Needle Segmentation in Volumetric Optical Coherence Tomography Images for Ophthalmic Microsurgery
Abstract
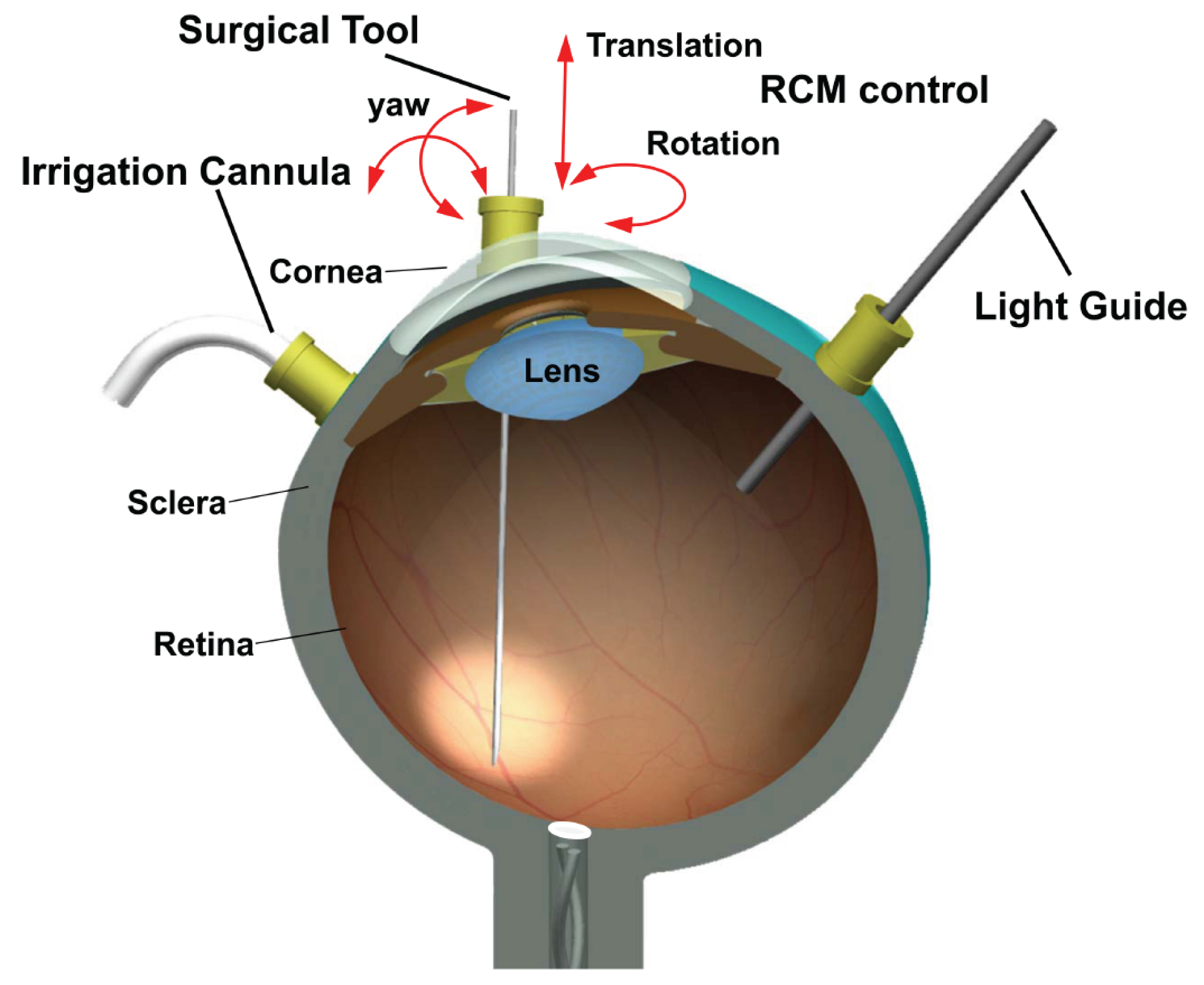
