Comparative Serum Proteome Profiling of Canine Benign Prostatic Hyperplasia before and after Castration
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Sample Collection
2.2. In-Solution Digestion by Trypsin
2.3. LC-MS/MS
2.4. Bioinformatics and Data Analysis
2.5. Statistical Analysis
3. Results
3.1. The Prostatic Size before and after Castration
3.2. LC-MS/MS Results
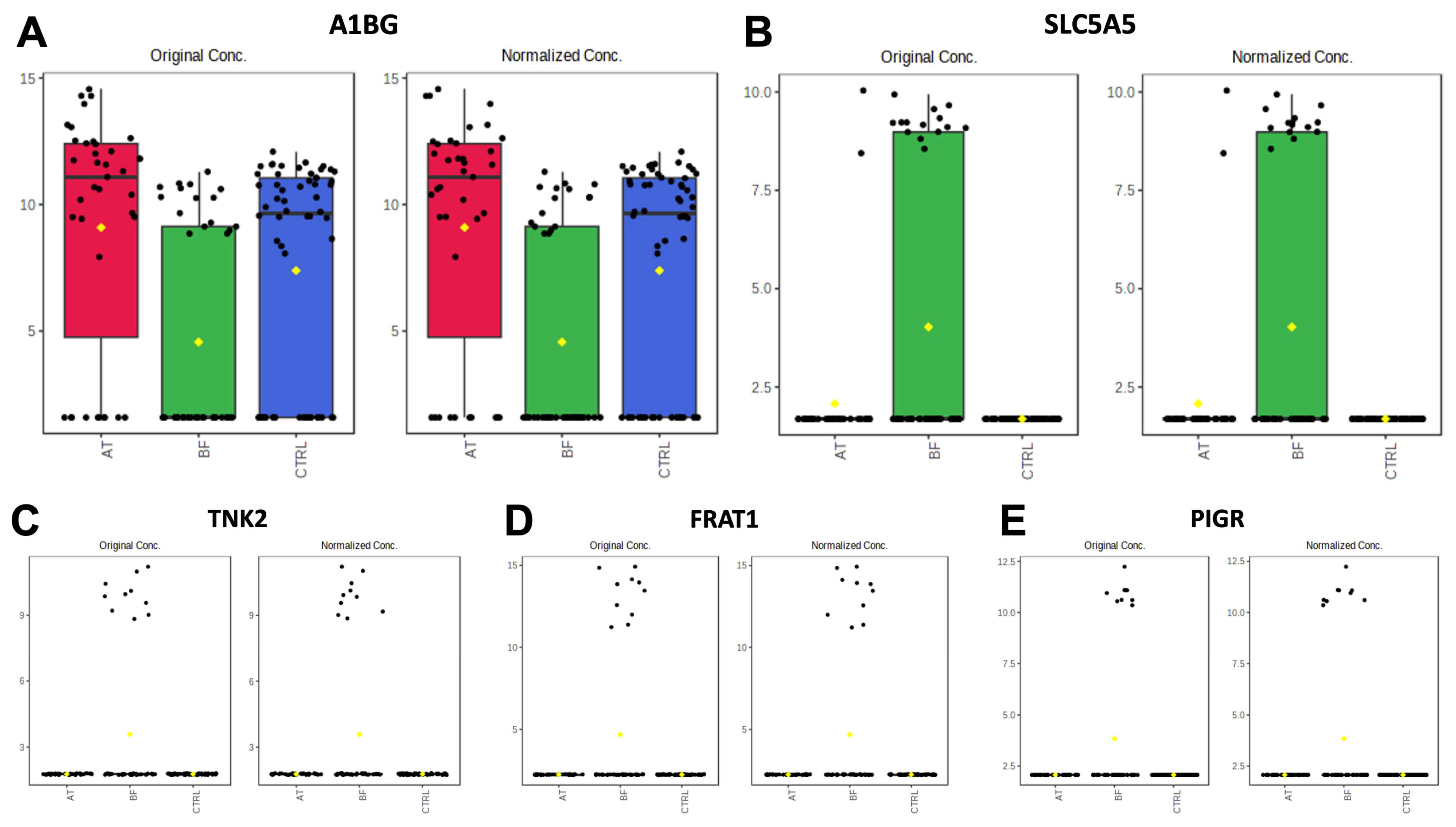
3.2.1. Comparisons of BF Subgroup versus Other Groups (BF vs. AT and BF vs. CTRL)
3.2.2. Comparisons of AT Subgroup versus Other Groups (BF vs. AT and AT vs. CTRL)
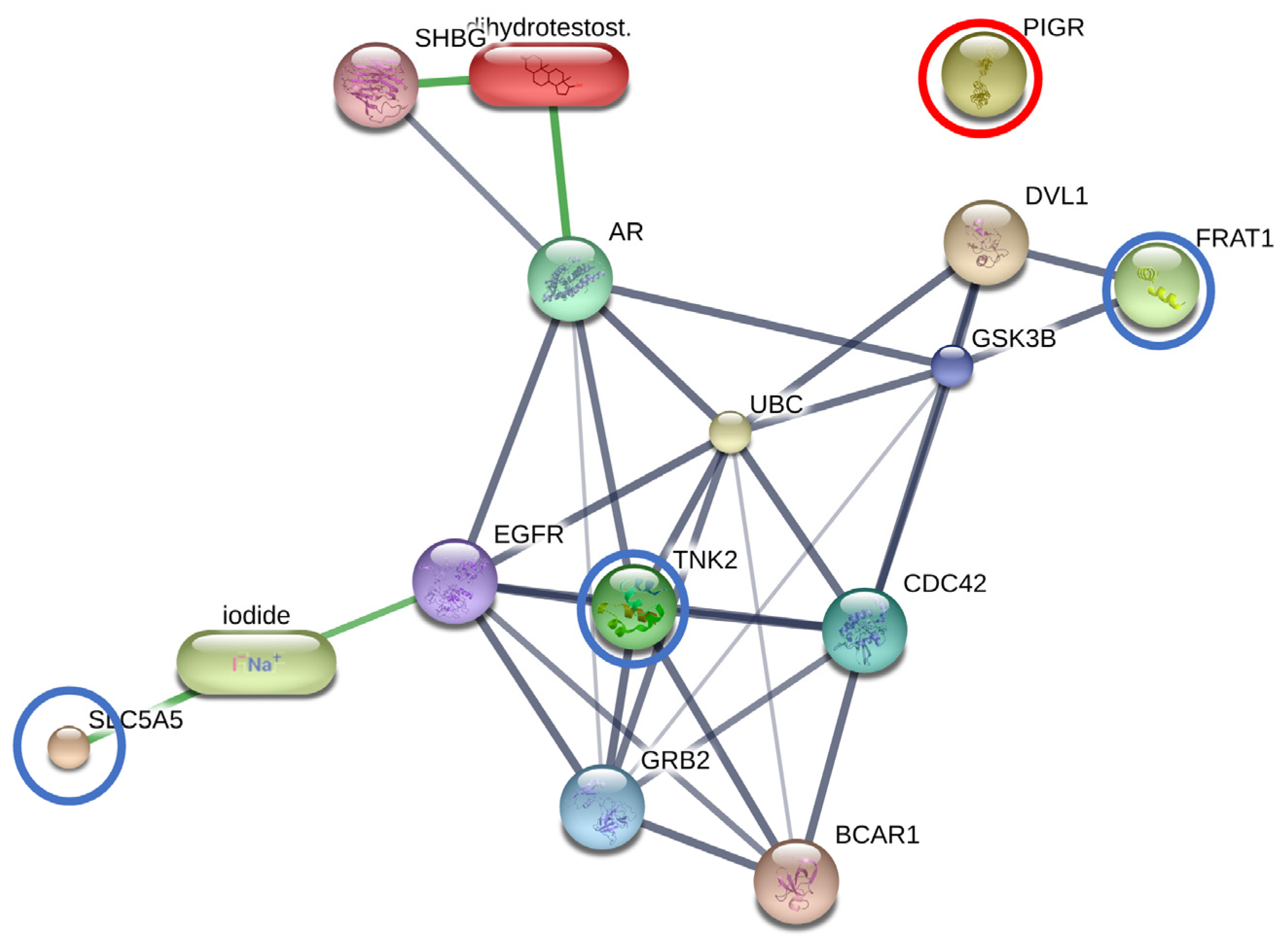
3.2.3. The Interactions between DHT and Overexpressed Proteins in the BF Subgroup
3.2.4. The Interactions between Finasteride and Overexpressed Proteins in the AT Subgroup
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Johnston, S.D.; Kamolpatana, K.; Root-Kustritz, M.V.; Johnston, G.R. Prostatic disorders in the dog. Anim. Reprod. Sci. 2000, 60–61, 405–415. [Google Scholar] [CrossRef] [PubMed]
- Berry, S.J.; Strandberg, J.D.; Saunders, W.J.; Coffey, D.S. Development of canine benign prostatic hyperplasia with age. Prostate 1986, 9, 363–373. [Google Scholar] [CrossRef] [PubMed]
- Krawiec, D.R.; Heflin, D. Study of prostatic disease in dogs: 177 cases (1981–1986). J. Am. Vet. Med. Assoc. 1992, 200, 1119–1122. [Google Scholar] [PubMed]
- Johnston, S.D.; Root, M.V.K.; Olson, P.N.S. Disorders of the Canine Prostate. In Canine and Feline Theriogenology; Johnston, S.D., Root, M.V.K., Olson, P.N.S., Eds.; WB Saunders: Philadelphia, PA, USA, 2001; pp. 337–355. [Google Scholar]
- Monti, S.; Di Silverio, F.; Iraci, R.; Martini, C.; Lanzara, S.; Falasca, P.; Poggi, M.; Stigliano, A.; Sciarra, F.; Toscano, V. Regional variations of insulin-like growth factor I (IGF-I), IGF-II, and receptor type I in benign prostatic hyperplasia tissue and their correlation with intraprostatic androgens. J. Clin. Endocrinol. Metab. 2001, 86, 1700–1706. [Google Scholar] [CrossRef] [PubMed]
- Soulitzis, N.; Karyotis, I.; Delakas, D.; Spandidos, D.A. Expression analysis of peptide growth factors VEGF, FGF2, TGFB1, EGF and IGF1 in prostate cancer and benign prostatic hyperplasia. Int. J. Oncol. 2006, 29, 305–314. [Google Scholar] [CrossRef] [PubMed]
- Levy, X.; Nizanski, W.; von Heimendahl, A.; Mimouni, P. Diagnosis of common prostatic conditions in dogs: An update. Reprod. Domest. Anim. 2014, 49 (Suppl. S2), 50–57. [Google Scholar] [CrossRef] [PubMed]
- Winter, M.L.; Liehr, J.G. Possible mechanism of induction of benign prostatic hyperplasia by estradiol and dihydrotestosterone in dogs. Toxicol. Appl. Pharmacol. 1996, 136, 211–219. [Google Scholar] [CrossRef] [PubMed]
- Lai, K.P.; Huang, C.K.; Fang, L.Y.; Izumi, K.; Lo, C.W.; Wood, R.; Kindblom, J.; Yeh, S.; Chang, C. Targeting stromal androgen receptor suppresses prolactin-driven benign prostatic hyperplasia (BPH). Mol. Endocrinol. 2013, 27, 1617–1631. [Google Scholar] [CrossRef]
- Smith, J. Canine prostatic disease: A review of anatomy, pathology, diagnosis, and treatment. Theriogenology 2008, 70, 375–383. [Google Scholar] [CrossRef]
- Gunzel-Apel, A.R.; Mohrke, C.; Poulsen Nautrup, C. Colour-coded and pulsed Doppler sonography of the canine testis, epididymis and prostate gland: Physiological and pathological findings. Reprod. Domest. Anim. 2001, 36, 236–240. [Google Scholar] [CrossRef]
- Atalan, G.; Holt, P.E.; Barr, F.J. Ultrasonographic estimation of prostate size in normal dogs and relationship to bodyweight and age. J. Small Anim. Pract. 1999, 40, 119–122. [Google Scholar] [CrossRef] [PubMed]
- Root Kustritz, M.V. Collection of tissue and culture samples from the canine reproductive tract. Theriogenology 2006, 66, 567–574. [Google Scholar] [CrossRef] [PubMed]
- Crawford, D.; Shore, N.; Cooperberg, M.; Dall’Era, M.; La Rosa, F. Clinical considerations after a negative prostate biopsy. J. Prostate Cancer 2017, 2, 1–5. [Google Scholar]
- Al-Kafaji, G.; Said, H.M.; Alam, M.A.; Al Naieb, Z.T. Blood-based microRNAs as diagnostic biomarkers to discriminate localized prostate cancer from benign prostatic hyperplasia and allow cancer-risk stratification. Oncol. Lett. 2018, 16, 1357–1365. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, M.; He, C.; Siddiqui, J.; Wei, J.T.; Macoska, J.A. CCL11 (eotaxin-1): A new diagnostic serum marker for prostate cancer. Prostate 2013, 73, 573–581. [Google Scholar] [CrossRef]
- Hicklin, D.J.; Ellis, L.M. Role of the vascular endothelial growth factor pathway in tumor growth and angiogenesis. J. Clin. Oncol. 2005, 23, 1011–1027. [Google Scholar] [CrossRef]
- Mado, C.O.; Lu, Y. Novel diagnostic biomarkers for prostate cancer. J. Cancer 2010, 1, 150–177. [Google Scholar] [CrossRef]
- Golchin-Rad, K.; Mogheiseh, A.; Nazifi, S.; Ahrari Khafi, M.S.; Derakhshandeh, N.; Abbaszadeh-Hasiri, M. Changes in specific serum biomarkers during the induction of prostatic hyperplasia in dogs. BMC Vet. Res. 2019, 15, 440. [Google Scholar] [CrossRef]
- Wieszczeczynski, M.; Krakowski, L.; Opielak, G.; Krakowska, I.; Furmaga, J.; Brodzki, P.; Bochniarz, M.; Dabrowski, R.; Piech, T.; Zdzisinska, B.; et al. MicroRNA and vascular endothelial growth factor (VEGF) as new useful markers in the diagnosis of benign prostatic hyperplasia in dogs. Theriogenology 2021, 171, 113–118. [Google Scholar] [CrossRef]
