Rumen Development of Tianhua Mutton Sheep Was Better than That of Gansu Alpine Fine Wool Sheep under Grazing Conditions
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. H&E Stain
2.3. Determination of Rumen VFAs
2.4. DNA Extraction from Gastrointestinal Contents
2.5. 16S rRNA Full-Length Sequencing
2.6. Sequencing Data Processing
2.7. Statistical Analysis
3. Results
3.1. Differences in Volatile Fatty Acids in Rumen Contents of Tianhua Mutton Sheep and Gansu Alpine Fine Wool Sheep
3.2. Differences in Rumen Histomorphology between Tianhua Mutton Sheep and Gansu Alpine Fine Wool Sheep
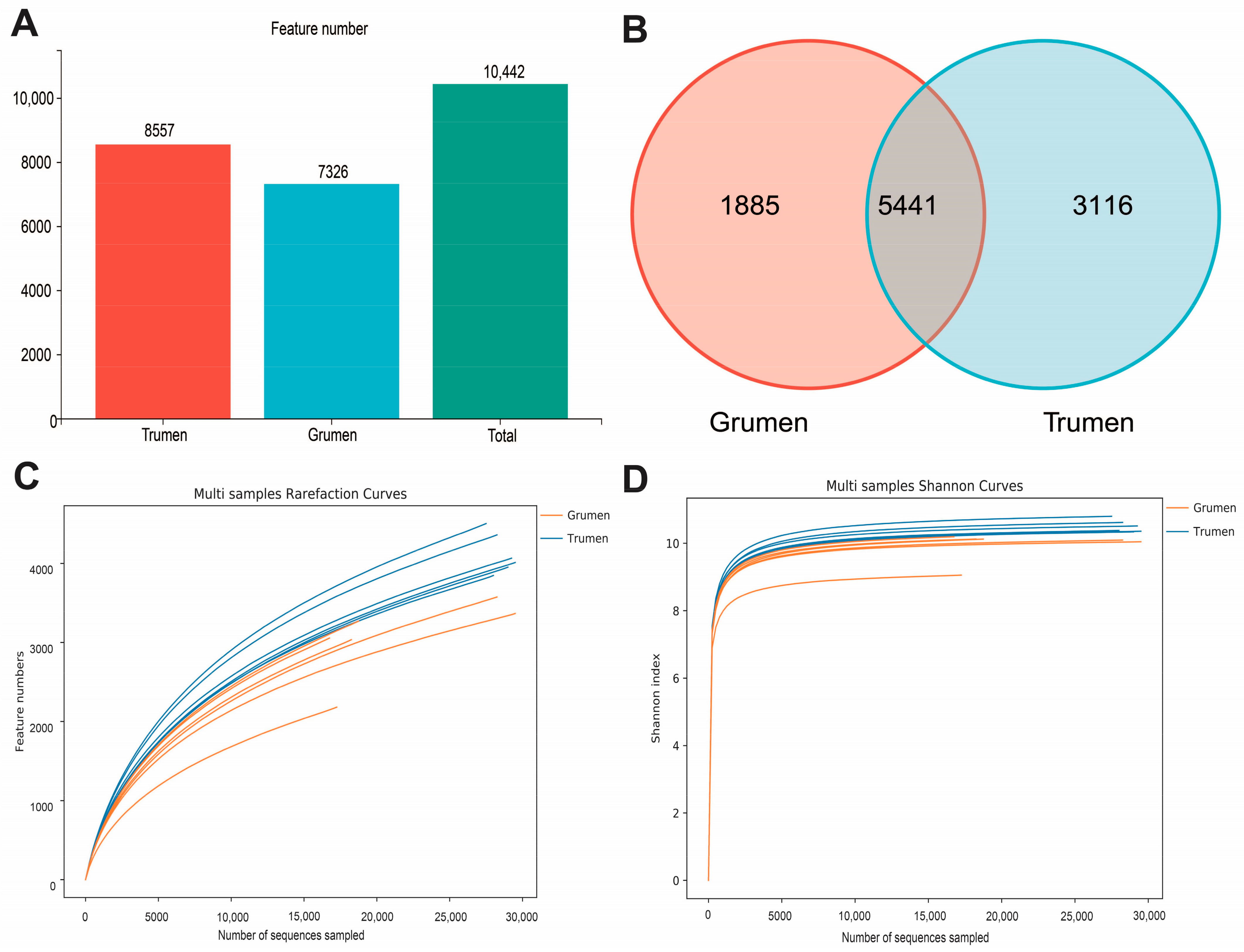
3.3. Alpha Diversity Analysis of Rumen Flora in Tianhua Mutton Sheep and Gansu Alpine Fine Wool Sheep
3.4. Beta Diversity Analysis of Rumen Flora in Tianhua Mutton Sheep and Gansu Alpine Fine Wool Sheep
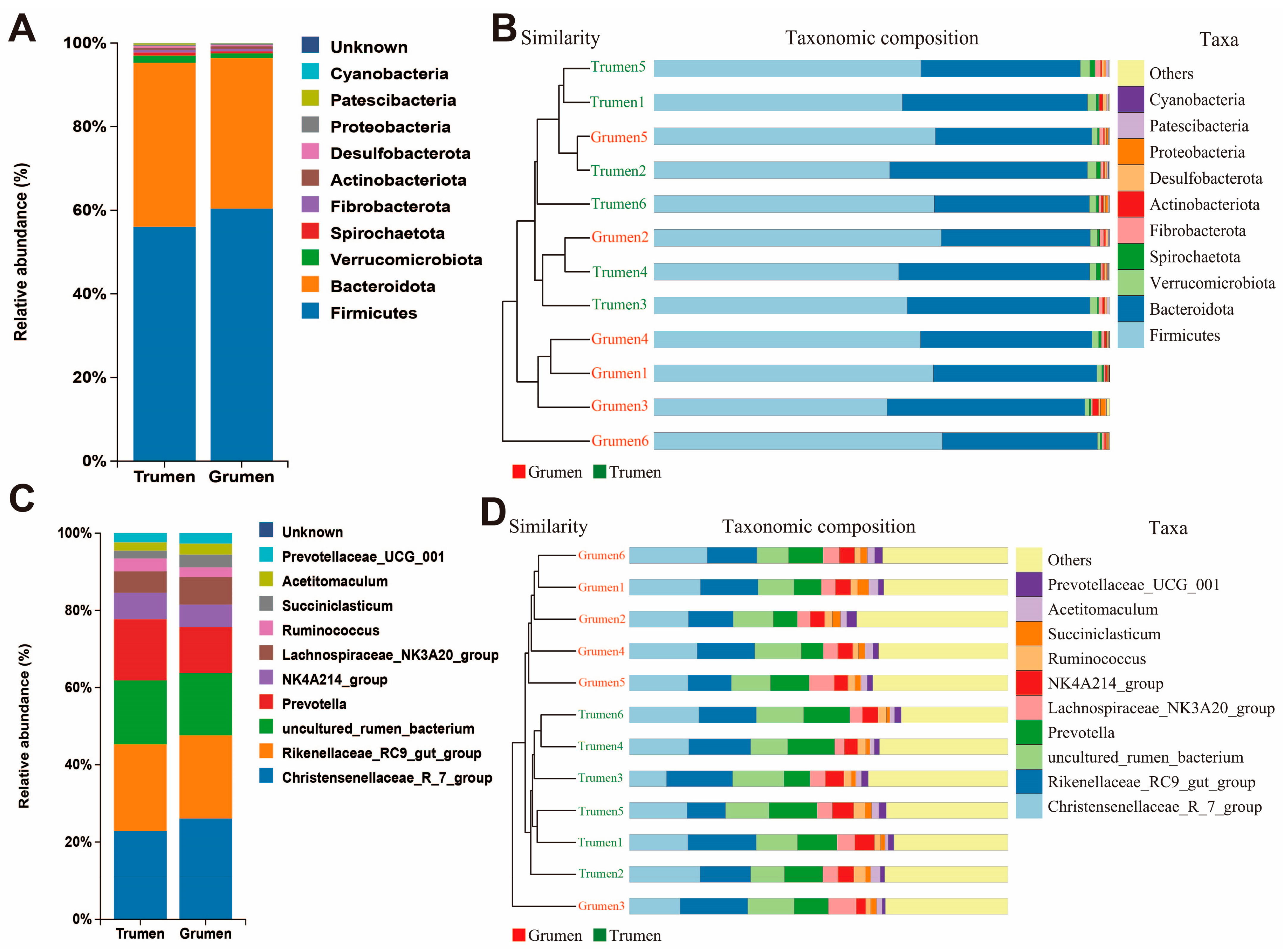
3.5. Analysis of the Difference in Species Composition of the Rumen Flora of Tianhua Mutton Sheep and Gansu Alpine Fine Wool Sheep
3.6. LEfSe Analysis of Rumen Flora in Tianhua Mutton Sheep and Gansu Alpine Fine Wool Sheep
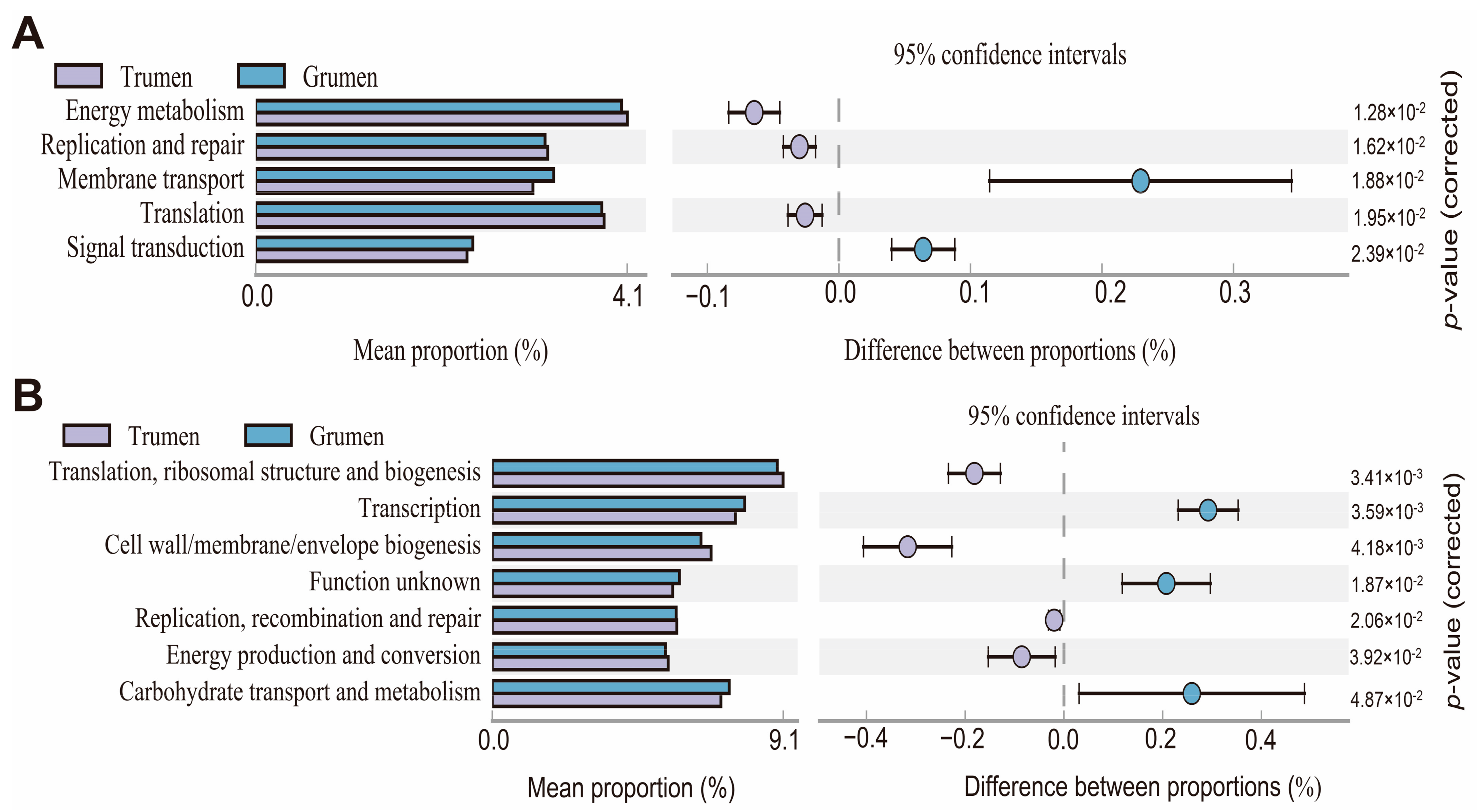
3.7. Predictive Analysis of Metabolic Pathways and Functions of Rumen Flora in Tianhua Mutton Sheep and Gansu Alpine Fine Wool Sheep
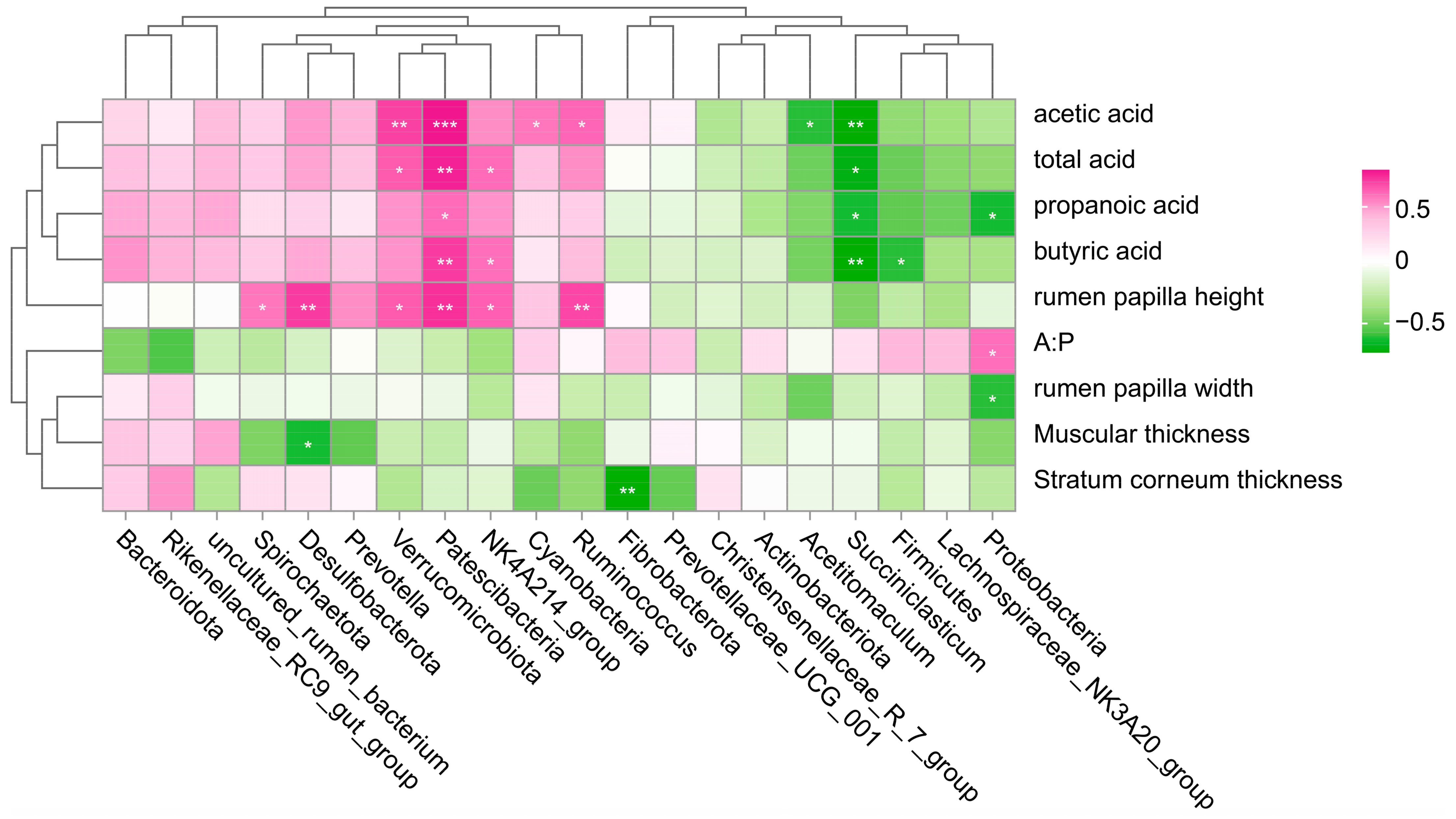
3.8. Correlation Alysis of Dominant Bacteria with Histological Morphology and VFAs Indexes
4. Discussion
4.1. Differences in Volatile Fatty Acids in the Rumen Contents of Tianhua Mutton Sheep and Gansu Alpine Fine Wool Sheep
4.2. Differences in Rumen Histomorphology between Tianhua Mutton Sheep and Gansu Alpine Fine Wool Sheep
4.3. Differences in the Rumen Flora Structure of Tianhua Mutton Sheep and Gansu Alpine Fine Wool Sheep
