Detection of Wood Mice (Apodemus sylvaticus) Carrying Non-Tuberculous Mycobacteria Able to Infect Cattle and Interfere with the Diagnosis of Bovine Tuberculosis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Area and Small Mammal Sampling
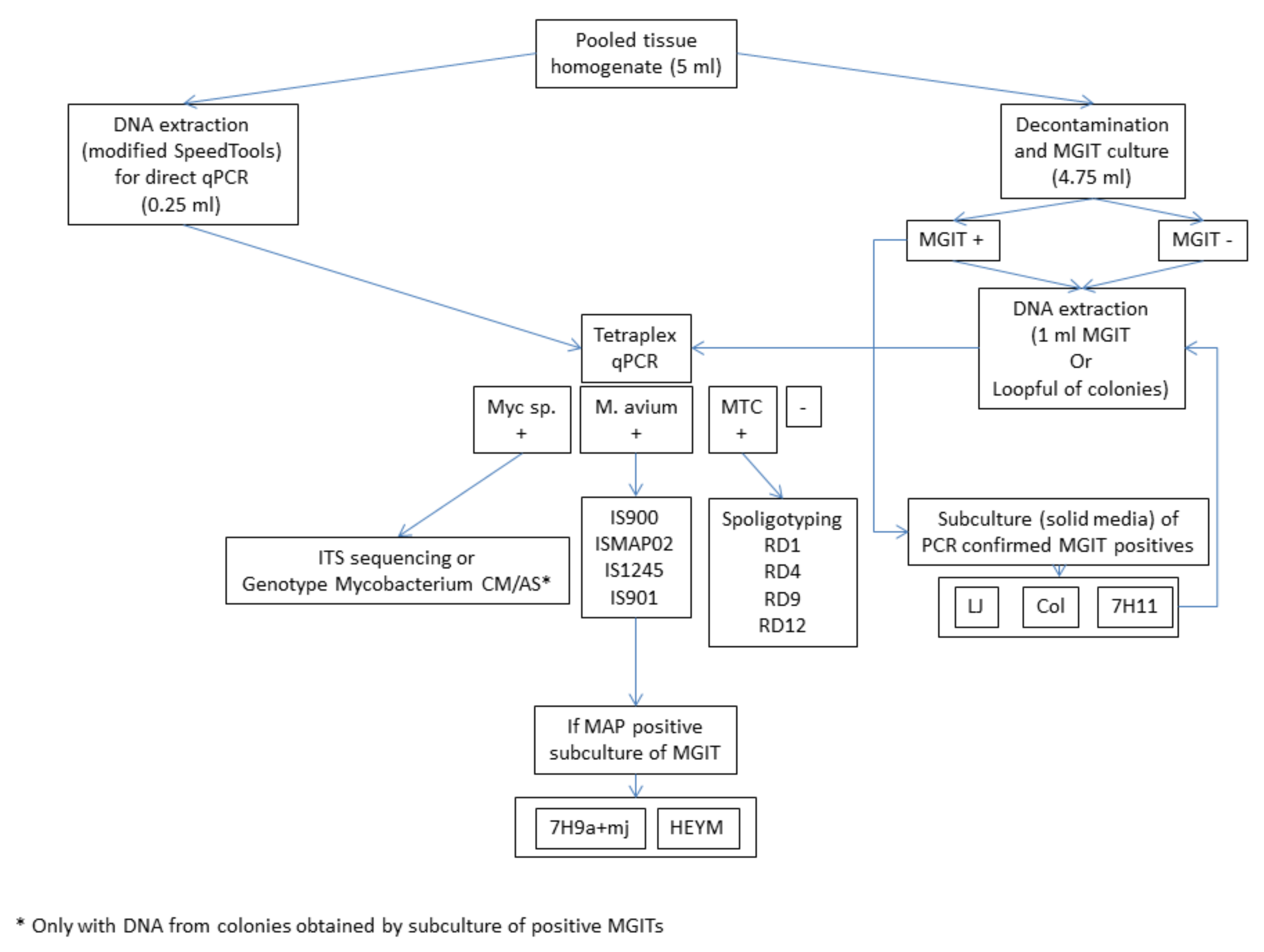
2.2. Processing of Small Mammals and Sample Preparation
2.3. Culture
2.4. DNA Extraction
2.5. Tetraplex Real-Time PCR for the Screening of Tissues and Cultures
2.6. Further Molecular Identification of Mycobacteria Detected by the Tetraplex Real-Time PCR
2.6.1. Identification of Mycobacterium sp.-Positive Samples
2.6.2. Identification of M. avium subsp.-Positive Samples
2.7. Identification of MTC-Positive Samples
2.8. Statistical Analyses
3. Results
3.1. Identification and Processing of Small Mammals
3.2. Mycobacteria Detection and Identification
3.3. Statistics
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Pavlik, I.; Machackova, M.; Yayo Ayele, W.; Lamka, J.; Parmova, I.; Melicharek, I.; Hanzlikova, M.; Körmendy, B.; Nagy, G.; Cvetnic, Z.; et al. Incidence of bovine tuberculosis in wild and domestic animals other than cattle in six Central European countries during 1990–1999. Vet. Med. 2002, 47, 122–131. [Google Scholar] [CrossRef] [Green Version]
- Bercovier, H.; Vincent, V. Mycobacterial infections in domestic and wild animals due to Mycobacterium marinum, M. fortuitum, M. chelonae, M. porcinum, M. farcinogenes, M. smegmatis, M. scrofulaceum, M. xenopi, M. kansasii, M. simiae and M. genavense. OIE Rev. Sci. Tech. 2001, 20, 265–290. [Google Scholar] [CrossRef] [PubMed]
- Ashford, D.A.; Whitney, E.; Raghunathan, P.; Cosivi, O. Epidemiology of selected mycobacteria humans and other animals. OIE Rev. Sci. Tech. 2001, 20, 325–337. [Google Scholar] [CrossRef] [PubMed]
- Hruska, K.; Kaevska, M. Mycobacteria in water, soil, plants and air: A review. Vet. Med. 2012, 57, 623–679. [Google Scholar] [CrossRef] [Green Version]
- World Health Organization. Global Tuberculosis Report; WHO: Geneva, Switzerland, 2019. [Google Scholar]
- Zinsstag, J.; Schelling, E.; Roth, F.; Kazwala, R.R. Economics of bovine tuberculosis. In Mycobacterium Bovis Infection in Animals and Humans, 2nd ed.; Blackwell Publishing Ltd.: Hoboken, NJ, USA, 2008; pp. 68–83. ISBN 0813809193. [Google Scholar]
- Kaneene, J.B.; Miller, R.A.; Kaplan, B.; Steele, J.H.; Thoen, C.O. Preventing and controlling zoonotic tuberculosis: A one health approach. Vet. Ital. 2014, 50, 7–22. [Google Scholar] [PubMed]
- Biet, F.; Boschiroli, M.L. Non-tuberculous mycobacterial infections of veterinary relevance. Res. Vet. Sci. 2014, 97, S69–S77. [Google Scholar] [CrossRef]
- Griffith, D.E.; Aksamit, T.; Brown-Elliott, B.A.; Catanzaro, A.; Daley, C.; Gordin, F.; Holland, S.M.; Horsburgh, R.; Huitt, G.; Iademarco, M.F.; et al. An official ATS/IDSA statement: Diagnosis, treatment, and prevention of nontuberculous mycobacterial diseases. Am. J. Respir. Crit. Care Med. 2007, 175, 367–416. [Google Scholar] [CrossRef]