- Cunto, M.; Mariani, E.; Anicito Guido, E.; Ballotta, G.; Zambelli, D. Clinical approach to prostatic diseases in the dog. Reprod. Domest. Anim. 2019, 54, 815–822. [Google Scholar] [CrossRef]
- Nizanski, W.; Levy, X.; Ochota, M.; Pasikowska, J. Pharmacological treatment for common prostatic conditions in dogs-benign prostatic hyperplasia and prostatitis: An update. Reprod. Domest. Anim. 2014, 49 (Suppl. S2), 8–15. [Google Scholar] [CrossRef] [PubMed]
- Christensen, B.W. Canine prostate disease. Vet. Clin. N. Am. Small Anim. Pract. 2018, 48, 701–719. [Google Scholar] [CrossRef]
- Cazzuli, G.; Damian, J.P.; Molina, E.; Pessina, P. Post-castration prostatic involution: A morphometric and endocrine study of healthy canines and those with benign prostatic hyperplasia. Reprod. Domest. Anim. 2022, 57, 157–164. [Google Scholar] [CrossRef] [PubMed]
- Angrimani, D.S.R.; Francischini, M.C.P.; Brito, M.M.; Vannucchi, C.I. Prostatic hyperplasia: Vascularization, hemodynamic and hormonal analysis of dogs treated with finasteride or orchiectomy. PLoS ONE 2020, 15, e0234714. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.; Alfajaro, M.M.; Cho, K.O.; Choi, U.S.; Je, H.; Jung, J.; Jang, Y.; Choi, J. Perfusion change in benign prostatic hyperplasia before and after castration in a canine model: Contrast enhanced ultrasonography and CT perfusion study. Theriogenology 2020, 156, 97–106. [Google Scholar] [CrossRef] [PubMed]
- Clarke, W.; Zhang, Z.; Chan, D.W. The application of clinical proteomics to cancer and other diseases. Clin. Chem. Lab. Med. 2003, 41, 1562–1570. [Google Scholar] [CrossRef] [PubMed]
- Adaszek, L.; Banach, T.; Bartnicki, M.; Winiarczyk, D.; Lyp, P.; Winiarczyk, S. Application the mass spectrometry MALDI-TOF technique for detection of Babesia canis canis infection in dogs. Parasitol. Res. 2014, 113, 4293–4295. [Google Scholar] [CrossRef]
- Franco-Martinez, L.; Horvatic, A.; Gelemanovic, A.; Samardzija, M.; Mrljak, V.; Contreras-Aguilar, M.D.; Martinez-Subiela, S.; Dabrowski, R.; Tvarijonaviciute, A. Changes in the Salivary Proteome Associated With Canine Pyometra. Front. Vet. Sci. 2020, 7, 277. [Google Scholar] [CrossRef]
- Kules, J.; Horvatic, A.; Guillemin, N.; Ferreira, R.F.; Mischke, R.; Mrljak, V.; Chadwick, C.C.; Eckersall, P.D. The plasma proteome and the acute phase protein response in canine pyometra. J. Proteom. 2020, 223, 103817. [Google Scholar] [CrossRef]
- Nizanski, W.; Ochota, M.; Fontaine, C.; Pasikowska, J. Comparison of Clinical Effectiveness of Deslorelin Acetate and Osaterone Acetate in Dogs with Benign Prostatic Hyperplasia. Animals 2020, 10, 1936. [Google Scholar] [CrossRef]
- Russo, M.; Vignoli, M.; England, G.C. B-mode and contrast-enhanced ultrasonographic findings in canine prostatic disorders. Reprod. Domest. Anim. 2012, 47 (Suppl. S6), 238–242. [Google Scholar] [CrossRef] [PubMed]
- Kamolpatana, K.; Johnston, G.R.; Johnston, S.D. Determination of canine prostatic volume using transabdominal ultrasonography. Vet. Radiol. Ultrasound 2000, 41, 73–77. [Google Scholar] [CrossRef] [PubMed]
- Tyanova, S.; Temu, T.; Cox, J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 2016, 11, 2301–2319. [Google Scholar] [CrossRef]
- Tyanova, S.; Temu, T.; Sinitcyn, P.; Carlson, A.; Hein, M.Y.; Geiger, T.; Mann, M.; Cox, J. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 2016, 13, 731–740. [Google Scholar] [CrossRef] [PubMed]
- Ishioka, N.; Takahashi, N.; Putnam, F.W. Amino acid sequence of human plasma alpha 1B-glycoprotein: Homology to the immunoglobulin supergene family. Proc. Natl. Acad. Sci. USA 1986, 83, 2363–2367. [Google Scholar] [CrossRef] [PubMed]
- Abdul-Rahman, P.S.; Lim, B.K.; Hashim, O.H. Expression of high-abundance proteins in sera of patients with endometrial and cervical cancers: Analysis using 2-DE with silver staining and lectin detection methods. Electrophoresis 2007, 28, 1989–1996. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.; Hou, C.R.; Jiang, H.W.; Xiang, C.Q.; Shi, N.; Yuan, H.C.; Ding, Q.; Zhang, Y.F. Serum protein profiling to identify biomarkers for small renal cell carcinoma. Indian J. Biochem. Biophys. 2010, 47, 211–218. [Google Scholar]
- Jeong, D.H.; Kim, H.K.; Prince, A.E.; Lee, D.S.; Kim, Y.N.; Han, J.; Kim, K.T. Plasma proteomic analysis of patients with squamous cell carcinoma of the uterine cervix. J. Gynecol. Oncol. 2008, 19, 173–180. [Google Scholar] [CrossRef]
- Piyaphanee, N.; Ma, Q.; Kremen, O.; Czech, K.; Greis, K.; Mitsnefes, M.; Devarajan, P.; Bennett, M.R. Discovery and initial validation of alpha 1-B glycoprotein fragmentation as a differential urinary biomarker in pediatric steroid-resistant nephrotic syndrome. Proteom. Clin. Appl. 2011, 5, 334–342. [Google Scholar] [CrossRef]
- Kules, J.; de Torre-Minguela, C.; Baric Rafaj, R.; Gotic, J.; Nizic, P.; Ceron, J.J.; Mrljak, V. Plasma biomarkers of SIRS and MODS associated with canine babesiosis. Res. Vet. Sci. 2016, 105, 222–228. [Google Scholar] [CrossRef]
- Dutta, M.; Subramani, E.; Taunk, K.; Gajbhiye, A.; Seal, S.; Pendharkar, N.; Dhali, S.; Ray, C.D.; Lodh, I.; Chakravarty, B.; et al. Investigation of serum proteome alterations in human endometriosis. J. Proteom. 2015, 114, 182–196. [Google Scholar] [CrossRef] [PubMed]