:1. Introduction
2. Method
2.1. Morphological Features Based Method
2.2. Full Convolution Neural Networks Based Method
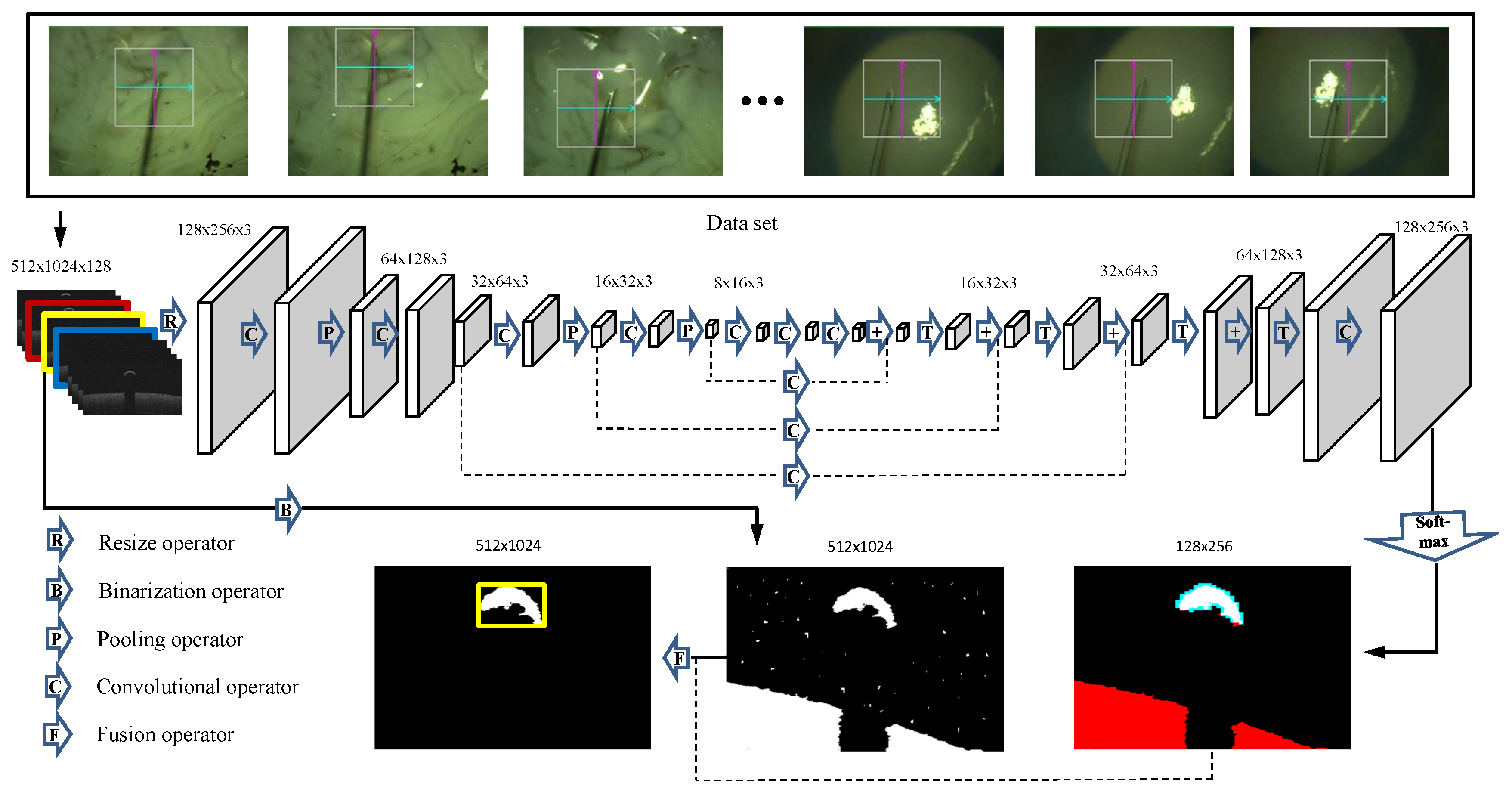
2.2.1. Network Description
2.2.2. Training
3. Experiments and Results
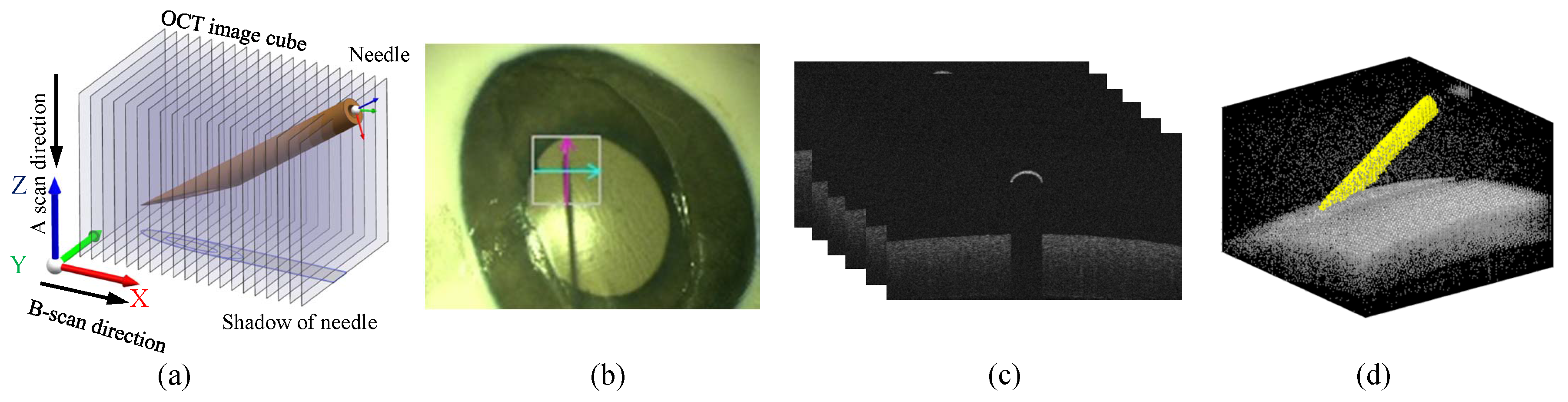