- Mendoza, J.L.; Lana, R.; Díaz-Rubio, M. Mycobacterium avium subspecies paratuberculosis and its relationship with Cronh’s disease. World J. Gastroenterol. 2009, 15, 417–422. [Google Scholar] [CrossRef]
- De la Rua-Domenech, R.; Goodchild, A.T.; Vordermeier, H.M.; Hewinson, R.G.; Christiansen, K.H.; Clifton-Hadley, R.S. Ante mortem diagnosis of tuberculosis in cattle: A review of the tuberculin tests, γ-interferon assay and other ancillary diagnostic techniques. Res. Vet. Sci. 2006, 81, 190–210. [Google Scholar] [CrossRef]
- Buddle, B.M.; Wards, B.J.; Aldwell, F.E.; Collins, D.M.; De Lisle, G.W. Influence of sensitisation to environmental mycobacteria on subsequent vaccination against bovine tuberculosis. Vaccine 2002, 20, 1126–1133. [Google Scholar] [CrossRef]
- Fischer, O.; Mátlová, L.; Bartl, J.; Dvorská, L.; Melichárek, I.; Pavlík, I. Findings of mycobacteria in insectivores and small rodents. Folia Microbiol. 2000, 45, 147–152. [Google Scholar] [CrossRef] [PubMed]
- Mathews, F.; Macdonald, D.W.; Taylor, G.M.; Gelling, M.; Norman, R.A.; Honess, P.E.; Foster, R.; Gower, C.M.; Varley, S.; Harris, A.; et al. Bovine tuberculosis (Mycobacterium bovis) in British farmland wildlife: The importance to agriculture. Proc. R. Soc. 2006, 273, 357–365. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Delahay, R.J.; De Leeuw, A.N.S.; Barlow, A.M.; Clifton-Hadley, R.S.; Cheeseman, C.L. The status of Mycobacterium bovis infection in UK wild mammals: A review. Vet. J. 2002, 164, 90–105. [Google Scholar] [CrossRef] [PubMed]
- Clarke, A.K.R.; Fitzgerald, S.D.; Zwick, L.S.; Church, S.V.; Kaneene, J.B. Experimental inoculation of meadow voles (Microtus pennsylvanicus), house mice (Mus musculus), and norway rats (Rattus norvegicus) with Mycobacterium bovis. J. Wildl. Dis. 2007, 43, 353–365. [Google Scholar] [CrossRef] [Green Version]
- Durnez, L.; Katakweba, A.; Sadiki, H.; Katholi, C.R.; Kazwala, R.R.; MacHang’U, R.R.; Portaels, F.; Leirs, H. Mycobacteria in terrestrial small mammals on cattle farms in Tanzania. Vet. Med. Int. 2011, 2011, 495074. [Google Scholar] [CrossRef] [Green Version]
- Kopecna, M.; Trcka, I.; Lamka, J.; Moravkova, M.; Koubek, P.; Heroldova, M.; Mrlik, V.; Kralova, A.; Pavlik, I. The wildlife hosts of Mycobacterium avium subsp. paratuberculosis in the Czech Republic during the years 2002–2007. Vet. Med. 2008, 53, 420–426. [Google Scholar] [CrossRef] [Green Version]
- Florou, M.; Leontides, L.; Kostoulas, P.; Billinis, C.; Sofia, M.; Kyriazakis, I.; Lykotrafitis, F. Isolation of Mycobacterium avium subspecies paratuberculosis from non-ruminant wildlife living in the sheds and on the pastures of Greek sheep and goats. Epidemiol. Infect. 2008, 136, 644–652. [Google Scholar] [CrossRef]