- Masood, A.; Benabdelkamel, H.; Ekhzaimy, A.A.; Alfadda, A.A. Plasma-based proteomics profiling of patients with hyperthyroidism after antithyroid treatment. Molecules 2020, 25, 2831. [Google Scholar] [CrossRef] [PubMed]
- Villar, M.; Rajbhandari, R.M.; Artigas-Jeronimo, S.; Contreras, M.; Sadaula, A.; Karmacharya, D.; Alves, P.C.; Gortazar, C.; de la Fuente, J. Differentially represented proteins in response to infection with mycobacterium tuberculosis identified by quantitative serum proteomics in asian elephants. Pathogens 2022, 11, 1010. [Google Scholar] [CrossRef] [PubMed]
- Cordeiro, Y.G.; Mulder, L.M.; van Zeijl, R.J.M.; Paskoski, L.B.; van Veelen, P.; de Ru, A.; Strefezzi, R.F.; Heijs, B.; Fukumasu, H. Proteomic analysis identifies FNDC1, A1BG, and antigen processing proteins associated with tumor heterogeneity and malignancy in a canine model of breast cancer. Cancers 2021, 13, 5901. [Google Scholar] [CrossRef] [PubMed]
- Dohan, O.; De la Vieja, A.; Paroder, V.; Riedel, C.; Artani, M.; Reed, M.; Ginter, C.S.; Carrasco, N. The sodium/iodide symporter (NIS): Characterization, regulation, and medical significance. Endocr. Rev. 2003, 24, 48–77. [Google Scholar] [CrossRef] [PubMed]
- De Morais, R.M.; Sobrinho, A.B.; de Souza Silva, C.M.; de Oliveira, J.R.; da Silva, I.C.R.; de Toledo Nobrega, O. The role of the NIS (SLC5A5) gene in papillary thyroid cancer: A systematic review. Int. J. Endocrinol. 2018, 2018, 9128754. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Gong, M.; Yuan, H.; Park, H.G.; Frierson, H.F.; Li, H. Chimeric transcript generated by cis-splicing of adjacent genes regulates prostate cancer cell proliferation. Cancer Discov. 2012, 2, 598–607. [Google Scholar] [CrossRef]
- Verma, S.; Shankar, E.; Chan, E.R.; Gupta, S. Metabolic reprogramming and predominance of solute carrier genes during acquired enzalutamide resistance in prostate cancer. Cells 2020, 9, 2535. [Google Scholar] [CrossRef]
- Kogai, T.; Taki, K.; Brent, G.A. Enhancement of sodium/iodide symporter expression in thyroid and breast cancer. Endocr. Relat. Cancer 2006, 13, 797–826. [Google Scholar] [CrossRef]
- Kan, Z.; Jaiswal, B.S.; Stinson, J.; Janakiraman, V.; Bhatt, D.; Stern, H.M.; Yue, P.; Haverty, P.M.; Bourgon, R.; Zheng, J.; et al. Diverse somatic mutation patterns and pathway alterations in human cancers. Nature 2010, 466, 869–873. [Google Scholar] [CrossRef]
- Kung, H.J. Targeting tyrosine kinases and autophagy in prostate cancer. Horm. Cancer 2011, 2, 38–46. [Google Scholar] [CrossRef] [PubMed]
- Drake, J.M.; Graham, N.A.; Stoyanova, T.; Sedghi, A.; Goldstein, A.S.; Cai, H.; Smith, D.A.; Zhang, H.; Komisopoulou, E.; Huang, J.; et al. Oncogene-specific activation of tyrosine kinase networks during prostate cancer progression. Proc. Natl. Acad. Sci. USA 2012, 109, 1643–1648. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Xiong, H.; Zou, Y.; Xu, S.; Quan, L.; Yuan, X.; Xu, N.; Wang, Y. Frequently rearranged in advanced T-cell lymphomas-1 demonstrates oncogenic properties in prostate cancer. Mol. Med. Rep. 2016, 14, 3551–3558. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Hewitt, S.M.; Liu, S.; Zhou, X.; Zhu, H.; Zhou, C.; Zhang, G.; Quan, L.; Bai, J.; Xu, N. Tissue microarray analysis of human FRAT1 expression and its correlation with the subcellular localisation of beta-catenin in ovarian tumours. Br. J. Cancer 2006, 94, 686–691. [Google Scholar] [CrossRef] [PubMed]
- Saitoh, T.; Katoh, M. FRAT1 and FRAT2, clustered in human chromosome 10q24.1 region, are up-regulated in gastric cancer. Int. J. Oncol. 2001, 19, 311–315. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Liu, S.; Zhu, H.; Zhang, W.; Zhang, G.; Zhou, X.; Zhou, C.; Quan, L.; Bai, J.; Xue, L.; et al. FRAT1 overexpression leads to aberrant activation of beta-catenin/TCF pathway in esophageal squamous cell carcinoma. Int. J. Cancer 2008, 123, 561–568. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Han, Y.; Zheng, R.; Yu, J.H.; Miao, Y.; Wang, L.; Wang, E.H. Expression of Frat1 correlates with expression of beta-catenin and is associated with a poor clinical outcome in human SCC and AC. Tumour Biol. 2012, 33, 1437–1444. [Google Scholar] [CrossRef] [PubMed]
- Claessens, F.; Verrijdt, G.; Schoenmakers, E.; Haelens, A.; Peeters, B.; Verhoeven, G.; Rombauts, W. Selective DNA binding by the androgen receptor as a mechanism for hormone-specific gene regulation. J. Steroid Biochem. Mol. Biol. 2001, 76, 23–30. [Google Scholar] [CrossRef]
- Kaetzel, C.S. The polymeric immunoglobulin receptor: Bridging innate and adaptive immune responses at mucosal surfaces. Immunol. Rev. 2005, 206, 83–99. [Google Scholar] [CrossRef]
- Phalipon, A.; Corthesy, B. Novel functions of the polymeric Ig receptor: Well beyond transport of immunoglobulins. Trends Immunol. 2003, 24, 55–58. [Google Scholar] [CrossRef]
- DeSouza, L.V.; Grigull, J.; Ghanny, S.; Dube, V.; Romaschin, A.D.; Colgan, T.J.; Siu, K.W. Endometrial carcinoma biomarker discovery and verification using differentially tagged clinical samples with multidimensional liquid chromatography and tandem mass spectrometry. Mol. Cell Proteom. 2007, 6, 1170–1182. [Google Scholar] [CrossRef] [PubMed]