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sun, H.X.; He, Z.H.; Zhao, F.F.; Hu, J.; Wang, J.Q.; Liu, X.; Zhao, Z.D.; Li, M.N.; Luo, Y.Z.; Li, S.B. Hoxc13Molecular Genetic Characteristics of the Gene and Association Analysis of Wool Traits. Int. J. Mol. Sci. 2024, 25, 1594. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.Q.; Li, S.B.; Zhou, H.T.; Zhao, F.F.; Hu, J.; Wang, J.Q.; Liu, X.; Li, M.N.; Zhao, Z.D.; Hao, Z.Y.; et al. Spatiotemporal Expression and Haplotypes Identification of KRT84 Gene and Their Association with Wool Traits in Gansu Alpine Fine-Wool Sheep. Genes 2024, 15, 248. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Bai, C.; Shi, L.; He, Y.; Hu, M.Y.; Sun, H.; Peng, H.Y.; Lai, W.N.; Jiao, S.Y.; Zhao, Z.L.; et al. Detection of selection signatures in South African Mutton Merino sheep using whole-genome sequencing data. Anim. Genet. 2022, 53, 224–229. [Google Scholar] [CrossRef]
- Baldwin, R.L.; Connor, E.E. Rumen Function and Development. Vet. Clin. N. Am. Food Anim. Pract. 2017, 33, 427. [Google Scholar] [CrossRef]
- Gao, Z.; Li, Y.J.; Xu, C.; Luo, D.; Qiu, Q.H.; Pan, K.; Xiong, X.W.; Qu, M.R.; Ouyang, K.H. Niacin mitigates rumen epithelial damage in vivo by inhibiting rumen epithelial cell apoptosis on a high concentrate diet. Vet. Res. Commun. 2022, 46, 699–709. [Google Scholar] [CrossRef]
- Ma, Y.; Zhang, Y.; Zhang, H.; Wang, H.; Elmhadi, M. Thiamine Alleviates High-Concentrate-Diet-Induced Oxidative Stress, Apoptosis, and Protects the Rumen Epithelial Barrier Function in Goats. Front. Vet. Sci. 2021, 8, 663698. [Google Scholar] [CrossRef] [PubMed]
- Sha, Y.Z.; He, Y.Y.; Liu, X.; Zhao, S.G.; Hu, J.; Wang, J.Q.; Li, S.B.; Li, W.H.; Shi, B.G.; Hao, Z.Y. Rumen Epithelial Development- and Metabolism-Related Genes Regulate Their Micromorphology and VFAs Mediating Plateau Adaptability at Different Ages in Tibetan Sheep. Int. J. Mol. Sci. 2022, 23, 16078. [Google Scholar] [CrossRef]
- Liu, X.; Sha, Y.Z.; Dingkao, R.Q.; Zhang, W.; Lv, W.B.; Wei, H.; Shi, H.; Hu, J.; Wang, J.Q.; Li, S.B.; et al. Interactions Between Rumen Microbes, VFAs, and Host Genes Regulate Nutrient Absorption and Epithelial Barrier Function During Cold Season Nutritional Stress in Tibetan Sheep. Front. Microbiol. 2020, 11, 593062. [Google Scholar] [CrossRef]