- Panzironi, C.; Cerone, G.; Mauro, C.; Amori, G. A method for the morphometric identification of southern Italian populations of Apodemus (Sylvaemus). Hystrix Ital. J. Mammal. 1994, 5, 1–16. [Google Scholar]
- Blanco, J.C. Mamíferos de España; Planeta: New York, NY, USA, 1998. [Google Scholar]
- Whittington, R.J.; Whittington, A.M.; Waldron, A.; Begg, D.J.; De Silva, K.; Purdie, A.C.; Plain, K.M. Development and validation of a liquid medium (M7H9C) for routine culture of Mycobacterium avium subsp. paratuberculosis to replace modified bactec 12B medium. J. Clin. Microbiol. 2013, 51, 3993–4000. [Google Scholar] [CrossRef] [Green Version]
- Sevilla, I.A.; Molina, E.; Elguezabal, N.; Pérez, V.; Garrido, J.M.; Juste, R.A. Detection of mycobacteria, Mycobacterium avium subspecies, and Mycobacterium tuberculosis complex by a novel tetraplex real-time PCR assay. J. Clin. Microbiol. 2015, 53, 930–940. [Google Scholar] [CrossRef] [Green Version]
- Sevilla, I.A.; Molina, E.; Tello, M.; Elguezabal, N.; Juste, R.A.; Garrido, J.M. Detection of mycobacteria by culture and DNA-based methods in animal-derived food products purchased at Spanish supermarkets. Front. Microbiol. 2017, 8, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Fyfe, J.A.; McCowan, C.; O’Brien, C.R.; Globan, M.; Birch, C.; Revill, P.; Barrs, V.R.D.; Wayne, J.; Hughes, M.S.; Holloway, S.; et al. Molecular characterization of a novel fastidious Mycobacterium causing lepromatous lesions of the skin, subcutis, cornea, and conjunctiva of cats living in Victoria, Australia. J. Clin. Microbiol. 2008, 46, 618–626. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sevilla, I.A.; Garrido, J.M.; Molina, E.; Geijo, M.V.; Elguezabal, N.; Vázquez, P.; Juste, R.A. Development and evaluation of a novel multicopy-element-targeting triplex PCR for detection of Mycobacterium avium subsp. paratuberculosis in feces. Appl. Environ. Microbiol. 2014, 80, 3757–3768. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Slana, I.; Kaevska, M.; Kralik, P.; Horvathova, A.; Pavlik, I. Distribution of Mycobacterium avium subsp. avium and M. a. hominissuis in artificially infected pigs studied by culture and IS901 and IS1245 quantitative real time PCR. Vet. Microbiol. 2010, 144, 437–443. [Google Scholar] [CrossRef] [PubMed]
- Kamerbeek, J.; Schouls, L.; Kolk, A.; Van Agterveld, M.; Van Soolingen, D.; Kuijper, S.; Bunschoten, A.; Molhuizen, H.; Shaw, R.; Goyal, M.; et al. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J. Clin. Microbiol. 1997, 35, 907–914. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Halse, T.A.; Escuyer, V.E.; Musser, K.A. Evaluation of a single-tube multiplex real-time PCR for differentiation of members of the Mycobacterium tuberculosis complex in clinical specimens. J. Clin. Microbiol. 2011, 49, 2562–2567. [Google Scholar] [CrossRef] [Green Version]
