localization of the pyoverdine biogenesis machinery of Pseudomonas aeruginosa: A membrane-associated “siderosome”. FEBS Lett. 2013, 587, 3387–3391. [Google Scholar] [CrossRef]
- Gasser, V.; Guillon, L.; Cunrath, O.; Schalk, I.J. Cellular organization of siderophore biosynthesis in Pseudomonas aeruginosa: Evidence for siderosomes. J. Inorg. Biochem. 2015, 148, 27–34. [Google Scholar] [CrossRef]
- Mouriño, S.; Rodríguez-Ares, I.; Osorio, C.R.; Lemos, M.L. Genetic variability of the heme uptake system among different strains of the fish pathogen Vibrio anguillarum: Identification of a new heme receptor. Appl. Environ. Microbiol. 2005, 71, 8434–8441. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Heilier, J.F.; Madalinski, G.; Genin, E.; Ezan, E.; Tabet, J.C.; Junot, C. Evaluation of accurate mass and relative isotopic abundance measurements in the LTQ-orbitrap mass spectrometer for further metabolomics database building. Anal. Chem. 2010, 82, 5490–5501. [Google Scholar] [CrossRef] [PubMed]
- Schiewe, M.H.; Trust, T.J.; Crosa, J.H. Vibrio ordalii sp. nov.: A causative agent of vibriosis in fish. Curr. Microbiol. 1981, 6, 343–348. [Google Scholar] [CrossRef]
- Ransom, D.P.; Lannan, C.N.; Rohovec, J.S.; Fryer, J.L. Comparison of histopathology caused by Vibrio anguillarum and Vibrio ordalii in three species of Pacific salmon. J. Fish. Dis. 1984, 7, 107–115. [Google Scholar] [CrossRef]
- Li, Y.; Ma, Q. Iron Acquisition Strategies of Vibrio anguillarum. Front. Cell. Infect. Microbiol. 2017, 7, 342. [Google Scholar] [CrossRef] [PubMed]
- Balado, M.; Osorio, C.R.; Lemos, M.L. FvtA is the receptor for the siderophore vanchrobactin in Vibrio anguillarum: Utility as a route of entry for vanchrobactin analogues. Appl. Environ. Microbiol. 2009, 75, 2775–2783. [Google Scholar] [CrossRef] [PubMed]
- Stork, M.; Di Lorenzo, M.; Welch, T.J.; Crosa, L.M.; Crosa, J.H. Plasmid-mediated iron uptake and virulence in Vibrio anguillarum. Plasmid 2002, 48, 222–228. [Google Scholar] [CrossRef]
- Actis, L.A.; Tolmasky, M.E.; Crosa, J.H. Vibriosis. In Fish Diseases and Disorders; Woo, P.T.K., Bruno, D.W., Eds.; CAB International: London, UK, 2011; Volume 3, pp. 570–604. [Google Scholar]
- Naka, H.; Lopez, C.S.; Crosa, J.H. Reactivation of the vanchrobactin siderophore system of Vibrio anguillarum by removal of a chromosomal insertion sequence originated in plasmid pJM1 encoding the anguibactin siderophore system. Environ. Microbiol. 2008, 10, 265–277. [Google Scholar] [CrossRef] [PubMed]

| Primers | Sequence (5′-3′) * | Amplified Fragment (bp) |
|---|---|---|
| Amplification of potential promoters | ||
| P1 | ||
| Promoter 1_F | GCGTCTAGACACTTTGCCACCCACCATTA | 879 |
| Promoter 1_R | GCGGGATCCACGAATCGTCGTGTTGGCAT | |
| P2 | ||
| Promoter 2_F | GCGTCTAGACCGCTTAGAGAAACCAACGT | 1165 |