- Langda, S.; Zhang, C.G.; Zhang, K.; Gui, B.; Ji, D.; Deji, C.; Cuoji, A.; Wang, X.L.; Wu, Y.J. Diversity and Composition of Rumen Bacteria, Fungi, and Protozoa in Goats and Sheep Living in the Same High-Altitude Pasture. Animals 2020, 10, 186. [Google Scholar] [CrossRef]
- Yu, C.C.; Luo, Q.J.; Chen, Y.; Liu, S.M.; Zang, C.J. Impact of docusate and fauna-free on feed intake, ruminal flora and digestive enzyme activities of sheep. J. Anim. Physiol. Anim. Nutr. 2020, 104, 1043–1051. [Google Scholar] [CrossRef]
- Cao, Y.R.; Zhu, B.Z.; Li, F.; Zhang, D.H.; Guo, T.Q.; Li, F.D.; Yang, G. The Destruction of the Anaerobic Environment Caused by Rumen Fistula Surgery Leads to Differences in the Rumen Microbial Diversity and Function of Sheep. Front. Vet. Sci. 2021, 8, 754195. [Google Scholar] [CrossRef] [PubMed]
- Shen, H.; Xu, Z.H.; Shen, Z.M.; Lu, Z.Y. The Regulation of Ruminal Short-Chain Fatty Acids on the Functions of Rumen Barriers. Front. Physiol. 2019, 10, 1305. [Google Scholar] [CrossRef] [PubMed]
- Zhen, Y.K.; Xi, Z.N.; Nasr, S.M.; He, F.Y.; Han, M.L.; Yin, J.L.; Ge, L.; Chen, Y.F.; Wang, Y.S.; Wei, W.J.; et al. Multi-Omics Reveals the Impact of Exogenous Short-Chain Fatty Acid Infusion on Rumen Homeostasis: Insights into Crosstalk between the Microbiome and the Epithelium in a Goat Model. Microbiol. Spectr. 2023, 11, e0534322. [Google Scholar] [CrossRef] [PubMed]
- Mani, S.; Aiyegoro, O.A.; Adeleke, M.A. Association between host genetics of sheep and the rumen microbial composition. Trop. Anim. Health Prod. 2022, 54, 109. [Google Scholar] [CrossRef] [PubMed]
- Stumpff, F.; Georgi, M.I.; Mundhenk, L.; Rabbani, I.; Fromm, M.; Martens, H.; Günzel, D. Sheep rumen and omasum primary cultures and source epithelia: Barrier function aligns with expression of tight junction proteins. J. Exp. Biol. 2011, 214, 2871–2872. [Google Scholar] [CrossRef] [PubMed]