- Berntsson, J.; Lundgren, S.; Nodin, B.; Uhlen, M.; Gaber, A.; Jirstrom, K. Expression and prognostic significance of the polymeric immunoglobulin receptor in epithelial ovarian cancer. J. Ovarian Res. 2014, 7, 26. [Google Scholar] [CrossRef] [PubMed]
- Mehta, A.; Shapiro, M.D. Apolipoproteins in vascular biology and atherosclerotic disease. Nat. Rev. Cardiol. 2022, 19, 168–179. [Google Scholar] [CrossRef] [PubMed]
- Ryan, R.O.; van der Horst, D.J. Lipid transport biochemistry and its role in energy production. Annu. Rev. Entomol. 2000, 45, 233–260. [Google Scholar] [CrossRef] [PubMed]
- Gliemann, J. Receptors of the low density lipoprotein (LDL) receptor family in man. Multiple functions of the large family members via interaction with complex ligands. Biol. Chem. 1998, 379, 951–964. [Google Scholar] [PubMed]
- Nicoll, J.A.; Zunarelli, E.; Rampling, R.; Murray, L.S.; Papanastassiou, V.; Stewart, J. Involvement of apolipoprotein E in glioblastoma: Immunohistochemistry and clinical outcome. Neuroreport 2003, 14, 1923–1926. [Google Scholar] [CrossRef] [PubMed]
- Venanzoni, M.C.; Giunta, S.; Muraro, G.B.; Storari, L.; Crescini, C.; Mazzucchelli, R.; Montironi, R.; Seth, A. Apolipoprotein E expression in localized prostate cancers. Int. J. Oncol. 2003, 22, 779–786. [Google Scholar] [CrossRef]
- Oue, N.; Hamai, Y.; Mitani, Y.; Matsumura, S.; Oshimo, Y.; Aung, P.P.; Kuraoka, K.; Nakayama, H.; Yasui, W. Gene expression profile of gastric carcinoma: Identification of genes and tags potentially involved in invasion, metastasis, and carcinogenesis by serial analysis of gene expression. Cancer Res. 2004, 64, 2397–2405. [Google Scholar] [CrossRef]
- Huvila, J.; Brandt, A.; Rojas, C.R.; Pasanen, S.; Talve, L.; Hirsimaki, P.; Fey, V.; Kytomaki, L.; Saukko, P.; Carpen, O.; et al. Gene expression profiling of endometrial adenocarcinomas reveals increased apolipoprotein E expression in poorly differentiated tumors. Int. J. Gynecol. Cancer 2009, 19, 1226–1231. [Google Scholar] [CrossRef]
- Su, W.P.; Chen, Y.T.; Lai, W.W.; Lin, C.C.; Yan, J.J.; Su, W.C. Apolipoprotein E expression promotes lung adenocarcinoma proliferation and migration and as a potential survival marker in lung cancer. Lung Cancer 2011, 71, 28–33. [Google Scholar] [CrossRef]
- Brawer, M.K.; Crawford, E.D.; Fowler, J.; Lucia, M.S.; Schroeder, F.H. Prostate cancer: Epidemiology and screening. Rev. Urol. 2000, 2 (Suppl. S4), S5–S9. [Google Scholar] [CrossRef] [PubMed]
- Vogel, T.; Guo, N.H.; Guy, R.; Drezlich, N.; Krutzsch, H.C.; Blake, D.A.; Panet, A.; Roberts, D.D. Apolipoprotein E: A potent inhibitor of endothelial and tumor cell proliferation. J. Cell. Biochem. 1994, 54, 299–308. [Google Scholar] [CrossRef] [PubMed]
- Mahley, R.W. Apolipoprotein E: Cholesterol transport protein with expanding role in cell biology. Science 1988, 240, 622–630. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Berglund, L.; Ramakrishnan, R.; Mayeux, R.; Ngai, C.; Holleran, S.; Tycko, B.; Leff, T.; Shachter, N.S. A common Hpa I RFLP of apolipoprotein C-I increases gene transcription and exhibits an ethnically distinct pattern of linkage disequilibrium with the alleles of apolipoprotein E. J. Lipid Res. 1999, 40, 50–58. [Google Scholar] [CrossRef] [PubMed]
- Leduc, V.; Jasmin-Belanger, S.; Poirier, J. APOE and cholesterol homeostasis in Alzheimer’s disease. Trends Mol. Med. 2010, 16, 469–477. [Google Scholar] [CrossRef]
- McKay, G.J.; Savage, D.A.; Patterson, C.C.; Lewis, G.; McKnight, A.J.; Maxwell, A.P.; Warren 3/UK GoKinD Study, G. Association analysis of dyslipidemia-related genes in diabetic nephropathy. PLoS ONE 2013, 8, e58472. [Google Scholar] [CrossRef] [PubMed]
- Bouillet, B.; Gautier, T.; Blache, D.; Pais de Barros, J.P.; Duvillard, L.; Petit, J.M.; Lagrost, L.; Verges, B. Glycation of apolipoprotein C1 impairs its CETP inhibitory property: Pathophysiological relevance in patients with type 1 and type 2 diabetes. Diabetes Care 2014, 37, 1148–1156. [Google Scholar] [CrossRef]
- Ki, C.S.; Na, D.L.; Kim, D.K.; Kim, H.J.; Kim, J.W. Genetic association of an apolipoprotein C-I (APOC1) gene polymorphism with late-onset Alzheimer’s disease. Neurosci. Lett. 2002, 319, 75–78. [Google Scholar] [CrossRef]
- Bus, P.; Pierneef, L.; Bor, R.; Wolterbeek, R.; van Es, L.A.; Rensen, P.C.; de Heer, E.; Havekes, L.M.; Bruijn, J.A.; Berbee, J.F.; et al. Apolipoprotein C-I plays a role in the pathogenesis of glomerulosclerosis. J. Pathol. 2017, 241, 589–599. [Google Scholar] [CrossRef]
- Sun, Y.; Zhang, J.; Guo, F.; Zhao, W.; Zhan, Y.; Liu, C.; Fan, Y.; Wang, J. Identification of apolipoprotein C-I peptides as a potential biomarker and its biological roles in breast cancer. Med. Sci. Monit. 2016, 22, 1152–1160. [Google Scholar] [CrossRef]
- Ko, H.L.; Wang, Y.S.; Fong, W.L.; Chi, M.S.; Chi, K.H.; Kao, S.J. Apolipoprotein C1 (APOC1) as a novel diagnostic and prognostic biomarker for lung cancer: A marker phase I trial. Thorac. Cancer 2014, 5, 500–508. [Google Scholar] [CrossRef] [PubMed]
- Takano, S.; Yoshitomi, H.; Togawa, A.; Sogawa, K.; Shida, T.; Kimura, F.; Shimizu, H.; Tomonaga, T.; Nomura, F.; Miyazaki, M. Apolipoprotein C-1 maintains cell survival by preventing from apoptosis in pancreatic cancer cells. Oncogene 2008, 27, 2810–2822. [Google Scholar] [CrossRef] [PubMed]