| Promoter 2_R | GCGGGATCCACGTTTCGGTAAGCGTATGG | |
| Transcriptional regulation of irp gene cluster | ||
| RT | TTTGGAGATGAGTGCGACAC | |
| PCR1 | ||
| ARC1ordalii_F | GATATGCGCTTTGACTGCCA | 196 |
| ARC1ordalii_R | CTGTGAGACGGCATACAAGC | |
| PCR2 | ||
| FrpA_ordalii_F | CGGTGGTAATGCTCAAGGTG | 204 |
| FrpA_ordalii_R | TGGCTCGGTAGGTGTTCAAT | |
| PCR3 | ||
| Irp2_ordalii_F | AGCAGGCAACAAAGAGTGAG | 413 |
| Irp1_ordalii_R | GGGCGAATAACCAAACAAGC | |
| Band in Gel (Figure 5) | Estimated Size (kDa) | Closest Homologues | Accession No. | Similarity (%) |
|---|---|---|---|---|
| Band I | 311 | VabF, V. anguillarum | CAJ45639.1 | 98 |
| Band II | 270 | Irp1, V. anguillarum | WP_019281879.1 | 98 |
| Irp1, P. damselae subsp. piscicida | AKQ52532.1 | 70 | ||
| Band III | 224 | Irp2, V. anguillarum | WP_019281878.1 | 98 |
| Irp2, P. damselae subsp. piscicida | AKQ52531.1 | 68 | ||
| Band IV | 79 | HuvS, V. anguillarum | CAJ14788.1 | 99 |
| Band V | 71 | FrpA, V. anguillarum | WP_019281876.1 | 96 |
| FrpA, P. damselae subsp. piscicida | AKQ52529.1 | 68 |
| Ion | Retention Time (min) | Detected [M+H]+ | ∆ m/z (ppm) | Ion Formula | Mean Intensity | δRIA (%) |
|---|---|---|---|---|---|---|
| Piscibactin-Ga(III) | 12.08 | 519.99280 | 3.1 | 12C19H2169GaN3O4S3+ | 5.8 × 106 | - |
| 520.99603 | 3.4 | 13C112C18H2169GaN3O4S3+ | 1.1 × 106 | −5.3 | ||
| 521.99127 | 4.4 | 12C19H2171GaN3O4S3+ | 4.6 × 106 | - | ||
| 522.99481 | 4.1 | 13C112C18H2171GaN3O4S3+ | 8.0 × 105 | 5.9 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ruiz, P.; Balado, M.; Fuentes-Monteverde, J.C.; Toranzo, A.E.; Rodríguez, J.; Jiménez, C.; Avendaño-Herrera, R.; Lemos, M.L. The Fish Pathogen Vibrio ordalii Under Iron Deprivation Produces the Siderophore Piscibactin. Microorganisms 2019, 7, 313. https://doi.org/10.3390/microorganisms7090313
Ruiz P, Balado M, Fuentes-Monteverde JC, Toranzo AE, Rodríguez J, Jiménez C, Avendaño-Herrera R, Lemos ML. The Fish Pathogen Vibrio ordalii Under Iron Deprivation Produces the Siderophore Piscibactin. Microorganisms. 2019; 7(9):313. https://doi.org/10.3390/microorganisms7090313
Chicago/Turabian StyleRuiz, Pamela, Miguel Balado, Juan Carlos Fuentes-Monteverde, Alicia E. Toranzo, Jaime Rodríguez, Carlos Jiménez, Ruben Avendaño-Herrera, and Manuel L. Lemos. 2019. "The Fish Pathogen Vibrio ordalii Under Iron Deprivation Produces the Siderophore Piscibactin" Microorganisms 7, no. 9: 313. https://doi.org/10.3390/microorganisms7090313
APA StyleRuiz, P., Balado, M., Fuentes-Monteverde, J. C., Toranzo, A. E., Rodríguez, J., Jiménez, C., Avendaño-Herrera, R., & Lemos, M. L. (2019). The Fish Pathogen Vibrio ordalii Under Iron Deprivation Produces the Siderophore Piscibactin. Microorganisms, 7(9), 313. https://doi.org/10.3390/microorganisms7090313