- Schofield, B.J.; Lachner, N.; Le, O.T.; McNeill, D.M.; Dart, P.; Ouwerkerk, D.; Hugenholtz, P.; Klieve, A.V. Beneficial changes in rumen bacterial community profile in sheep and dairy calves as a result of feeding the probiotic Bacillus amyloliquefaciens H57. J. Appl. Microbiol. 2018, 124, 855–866. [Google Scholar] [CrossRef] [PubMed]
- Bharanidharan, R.; Lee, C.H.; Thirugnanasambantham, K.; Ibidhi, R.; Woo, Y.W.; Lee, H.G.; Kim, J.G.; Kim, K.H. Feeding Systems and Host Breeds Influence Ruminal Fermentation, Methane Production, Microbial Diversity and Metagenomic Gene Abundance. Front. Microbiol. 2021, 12, 701081. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.Y.; Sha, Y.Z.; Lv, W.B.; Pu, X.N.; Liu, X.; Luo, Y.Z.; Hu, J.; Wang, J.Q.; Li, S.B.; Zhao, Z.D. Sex differences in rumen fermentation and microbiota of Tibetan goat. Microb. Cell Fact. 2022, 21, 55. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.Y.; Ju, L.; Cheng, Q.J.; Jiang, Y.; Hou, Q.L.; Hu, Z.Y.; Wang, Y.; Wang, Z.H. Comparison of growth performance and rumen metabolic pathways in sheep and goats under the same feeding pattern. Front. Vet. Sci. 2023, 10, 1013252. [Google Scholar] [CrossRef]
- King, E.E.; Smith, R.P.; St-Pierre, B.; Wright, A.G. Differences in the rumen methanogen populations of lactating Jersey and Holstein dairy cows under the same diet regimen. Appl. Environ. Microbiol. 2011, 77, 5682–5687. [Google Scholar] [CrossRef]
- Huang, J.Q.; Li, Y.J.; Luo, Y.Z. Bacterial community in the rumen of Tibetan sheep and Gansu alpine fine-wool sheep grazing on the Qinghai-Tibetan Plateau, China. J. Gen. Appl. Microbiol. 2017, 63, 122–130. [Google Scholar] [CrossRef]
- Chang, J.J.; Yao, X.T.; Zuo, C.X.; Qi, Y.X.; Chen, D.K.; Ma, W.T. The gut bacterial diversity of sheep associated with different breeds in Qinghai province. BMC Vet. Res. 2020, 16, 254. [Google Scholar] [CrossRef]