- Su, W.P.; Sun, L.N.; Yang, S.L.; Zhao, H.; Zeng, T.Y.; Wu, W.Z.; Wang, D. Apolipoprotein C1 promotes prostate cancer cell proliferation in vitro. J. Biochem. Mol. Toxicol. 2018, 32, e22158. [Google Scholar] [CrossRef] [PubMed]
- Lai, C.Q.; Smith, C.E.; Parnell, L.D.; Lee, Y.C.; Corella, D.; Hopkins, P.; Hidalgo, B.A.; Aslibekyan, S.; Province, M.A.; Absher, D.; et al. Epigenomics and metabolomics reveal the mechanism of the APOA2-saturated fat intake interaction affecting obesity. Am. J. Clin. Nutr. 2018, 108, 188–200. [Google Scholar] [CrossRef] [PubMed]
- Honda, K.; Katzke, V.A.; Husing, A.; Okaya, S.; Shoji, H.; Onidani, K.; Olsen, A.; Tjonneland, A.; Overvad, K.; Weiderpass, E.; et al. CA19-9 and apolipoprotein-A2 isoforms as detection markers for pancreatic cancer: A prospective evaluation. Int. J. Cancer 2019, 144, 1877–1887. [Google Scholar] [CrossRef] [PubMed]
- Yoon, H.I.; Kwon, O.R.; Kang, K.N.; Shin, Y.S.; Shin, H.S.; Yeon, E.H.; Kwon, K.Y.; Hwang, I.; Jeon, Y.K.; Kim, Y.; et al. Diagnostic value of combining tumor and inflammatory markers in lung cancer. J. Cancer Prev. 2016, 21, 187–193. [Google Scholar] [CrossRef] [PubMed]
- Voronova, V.; Glybochko, P.; Svistunov, A.; Fomin, V.; Kopylov, P.; Tzarkov, P.; Egorov, A.; Gitel, E.; Ragimov, A.; Boroda, A.; et al. Diagnostic value of combinatorial markers in colorectal carcinoma. Front. Oncol. 2020, 10, 832. [Google Scholar] [CrossRef]
- Vermaat, J.S.; Gerritse, F.L.; van der Veldt, A.A.; Roessingh, W.M.; Niers, T.M.; Oosting, S.F.; Sleijfer, S.; Roodhart, J.M.; Beijnen, J.H.; Schellens, J.H.; et al. Validation of serum amyloid alpha as an independent biomarker for progression-free and overall survival in metastatic renal cell cancer patients. Eur. Urol. 2012, 62, 685–695. [Google Scholar] [CrossRef]
- Lima, T.; Henrique, R.; Vitorino, R.; Fardilha, M. Bioinformatic analysis of dysregulated proteins in prostate cancer patients reveals putative urinary biomarkers and key biological pathways. Med. Oncol. 2021, 38, 9. [Google Scholar] [CrossRef]
- Bao, M.; Xu, J.; Liu, J. Overexpression of APOA2 is Associated with Progression and Poor Prognosis in Gastric Cancer. Res. Artic. 2022, 1, 1–21. [Google Scholar] [CrossRef]
- Amah, U.; Mbaeri, T.; Onuegbu, J.; Olisekodiaka, J.; Okwara, J.; Onyema-Iloh, O.; Nnamdi, J.; Onah, C.; Ahaneku, J. Assessment of serum lipid profile and apolipoproteins in patients with prostate disorders in Nnewi, Anambra, South east Nigeria. Trop. J. Med. Res. 2022, 21, 205–212. [Google Scholar]
- Kelber, J.A.; Klemke, R.L. PEAK1, a novel kinase target in the fight against cancer. Oncotarget 2010, 1, 219–223. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Lapek, J.; Fujimura, K.; Strnadel, J.; Liu, B.; Gonzalez, D.J.; Zhang, W.; Watson, F.; Yu, V.; Liu, C.; et al. Pseudopodium-enriched atypical kinase 1 mediates angiogenesis by modulating GATA2-dependent VEGFR2 transcription. Cell Discov. 2018, 4, 26. [Google Scholar] [CrossRef] [PubMed]
- Skingen, V.E.; Hompland, T.; Fjeldbo, C.S.; Salberg, U.B.; Helgeland, H.; Ragnum, H.B.; Aarnes, E.K.; Vlatkovic, L.; Hole, K.H.; Seierstad, T.; et al. Prostate cancer radiogenomics reveals proliferative gene expression programs associated with distinct MRI-based hypoxia levels. Radiother. Oncol. 2023, 188, 109875. [Google Scholar] [CrossRef] [PubMed]
- Huynh, H. Induction of apoptosis in rat ventral prostate by finasteride is associated with alteration in MAP kinase pathways and Bcl-2 related family of proteins. Int. J. Oncol. 2002, 20, 1297–1303. [Google Scholar] [CrossRef] [PubMed]
- Brognard, J.; Sierecki, E.; Gao, T.; Newton, A.C. PHLPP and a second isoform, PHLPP2, differentially attenuate the amplitude of Akt signaling by regulating distinct Akt isoforms. Mol. Cell 2007, 25, 917–931. [Google Scholar] [CrossRef] [PubMed]
- Gao, T.; Brognard, J.; Newton, A.C. The phosphatase PHLPP controls the cellular levels of protein kinase C. J. Biol. Chem. 2008, 283, 6300–6311. [Google Scholar] [CrossRef] [PubMed]
- Gao, T.; Furnari, F.; Newton, A.C. PHLPP: A phosphatase that directly dephosphorylates Akt, promotes apoptosis, and suppresses tumor growth. Mol. Cell 2005, 18, 13–24. [Google Scholar] [CrossRef]
- Chen, M.; Pratt, C.P.; Zeeman, M.E.; Schultz, N.; Taylor, B.S.; O’Neill, A.; Castillo-Martin, M.; Nowak, D.G.; Naguib, A.; Grace, D.M.; et al. Identification of PHLPP1 as a tumor suppressor reveals the role of feedback activation in PTEN-mutant prostate cancer progression. Cancer Cell 2011, 20, 173–186. [Google Scholar] [CrossRef]
- Cuevas, B.D.; Abell, A.N.; Johnson, G.L. Role of mitogen-activated protein kinase kinase kinases in signal integration. Oncogene 2007, 26, 3159–3171. [Google Scholar] [CrossRef]
- Morrison, D.K. MAP kinase pathways. Cold Spring Harb. Perspect. Biol. 2012, 4, a011254. [Google Scholar] [CrossRef] [PubMed]
- Yustein, J.T.; Xia, L.; Kahlenburg, J.M.; Robinson, D.; Templeton, D.; Kung, H.J. Comparative studies of a new subfamily of human Ste20-like kinases: Homodimerization, subcellular localization, and selective activation of MKK3 and p38. Oncogene 2003, 22, 6129–6141. [Google Scholar] [CrossRef]
- Zhu, L.; Yu, Q.; Gao, P.; Liu, Q.; Luo, X.; Jiang, G.; Ji, R.; Yang, R.; Ma, X.; Xu, J.; et al. TAOK1 positively regulates TLR4-induced inflammatory responses by promoting ERK1/2 activation in macrophages. Mol. Immunol. 2020, 122, 124–131. [Google Scholar] [CrossRef] [PubMed]