- Torre, I.; Arrizabalaga, A.; Díaz, M. Ratón de campo Apodemus sylvaticus (Linnaeus, 1758). Galemys 2002, 14, 1–26. [Google Scholar]
- Falkinham, J.O., III; Norton, C.D.; Lechevallier, M.W. Factors Influencing Numbers of Mycobacterium avium, Mycobacterium intracellulare, and Other Mycobacteria in Drinking Water Distribution Systems. Society 2001, 67, 1225–1231. [Google Scholar] [CrossRef] [Green Version]
- Rastogi, N.; Legrand, E.; Sola, C. The Mycobacteria: An introduction to nomenclature and pathogenesis. OIE Rev. Sci. Tech. 2001, 20, 21–54. [Google Scholar] [CrossRef]
- Koets, A.P.; Rutten, V.P.M.G.; Bakker, D.; Van Der Hage, M.H.; Van Eden, W. Lewis rats are not susceptible to oral infection with Mycobacterium avium subsp. paratuberculosis. Vet. Microbiol. 2000, 77, 487–495. [Google Scholar] [CrossRef]
- Beard, P.M.; Daniels, M.J.; Henderson, D.; Pirie, A.; Rudge, K.; Buxton, D.; Rhind, S.; Greig, A.; Hutchings, M.R.; McKendrick, I.; et al. Paratuberculosis infection of nonruminant wildlife in Scotland. J. Clin. Microbiol. 2001, 39, 1517–1521. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jenkins, A.O.; Gormley, E.; Gcebe, N.; Fosgate, G.T.; Conan, A.; Aagaard, C.; Michel, A.L.; Rutten, V.P.M.G. Cross reactive immune responses in cattle arising from exposure to Mycobacterium bovis and non-tuberculous mycobacteria. Prev. Vet. Med. 2018, 152, 16–22. [Google Scholar] [CrossRef] [Green Version]
- Wolinsky, E. Nontuberculous mycobacteria and associated diseases. Am. Rev. Respir. Dis. 1979, 119, 107–159. [Google Scholar] [PubMed]
- Berg, S.; Firdessa, R.; Habtamu, M.; Gadisa, E.; Mengistu, A.; Yamuah, L.; Ameni, G.; Vordermeier, M.; Robertson, B.D.; Smith, N.H.; et al. The burden of mycobacterial disease in Ethiopian cattle: Implications for public health. PLoS ONE 2009, 4, e5068. [Google Scholar] [CrossRef]
- Honda, J.R.; Virdi, R.; Chan, E.D. Global environmental nontuberculous mycobacteria and their contemporaneous man-made and natural niches. Front. Microbiol. 2018, 9, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vordermeier, H.M.; Brown, J.; Cockle, P.J.; Franken, W.P.J.; Arend, S.M.; Ottenhoff, T.H.M.; Jahans, K.; Hewinson, R.G. Assessment of cross-reactivity between Mycobacterium bovis and M. kansasii ESAT-6 and CFP-10 at the T-cell epitope level. Clin. Vaccine Immunol. 2007, 14, 1203–1209. [Google Scholar] [CrossRef] [Green Version]
- Pate, M.; Žolnir-Dovč, M.; Kušar, D.; Krt, B.; Špičić, S.; Cvetni, Ž.; Ocepek, M. The first report of Mycobacterium celatum isolation from domestic pig (Sus scrofa domestica) and roe deer (Capreolus capreolus) and an overview of human infections in Slovenia. Vet. Med. Int. 2011, 2011, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Fattorini, L.; Baldassarri, L.; Li, Y.J.; Ammendolia, M.G.; Fan, Y.; Recchia, S.; Iona, E.; Orefici, G. Virulence and drug susceptibility of Mycobacterium celatum. Microbiology 2000, 146, 2733–2742. [Google Scholar] [CrossRef] [Green Version]