- Wang, H.H.; Su, M.C.; Wang, C.H.; Li, D.P.; Li, Q.; Liu, Z.L.; Qi, X.C.; Wu, Y.; Zhao, Y.J.; Li, T.T.; et al. Yeast culture repairs rumen epithelial injury by regulating microbial communities and metabolites in sheep. Front. Microbiol. 2023, 14, 1305772. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Wang, Y.; Sheng, H.F.; He, Y.; Wu, J.Y.; Jiang, Y.X.; Tam, N.F.Y.; Zhou, H.W. Comparison of the levels of bacterial diversity in freshwater, intertidal wetland, and marine sediments by using millions of illumina tags. Appl. Environ. Microbiol. 2012, 78, 8264–8271. [Google Scholar] [CrossRef]
- Gower, J.C. Some Distance Properties of Latent Root and Vector Methods Used in Multivariate Analysis. Biometrika 1966, 53, 325–338. [Google Scholar] [CrossRef]
- Looft, T.; Johnson, T.A.; Allen, H.K.; Bayles, D.O.; Alt, D.P.; Stedtfeld, R.D.; Sul, W.J.; Stedtfeld, T.M.; Chai, B.; Cole, J.R.; et al. In-feed antibiotic effects on the swine intestinal microbiome. Proc. Natl. Acad. Sci. USA 2012, 109, 1691–1696. [Google Scholar] [CrossRef]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef] [PubMed]
- Parks1, D.H.; Tyson, G.W.; Hugenholtz, P.; Beiko, R.G. STAMP: Statistical analysis of taxonomic and functional profiles. Bioinformatics 2014, 30, 3123–3124. [Google Scholar] [CrossRef] [PubMed]
- Dixit, S.; Kumar, S.; Sharma, R.; Banakar, P.S.; Singh, M.; Keshri, A.; Tyagi, A.K. Rumen multi-omics addressing diet-host-microbiome interplay in farm animals: A review. Anim. Biotechnol. 2023, 34, 3187–3205. [Google Scholar] [CrossRef]
- Li, C.; Wang, G.X.; Zhang, Q.; Huang, Y.; Li, F.D.; Wang, W.M. Developmental changes of nutrient digestion in young lambs are influenced by weaning and associated with intestinal microbiota. Anim. Biotechnol. 2023, 34, 1362–1376. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.C.; Wang, X.; Li, J.Z.; Huang, P.F.; Li, Y.L.; Ding, X.Q.; Huang, J.; Zhu, M.Z.; Yin, J.; Dai, C.P.; et al. The impact of lactating Hu sheep’s dietary protein levels on lactation performance, progeny growth and rumen development. Anim. Biotechnol. 2023, 34, 1919–1930. [Google Scholar] [CrossRef] [PubMed]
- Qianrige, R.S.; Kim, D.H.; Shishido, T.; Ogura, S.I. Effects of Rumen Fermentation Characteristics on Stress-Related Hormones and Behavior in Sheep. Animals 2023, 13, 3701. [Google Scholar] [CrossRef] [PubMed]