- Zihni, C.; Mitsopoulos, C.; Tavares, I.A.; Ridley, A.J.; Morris, J.D. Prostate-derived sterile 20-like kinase 2 (PSK2) regulates apoptotic morphology via C-Jun N-terminal kinase and Rho kinase-1. J. Biol. Chem. 2006, 281, 7317–7323. [Google Scholar] [CrossRef] [PubMed]
- Cinar, B.; Collak, F.K.; Lopez, D.; Akgul, S.; Mukhopadhyay, N.K.; Kilicarslan, M.; Gioeli, D.G.; Freeman, M.R. MST1 is a multifunctional caspase-independent inhibitor of androgenic signaling. Cancer Res. 2011, 71, 4303–4313. [Google Scholar] [CrossRef] [PubMed]
- Song, L.; Zhang, J.; Li, C.; Yao, J.; Jiang, C.; Li, Y.; Liu, S.; Liu, Z. Genome-wide identification of hsp40 genes in channel catfish and their regulated expression after bacterial infection. PLoS ONE 2014, 9, e115752. [Google Scholar] [CrossRef]
- Laybutt, D.R.; Preston, A.M.; Akerfeldt, M.C.; Kench, J.G.; Busch, A.K.; Biankin, A.V.; Biden, T.J. Endoplasmic reticulum stress contributes to beta cell apoptosis in type 2 diabetes. Diabetologia 2007, 50, 752–763. [Google Scholar] [CrossRef]
- Chien, V.; Aitken, J.F.; Zhang, S.; Buchanan, C.M.; Hickey, A.; Brittain, T.; Cooper, G.J.; Loomes, K.M. The chaperone proteins HSP70, HSP40/DnaJ and GRP78/BiP suppress misfolding and formation of beta-sheet-containing aggregates by human amylin: A potential role for defective chaperone biology in Type 2 diabetes. Biochem. J. 2010, 432, 113–121. [Google Scholar] [CrossRef]
- Walsh, P.; Bursac, D.; Law, Y.C.; Cyr, D.; Lithgow, T. The J-protein family: Modulating protein assembly, disassembly and translocation. EMBO Rep. 2004, 5, 567–571. [Google Scholar] [CrossRef]
- Yamamoto, Y.H.; Kasai, A.; Omori, H.; Takino, T.; Sugihara, M.; Umemoto, T.; Hamasaki, M.; Hatta, T.; Natsume, T.; Morimoto, R.I.; et al. ERdj8 governs the size of autophagosomes during the formation process. J. Cell Biol. 2020, 219, e201903127. [Google Scholar] [CrossRef]
- Chang, Y.S.; Lin, C.Y.; Liu, T.Y.; Huang, C.M.; Chung, C.C.; Chen, Y.C.; Tsai, F.J.; Chang, J.G.; Chang, S.J. Polygenic risk score trend and new variants on chromosome 1 are associated with male gout in genome-wide association study. Arthritis Res. Ther. 2022, 24, 229. [Google Scholar] [CrossRef] [PubMed]
- Moses, M.A.; Kim, Y.S.; Rivera-Marquez, G.M.; Oshima, N.; Watson, M.J.; Beebe, K.E.; Wells, C.; Lee, S.; Zuehlke, A.D.; Shao, H.; et al. Targeting the Hsp40/Hsp70 chaperone axis as a novel strategy to treat castration-resistant prostate cancer. Cancer Res. 2018, 78, 4022–4035. [Google Scholar] [CrossRef] [PubMed]
- Mei, L.; Xiong, W.C. Neuregulin 1 in neural development, synaptic plasticity and schizophrenia. Nat. Rev. Neurosci. 2008, 9, 437–452. [Google Scholar] [CrossRef] [PubMed]
- Anton, E.S.; Marchionni, M.A.; Lee, K.F.; Rakic, P. Role of GGF/neuregulin signaling in interactions between migrating neurons and radial glia in the developing cerebral cortex. Development 1997, 124, 3501–3510. [Google Scholar] [CrossRef]
- Wang, X.; Zhuo, X.; Gao, J.; Liu, H.; Lin, F.; Ma, A. Neuregulin-1beta partially improves cardiac function in volume-overload heart failure through regulation of abnormal calcium handling. Front. Pharmacol. 2019, 10, 616. [Google Scholar] [CrossRef]
- Jie, B.; Zhang, X.; Wu, X.; Xin, Y.; Liu, Y.; Guo, Y. Neuregulin-1 suppresses cardiomyocyte apoptosis by activating PI3K/Akt and inhibiting mitochondrial permeability transition pore. Mol. Cell. Biochem. 2012, 370, 35–43. [Google Scholar] [CrossRef]
- Vermeulen, Z.; Hervent, A.S.; Dugaucquier, L.; Vandekerckhove, L.; Rombouts, M.; Beyens, M.; Schrijvers, D.M.; De Meyer, G.R.Y.; Maudsley, S.; De Keulenaer, G.W.; et al. Inhibitory actions of the NRG-1/ErbB4 pathway in macrophages during tissue fibrosis in the heart, skin, and lung. Am. J. Physiol. Heart Circ. Physiol. 2017, 313, H934–H945. [Google Scholar] [CrossRef]
- Schumacher, M.A.; Hedl, M.; Abraham, C.; Bernard, J.K.; Lozano, P.R.; Hsieh, J.J.; Almohazey, D.; Bucar, E.B.; Punit, S.; Dempsey, P.J.; et al. ErbB4 signaling stimulates pro-inflammatory macrophage apoptosis and limits colonic inflammation. Cell Death Dis. 2017, 8, e2622. [Google Scholar] [CrossRef]
- Umehara, T.; Kawashima, I.; Kawai, T.; Hoshino, Y.; Morohashi, K.I.; Shima, Y.; Zeng, W.; Richards, J.S.; Shimada, M. Neuregulin 1 regulates proliferation of Leydig cells to support spermatogenesis and sexual behavior in adult mice. Endocrinology 2016, 157, 4899–4913. [Google Scholar] [CrossRef]
- Olivier, E.; Soury, E.; Ruminy, P.; Husson, A.; Parmentier, F.; Daveau, M.; Salier, J.P. Fetuin-B, a second member of the fetuin family in mammals. Biochem. J. 2000, 350 Pt 2, 589–597. [Google Scholar] [CrossRef]
- Cayatte, A.J.; Kumbla, L.; Subbiah, M.T. Marked acceleration of exogenous fatty acid incorporation into cellular triglycerides by fetuin. J. Biol. Chem. 1990, 265, 5883–5888. [Google Scholar] [CrossRef] [PubMed]
- Ombrellino, M.; Wang, H.; Yang, H.; Zhang, M.; Vishnubhakat, J.; Frazier, A.; Scher, L.A.; Friedman, S.G.; Tracey, K.J. Fetuin, a negative acute phase protein, attenuates TNF synthesis and the innate inflammatory response to carrageenan. Shock 2001, 15, 181–185. [Google Scholar] [CrossRef] [PubMed]