- Pate, M.; Zajc, U.; Kušar, D.; Žele, D.; Vengušt, G.; Pirš, T.; Ocepek, M. Mycobacterium spp. in wild game in Slovenia. Vet. J. 2016, 208, 93–95. [Google Scholar] [CrossRef]
- Delahay, R.J.; Smith, G.C.; Barlow, A.M.; Walker, N.; Harris, A.; Clifton-Hadley, R.S.; Cheeseman, C.L. Bovine tuberculosis infection in wild mammals in the South-West region of England: A survey of prevalence and a semi-quantitative assessment of the relative risks to cattle. Vet. J. 2007, 173, 287–301. [Google Scholar] [CrossRef]
- Smith, N.H.; Crawshaw, T.; Parry, J.; Birtles, R.J. Mycobacterium microti: More diverse than previously thought. J. Clin. Microbiol. 2009, 47, 2551–2559. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kipar, A.; Burthe, S.J.; Hetzel, U.; Abo Rokia, M.; Telfer, S.; Lambin, X.; Birtles, R.J.; Begon, M.; Bennett, M. Mycobacterium microti Tuberculosis in Its Maintenance Host, the Field Vole (Microtus agrestis): Characterization of the Disease and Possible Routes of Transmission. Vet. Pathol. 2014, 51, 903–914. [Google Scholar] [CrossRef] [Green Version]
- Pérez de Val, B.; Sanz, A.; Soler, M.; Allepuz, A.; Michelet, L.; Boschiroli, M.L.; Vidal, E. Mycobacterium microti infection in Free-Ranging Wild Boar, Spain, 2017-2019. Emerg. Infect. Dis. 2019, 25, 2152–2154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Michelet, L.; de Cruz, K.; Phalente, Y.; Karoui, C.; Hénault, S.; Beral, M.; Boschiroli, M.L. Mycobacterium microti Infection in Dairy Goats, France. Emerg. Infect. Dis. 2016, 22, 569–570. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Michelet, L.; De Cruz, K.; Zanella, G.; Aaziz, R.; Bulach, T.; Karoui, C.; Hénault, S.; Joncour, G.; Boschiroli, M.L. Infection with Mycobacterium microti in animals in France. J. Clin. Microbiol. 2015, 53, 981–985. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Corner, L.A.L.; Murphy, D.; Gormley, E. Mycobacterium bovis Infection in the Eurasian Badger (Meles meles): The Disease, Pathogenesis, Epidemiology and Control. J. Comp. Pathol. 2011, 144, 1–24. [Google Scholar] [CrossRef]
| Farm Locality | Mycobacterium Species Identified in Cattle (2014–2017) | Small Mammal Species (N) | Total Number Trapped | Mycobacteria Prevalence (%) in Small Mammals (95% CI) | Mycobacterium Species Identified in Small Mammals |
|---|---|---|---|---|---|
| Deba | 34 | 11.8 (4.7–26.6) | |||
| M. bovis | Apodemus sylvaticus (29) | M. intracellulare | |||
| Map | Mus domesticus (2) | Map | |||
| Microtus agrestis (1) | M. fortuitum | ||||
| Microtus gerbei (1) | M. gordonae | ||||
| Apodemus sp. (1) | |||||
| Kortezubi | 34 | 8.8 (3.0–23.0) | |||