- Rosenboom, R.W. Rumen Transition from Weaning to 400 Pounds. Vet. Clin. N. Am. Food Anim. Pract. 2022, 38, 153–164. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.H.; Li, H.W.; Zhu, W.Y.; Mao, S.Y. Dynamic changes in rumen fermentation and bacterial community following rumen fluid transplantation in a sheep model of rumen acidosis: Implications for rumen health in ruminants. FASEB J. 2019, 33, 8453–8467. [Google Scholar] [CrossRef] [PubMed]
- Ørskov, E.R.; Grubb, D.A.; Smith, J.S.; Webster, A.J.F.; Corrigall, W. Efficiency of utilization of volatile fatty acids for maintenance and energy retention by sheep. Br. J. Nutr. 1979, 41, 541–551. [Google Scholar] [CrossRef]
- Wang, L.J.; Zhang, G.N.; Li, Y.; Zhang, Y.G. Effects of High Forage/Concentrate Diet on Volatile Fatty Acid Production and the Microorganisms Involved in VFA Production in Cow Rumen. Animals 2020, 10, 223. [Google Scholar] [CrossRef] [PubMed]
- Na, S.W.; Guan, L.L. Understanding the role of rumen epithelial host-microbe interactions in cattle feed efficiency. Anim. Nutr. 2022, 10, 41–53. [Google Scholar] [CrossRef]
- Herath, H.; Pain, S.J.; Kenyon, P.R.; Blair, H.T.; Morel, P.C.H. Rumen Development of Artificially-Reared Lambs Exposed to Three Different Rearing Regimens. Animals 2021, 11, 3606. [Google Scholar] [CrossRef]
- Leite, E.R.; Conde, J.A.M.; Fonseca, C.M.B.; de Almeida, M.H.M.; de Carvalho, M.A.M.; de Melo, W.G.G.; Teodoro, A.; de Jesus, S.D.; da Silva Oliveira, L.S.; Ferraz, J.C.B.; et al. Impact of feeding native Caatinga pasture on the rumen histomorphometry of sheep raised in semi-extensive management. Anat. Histol. Embryol. 2024, 53, e13029. [Google Scholar] [CrossRef]
- Cheng, J.B.; Zhang, X.X.; Xu, D.; Zhang, D.Y.; Zhang, Y.K.; Song, Q.Z.; Li, X.L.; Zhao, Y.; Zhao, L.M.; Li, W.X.; et al. Relationship between rumen microbial differences and traits among Hu sheep, Tan sheep, and Dorper sheep. J. Anim. Sci. 2022, 100, skac261. [Google Scholar] [CrossRef] [PubMed]