- Denecke, B.; Graber, S.; Schafer, C.; Heiss, A.; Woltje, M.; Jahnen-Dechent, W. Tissue distribution and activity testing suggest a similar but not identical function of fetuin-B and fetuin-A. Biochem. J. 2003, 376, 135–145. [Google Scholar] [CrossRef] [PubMed]
- Jung, S.H.; Won, K.J.; Lee, K.P.; Kim, H.J.; Seo, E.H.; Lee, H.M.; Park, E.S.; Lee, S.H.; Kim, B. The serum protein fetuin-B is involved in the development of acute myocardial infarction. Clin. Sci. 2015, 129, 27–38. [Google Scholar] [CrossRef] [PubMed]
- Borlak, J.; Langer, F.; Chatterji, B. Serum proteome mapping of EGF transgenic mice reveal mechanistic biomarkers of lung cancer precursor lesions with clinical significance for human adenocarcinomas. Biochim. Biophys. Acta Mol. Basis Dis. 2018, 1864, 3122–3144. [Google Scholar] [CrossRef] [PubMed]
- Dietzel, E.; Wessling, J.; Floehr, J.; Schafer, C.; Ensslen, S.; Denecke, B.; Rosing, B.; Neulen, J.; Veitinger, T.; Spehr, M.; et al. Fetuin-B, a liver-derived plasma protein is essential for fertilization. Dev. Cell 2013, 25, 106–112. [Google Scholar] [CrossRef] [PubMed]
- Peter, A.; Kovarova, M.; Staiger, H.; Machann, J.; Schick, F.; Konigsrainer, A.; Konigsrainer, I.; Schleicher, E.; Fritsche, A.; Haring, H.U.; et al. The hepatokines fetuin-A and fetuin-B are upregulated in the state of hepatic steatosis and may differently impact on glucose homeostasis in humans. Am. J. Physiol. Endocrinol. Metab. 2018, 314, E266–E273. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.W.; Choi, J.W.; Lee, D.S.; Yun, J.W. Sex hormones regulate hepatic fetuin expression in male and female rats. Cell Physiol. Biochem. 2014, 34, 554–564. [Google Scholar] [CrossRef]
- Zhan, K.; Liu, R.; Tong, H.; Gao, S.; Yang, G.; Hossain, A.; Li, T.; He, W. Fetuin B overexpression suppresses proliferation, migration, and invasion in prostate cancer by inhibiting the PI3K/AKT signaling pathway. Biomed. Pharmacother. 2020, 131, 110689. [Google Scholar] [CrossRef]
- Kenmochi, N.; Suzuki, T.; Uechi, T.; Magoori, M.; Kuniba, M.; Higa, S.; Watanabe, K.; Tanaka, T. The human mitochondrial ribosomal protein genes: Mapping of 54 genes to the chromosomes and implications for human disorders. Genomics 2001, 77, 65–70. [Google Scholar] [CrossRef]
- Pang, S.T.; Weng, W.H.; Flores-Morales, A.; Johansson, B.; Pourian, M.R.; Nilsson, P.; Pousette, A.; Larsson, C.; Norstedt, G. Cytogenetic and expression profiles associated with transformation to androgen-resistant prostate cancer. Prostate 2006, 66, 157–172. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Ballinger, D.G.; Dai, J.Y.; Peters, U.; Hinds, D.A.; Cox, D.R.; Beilharz, E.; Chlebowski, R.T.; Rossouw, J.E.; McTiernan, A.; et al. Genetic variants in the MRPS30 region and postmenopausal breast cancer risk. Genome Med. 2011, 3, 42. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Morito, D.; Takashima, S.; Mineharu, Y.; Kobayashi, H.; Hitomi, T.; Hashikata, H.; Matsuura, N.; Yamazaki, S.; Toyoda, A.; et al. Identification of RNF213 as a susceptibility gene for moyamoya disease and its possible role in vascular development. PLoS ONE 2011, 6, e22542. [Google Scholar] [CrossRef] [PubMed]
- Morito, D.; Nishikawa, K.; Hoseki, J.; Kitamura, A.; Kotani, Y.; Kiso, K.; Kinjo, M.; Fujiyoshi, Y.; Nagata, K. Moyamoya disease-associated protein mysterin/RNF213 is a novel AAA+ ATPase, which dynamically changes its oligomeric state. Sci. Rep. 2014, 4, 4442. [Google Scholar] [CrossRef] [PubMed]
- Thery, F.; Martina, L.; Asselman, C.; Zhang, Y.; Vessely, M.; Repo, H.; Sedeyn, K.; Moschonas, G.D.; Bredow, C.; Teo, Q.W.; et al. Ring finger protein 213 assembles into a sensor for ISGylated proteins with antimicrobial activity. Nat. Commun. 2021, 12, 5772. [Google Scholar] [CrossRef] [PubMed]
- Raj, S.; Scott, D.R.; Nguyen, T.; Sachs, G.; Kraut, J.A. Acid stress increases gene expression of proinflammatory cytokines in Madin-Darby canine kidney cells. Am. J. Physiol. Renal Physiol. 2013, 304, F41–F48. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.L.; Li, B.; Luo, Y.X.; Lin, Q.; Liu, S.R.; Zhang, X.Q.; Zhou, H.; Yang, J.H.; Qu, L.H. Comprehensive genomic characterization of RNA-binding proteins across human cancers. Cell Rep. 2018, 22, 286–298. [Google Scholar] [CrossRef]
- Kim, M.Y.; Hur, J.; Jeong, S. Emerging roles of RNA and RNA-binding protein network in cancer cells. BMB Rep. 2009, 42, 125–130. [Google Scholar] [CrossRef]
- Lukong, K.E.; Chang, K.W.; Khandjian, E.W.; Richard, S. RNA-binding proteins in human genetic disease. Trends Genet. 2008, 24, 416–425. [Google Scholar] [CrossRef]


| Dog | Breed | Weight | Age | Estimated Volume | Prostatic Volume before Castration | Prostatic Volume after Castration |
|---|---|---|---|---|---|---|
| (kg) | (Years) | (cm3) | (cm3) | (cm3) | ||
| 1 | American Bully | 30.00 | 2 | 13.18 | 33.80 | 7.70 |
| 2 | Boston Terrier | 12.00 | 5 | 7.24 | 9.12 | 7.20 |
| 3 | Chihuahua | 3.20 | 7 | 4.34 | 6.80 | 4.37 |
| 4 | Chihuahua | 2.60 | 3 | 4.14 | 5.80 | 3.20 |
| 5 | Chihuahua | 4.90 | 6 | 4.90 | 7.20 | 3.56 |
| 6 | Golden Retriever | 45.00 | 8 | 18.13 | 24.70 | 9.47 |
| 7 | Jack Russell Terrier | 8.50 | 7 | 6.09 | 7.80 | 3.60 |
| 8 | Mongrel | 24.00 | 7 | 11.20 | 43.60 | 9.30 |
| 9 | Pomeranian | 4.00 | 6 | 4.60 | 6.30 | 4.30 |
| 10 | Pomeranian | 4.30 | 10 | 4.69 | 9.04 | 4.11 |