| M. avium subsp. avium | A. sylvaticus (32) | Map | |||
| Crocidura russula (2) | Mycobacterium sp.¥ | ||||
| M. celatum | |||||
| Kexaa | 40 | 0.0 (0.0–8.7) | |||
| M. bovis | A. sylvaticus (33) | ||||
| Map | Apodemus flavicollis (3) | ||||
| M. avium subsp. avium | Myodes glareolus (1) | ||||
| Mycobacterium sp.* | M. gerbei (1) | ||||
| M. domesticus (1) | |||||
| C. russula (1) |
| Rodent Species | Mycobacterium Isolation | MGIT PCR Result | Direct PCR Result (Tissue Homogenate) | Mycobacterium Identification Method | Final Identification |
|---|---|---|---|---|---|
| A. sylvaticus | Yes | Positive (Mycobacterium sp.) | Negative | Reverse hybridization and ITS sequencing | M. fortuitum |
| Apodemus sp. | Yes | Positive (Mycobacterium sp.) | Negative | Reverse hybridization | M. intracellulare |
| A. sylvaticus | Yes | Positive (Mycobacterium sp.) | Negative | Reverse hybridization | M. gordonae |
| A. sylvaticus | Yes | Positive (Mycobacterium sp.) | Negative | Reverse hybridization | M. celatum |
| A. sylvaticus | No | Negative | Positive (M. avium) | IS900, ISMap02, IS1245 and IS901 | Map |
| A. sylvaticus | No | Negative | Positive (M. avium) | IS900, ISMap02, IS1245 and IS901 | Map |
| A. sylvaticus | No | Negative | Positive (Mycobacterium sp.) | ITS sequencing | Mycobacterium sp.* |
| Variable | Number Tested | % Positives (95% CI) | p Value |
|---|---|---|---|
| Sex | 1 | ||
| Female | 42 | 7.1 (2.5–19.0) | |
| Male | 52 | 5.8 (2.0–15.6) | |
| Age | 1 | ||
| Juvenile | 40 | 7.5 (2.6–19.9) | |
| Adult | 50 | 6.0 (2.1–16.2) | |
| Season | 0.3 | ||
| Autumn | 23 | 0.0 (0.0–14.3) | |
| Winter | 36 | 11.1 (4.4–25.3) | |
| Spring | 26 | 3.8 (0.7–18.9) | |
| Summer | 9 | 11.1 (2.0–43.4) | |
| Farm Locality | 0.1 | ||
| Deba | 29 | 10.3 (3.6–26.4) | |
| Kortezubi | 32 | 9.4 (3.2–24.2) | |
| Kexaa | 33 | 0.0 (0.0–10.4) |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Varela-Castro, L.; Torrontegi, O.; Sevilla, I.A.; Barral, M. Detection of Wood Mice (Apodemus sylvaticus) Carrying Non-Tuberculous Mycobacteria Able to Infect Cattle and Interfere with the Diagnosis of Bovine Tuberculosis. Microorganisms 2020, 8, 374. https://doi.org/10.3390/microorganisms8030374
Varela-Castro L, Torrontegi O, Sevilla IA, Barral M. Detection of Wood Mice (Apodemus sylvaticus) Carrying Non-Tuberculous Mycobacteria Able to Infect Cattle and Interfere with the Diagnosis of Bovine Tuberculosis. Microorganisms. 2020; 8(3):374. https://doi.org/10.3390/microorganisms8030374
Chicago/Turabian StyleVarela-Castro, Lucía, Olalla Torrontegi, Iker A. Sevilla, and Marta Barral. 2020. "Detection of Wood Mice (Apodemus sylvaticus) Carrying Non-Tuberculous Mycobacteria Able to Infect Cattle and Interfere with the Diagnosis of Bovine Tuberculosis" Microorganisms 8, no. 3: 374. https://doi.org/10.3390/microorganisms8030374