- Cholewińska, P.; Wołoszyńska, M.; Michalak, M.; Czyż, K.; Rant, W.; Janczak, M. Evaluation of Changes in the Levels of Firmicutes and Bacteroidetes Phyla of Sheep Feces Depending on the Breed. Animals 2020, 10, 1901. [Google Scholar] [CrossRef] [PubMed]
- Szeligowska, N.; Cholewińska, P.; Czyż, K.; Wojnarowski, K.; Janczak, M. Inter and intraspecies comparison of the level of selected bacterial phyla in in cattle and sheep based on feces. BMC Vet. Res. 2021, 17, 224. [Google Scholar] [CrossRef] [PubMed]
- Fan, Q.S.; Cui, X.X.; Wang, Z.F.; Chang, S.H.; Wanapat, M.; Yan, T.H.; Hou, F.J. Rumen Microbiota of Tibetan Sheep (Ovis aries) Adaptation to Extremely Cold Season on the Qinghai-Tibetan Plateau. Front. Vet. Sci. 2021, 8, 673822. [Google Scholar] [CrossRef] [PubMed]
- Cholewińska, P.; Szeligowska, N.; Wojnarowski, K.; Nazar, P.; Greguła-Kania, M.; Junkuszew, A.; Rant, W.; Radzik-Rant, A.; Marcinkowska, A.; Bodkowski, R. Selected bacteria in sheep stool depending on breed and physiology state. Sci. Rep. 2023, 13, 11739. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.R.; Xie, W.; Zhou, S.D.; Ma, N.N.; Wang, Y.; Huang, J.; Shen, X.Z.; Chang, G.J. A high-concentrate diet induces colonic inflammation and barrier damage in Hu sheep. J. Dairy Sci. 2023, 106, 9644–9662. [Google Scholar] [CrossRef] [PubMed]
- Schirmer, M.; Garner, A.; Vlamakis, H.; Xavier, R.J. Microbial genes and pathways in inflammatory bowel disease. Nat. Rev. Microbiol. 2019, 17, 497–511. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.L.; Ma, H.L.; Huang, Y.T.; Li, J.C.; Li, W.D. Ruminococcaceae_UCG-013 Promotes Obesity Resistance in Mice. Biomedicines 2022, 10, 3272. [Google Scholar] [CrossRef]
- Sinha, A.K.; Løbner-Olesen, A.; Riber, L. Bacterial Chromosome Replication and DNA Repair During the Stringent Response. Front. Microbiol. 2020, 11, 582113. [Google Scholar] [CrossRef]


| Items | Tianhua Mutton Sheep | Gansu Alpine Fine Wool Sheep | p-Value |
|---|---|---|---|