| 11 | Pomeranian | 7.30 | 6 | 5.69 | 10.60 | 3.66 |
| 12 | Shih Tzu | 5.70 | 7 | 5.16 | 5.35 | 2.57 |
| 13 | Shih Tzu | 6.90 | 4 | 5.56 | 8.97 | 4.50 |
| 14 | Shih Tzu | 8.50 | 10 | 6.09 | 7.60 | 4.10 |
| 15 | Thai Bangkaew | 16.50 | 4 | 8.73 | 9.09 | 3.00 |
| Protein Names | Gene Names | −log10(p) | FDR | Fisher’s LSD | Biological Process | Cellular Component | Molecular Function |
|---|---|---|---|---|---|---|---|
| Regulator of G protein signaling 22 | RGS22 | 14.802 | 6.07 × 10−12 | AT—BF; AT—CTRL; BF—CTRL | Regulation of signal transduction | N/A | G-protein alpha-subunit binding |
| Apolipoprotein E | APOE | 9.3962 | 7.73 × 10−7 | AT—BF; AT—CTRL | AMPA glutamate receptor clustering, cGMP-mediated signaling, cholesterol catabolic process, G protein-coupled receptor signaling pathway, gene expression | Chylomicron, endoplasmic reticulum, extracellular exosome, extracellular matrix, extracellular space, Golgi apparatus, plasma membrane | Amyloid-beta binding, antioxidant activity, cholesterol transfer activity, enzyme binding, heparin binding, lipoprotein particle binding, phospholipid binding |
| E3 ubiquitin-protein ligase | TRIP12 | 8.341 | 5.85 × 10−6 | AT—BF; AT—CTRL; BF—CTRL | DNA repair, protein ubiquitination, ubiquitin-dependent protein catabolic process | Nucleoplasm | Ubiquitin protein ligase activity, zinc ion binding |
| Ubiquitin-associated domain-containing protein 1 | UBAC1 | 8.1622 | 6.62 × 10−6 | AT—BF; AT—CTRL; BF—CTRL | N/A | N/A | N/A |
| Apolipoprotein C-I | APOC1 | 6.7248 | 1.31 × 10−4 | AT—BF; AT—CTRL | Lipid transport, lipoprotein metabolic process, negative regulation of cholesterol transport, triglyceride metabolic process | High-density lipoprotein particle, very-low-density lipoprotein particle | Fatty acid binding, phospholipase inhibitor activity |
| Apolipoprotein A-II | APOA2 | 6.6902 | 1.31 × 10−4 | AT—BF; AT—CTRL | Lipid transport, lipoprotein metabolic process | Extracellular region | Lipid binding |
| Solute carrier family 5 member 5 | SLC5A5 | 6.3569 | 2.42 × 10−4 | BF—AT; BF—CTRL | N/A | Membrane | Transmembrane transporter activity |
| Mitochondrial ribosomal protein S30 | MRPS30 | 5.6704 | 0.001028 | AT—BF; AT—CTRL | Translation | Mitochondrial large ribosomal subunit | Structural constituent of ribosome |
| Pseudopodium enriched atypical kinase 1 | PEAK1 | 5.6141 | 0.0010402 | AT—BF; AT—CTRL | Protein phosphorylation | N/A | ATP binding, protein kinase activity |
| Tyrosine-protein kinase | TNK2 | 5.4357 | 0.001384 | BF—AT; BF—CTRL | Adaptive immune response, intracellular signal transduction, protein phosphorylation | Plasma membrane | ATP binding, metal ion binding protein, tyrosine kinase activity |
| FRAT regulator of WNT signaling pathway 1 | FRAT1 | 5.4029 | 0.001384 | BF—AT; BF—CTRL | N/A | N/A | N/A |
| TAO kinase 1 | TAOK1 | 5.2529 | 0.0017921 | AT—BF; AT—CTRL | Protein phosphorylation | N/A | ATP binding, protein kinase activity |
| RNA-binding protein 18 | RBM18 | 4.9606 | 0.0032425 | AT—BF; AT—CTRL | N/A | N/A | RNA binding |
| DnaJ homolog subfamily C member 16 | DNAJC16 | 4.8531 | 0.0037381 | AT—BF; AT—CTRL | N/A | Membrane | N/A |
| Polymeric immunoglobulin receptor | PIGR | 4.8367 | 0.0037381 | BF—AT; BF—CTRL | N/A | Membrane | N/A |
| Alpha-1-B glycoprotein | A1BG | 4.6873 | 0.0049434 | AT—BF; CTRL—BF | N/A | Membrane | N/A |
| Fetuin B | FETUB | 4.4265 | 0.008483 | AT—BF; AT—CTRL | Binding of sperm to zona pellucida, negative regulation of endopeptidase activity | Extracellular space | Cysteine-type endopeptidase inhibitor activity, metalloendopeptidase inhibitor activity |
| PH domain and leucine rich repeat protein phosphatase 1 | PHLPP1 | 4.263 | 0.011673 | AT—BF; AT—CTRL | N/A | N/A | N/A |
| Ig-like domain-containing protein | N/A | 4.2288 | 0.011965 | AT—CTRL; BF—CTRL | N/A | N/A | N/A |
| Neuregulin 1 | NRG1 | 4.1774 | 0.012795 | AT—BF; AT—CTRL | Nervous system development | Cellular anatomical entity | Signaling receptor binding |
| Ring finger protein 213 | RNF213 | 4.1374 | 0.013005 | AT—BF; AT—CTRL | N/A | Cytoplasm | ATP hydrolysis activity, metal ion binding, ubiquitin-protein transferase activity |
| Voltage-dependent L-type calcium channel subunit alpha | CACNA1D | 4.1289 | 0.013005 | AT—BF; AT—CTRL; CTRL—BF | Regulation of monoatomic ion transmembrane transport | Voltage-gated calcium channel complex | Metal ion binding, voltage-gated calcium channel activity |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ploypetch, S.; Wongbandue, G.; Roytrakul, S.; Phaonakrop, N.; Prapaiwan, N. Comparative Serum Proteome Profiling of Canine Benign Prostatic Hyperplasia before and after Castration. Animals 2023, 13, 3853. https://doi.org/10.3390/ani13243853
Ploypetch S, Wongbandue G, Roytrakul S, Phaonakrop N, Prapaiwan N. Comparative Serum Proteome Profiling of Canine Benign Prostatic Hyperplasia before and after Castration. Animals. 2023; 13(24):3853. https://doi.org/10.3390/ani13243853
Chicago/Turabian StylePloypetch, Sekkarin, Grisnarong Wongbandue, Sittiruk Roytrakul, Narumon Phaonakrop, and Nawarus Prapaiwan. 2023. "Comparative Serum Proteome Profiling of Canine Benign Prostatic Hyperplasia before and after Castration" Animals 13, no. 24: 3853. https://doi.org/10.3390/ani13243853