| acetic acid, mmol/L | 43.47 ± 1.51 a | 36.38 ± 1.55 b | 0.010 |
| Propanoic acid, mmol/L | 15.74 ± 1.55 | 11.85 ± 2.35 | 0.123 |
| Butyric acid, mmol/L | 14.88 ± 0.35 | 12.07 ± 2.22 | 0.151 |
| Total acid 1, mmol/L | 84.53 ± 3.82 a | 71.56 ± 5.03 b | 0.044 |
| A:P 2 | 2.74 ± 0.20 | 3.17 ± 0.51 | 0.329 |
| Items | Tianhua Mutton Sheep | Gansu Alpine Fine Wool Sheep | p-Value |
|---|---|---|---|
| Rumen papilla height, μm | 1715.17 ± 43.28 a | 1279.80 ± 144.79 b | 0.015 |
| Rumen papilla width, μm | 522.97 ± 145.39 | 462.03 ± 39.98 | 0.598 |
| Stratum corneum thickness, μm | 91.53 ± 15.80 | 84.17 ± 5.25 | 0.565 |
| Muscular thickness, μm | 1246.00 ± 180.92 | 1240.23 ± 11.95 | 0.966 |
| Items | Tianhua Mutton Sheep | Gansu Alpine Fine Wool Sheep | p-Value |
|---|---|---|---|
| Feature | 4134.00 ± 234.11 a | 3094.17 ± 443.57 b | 0.001 |
| ACE | 5754.15 ± 286.24 a | 4788.52 ± 431.60 b | 0.002 |
| Chao1 | 5858.27 ± 318.77 a | 4578.07 ± 583.13 b | 0.002 |
| Simpson | 1.00 ± 0.01 | 1.00 ± 0.01 | 0.141 |
| Shannon | 10.50 ± 0.17 a | 9.94 ± 0.40 b | 0.016 |
| PD_whole_tree | 230.18 ± 8.12 a | 185.54 ± 14.68 b | 0.001 |
| Coverage | 0.95 ± 0.01 | 0.94 ± 0.01 | 0.279 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, D.; Liu, Z.; Duan, X.; Wang, C.; Chen, Z.; Zhang, M.; Li, X.; Ma, Y. Rumen Development of Tianhua Mutton Sheep Was Better than That of Gansu Alpine Fine Wool Sheep under Grazing Conditions. Animals 2024, 14, 1259. https://doi.org/10.3390/ani14091259
Li D, Liu Z, Duan X, Wang C, Chen Z, Zhang M, Li X, Ma Y. Rumen Development of Tianhua Mutton Sheep Was Better than That of Gansu Alpine Fine Wool Sheep under Grazing Conditions. Animals. 2024; 14(9):1259. https://doi.org/10.3390/ani14091259
Chicago/Turabian StyleLi, Dengpan, Zhanjing Liu, Xinming Duan, Chunhui Wang, Zengping Chen, Muyang Zhang, Xujie Li, and Youji Ma. 2024. "Rumen Development of Tianhua Mutton Sheep Was Better than That of Gansu Alpine Fine Wool Sheep under Grazing Conditions" Animals 14, no. 9: 1259. https://doi.org/10.3390/ani14091259