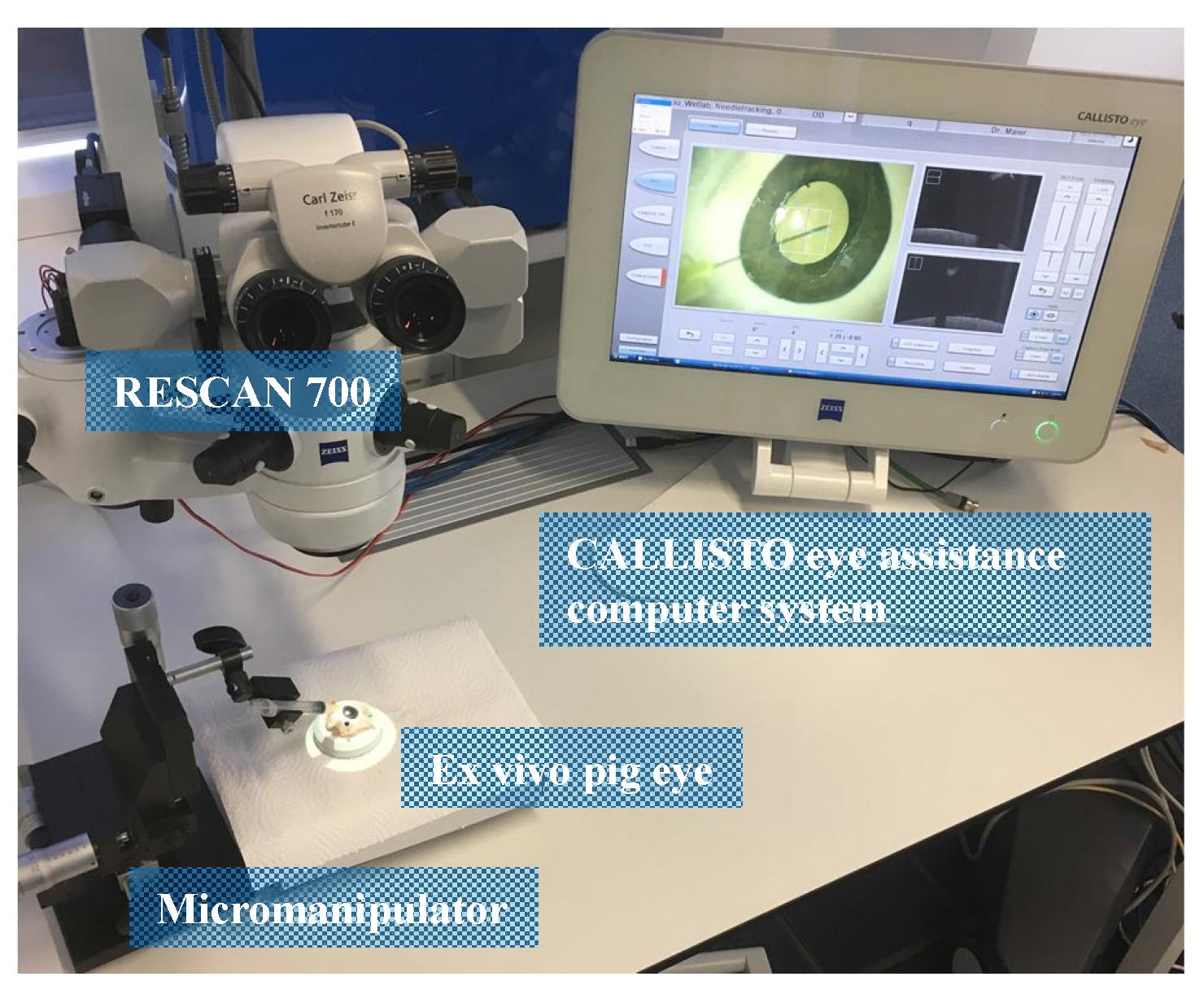
3.1. Experimental Setup and Evaluation Metrics
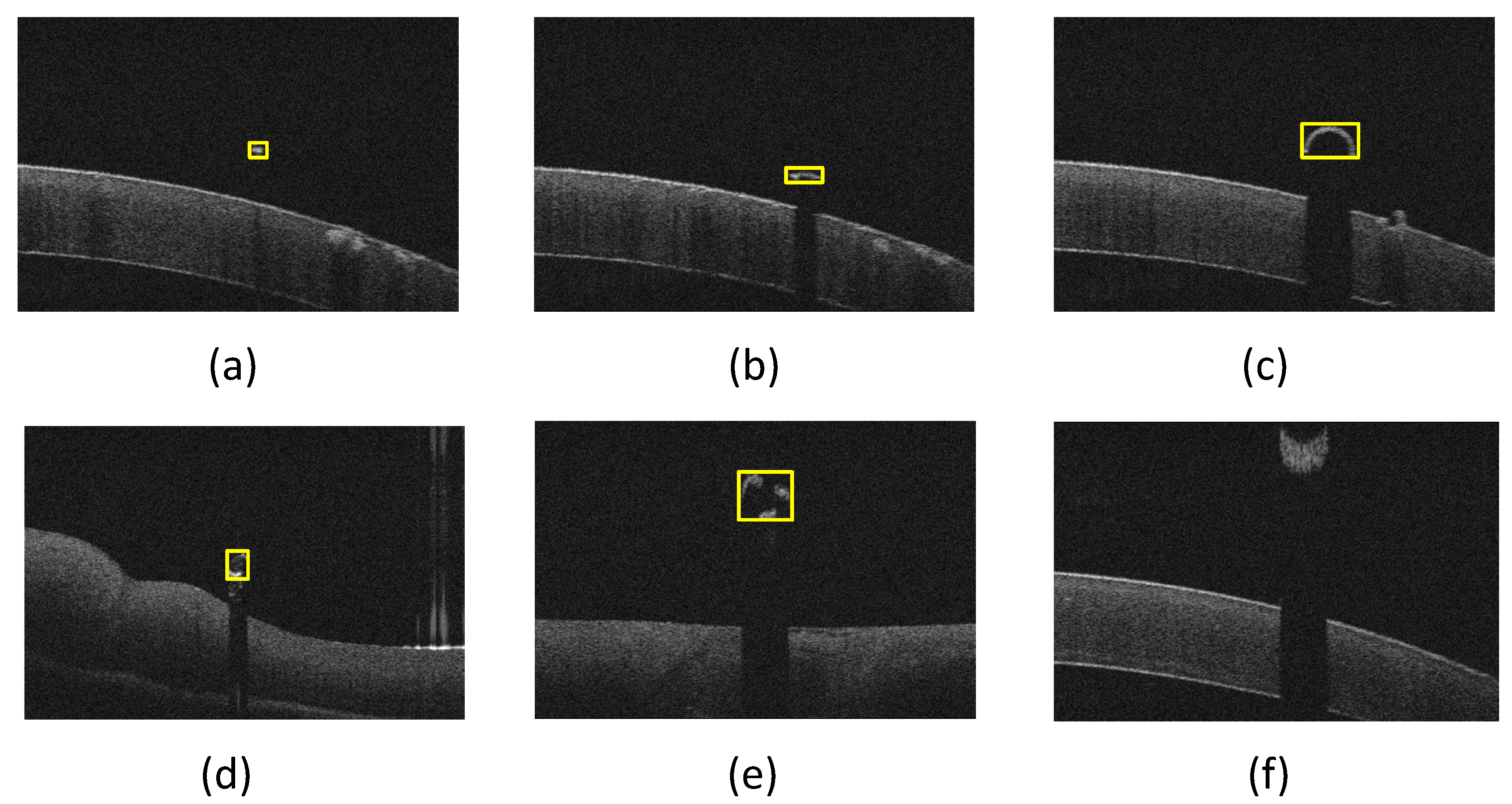
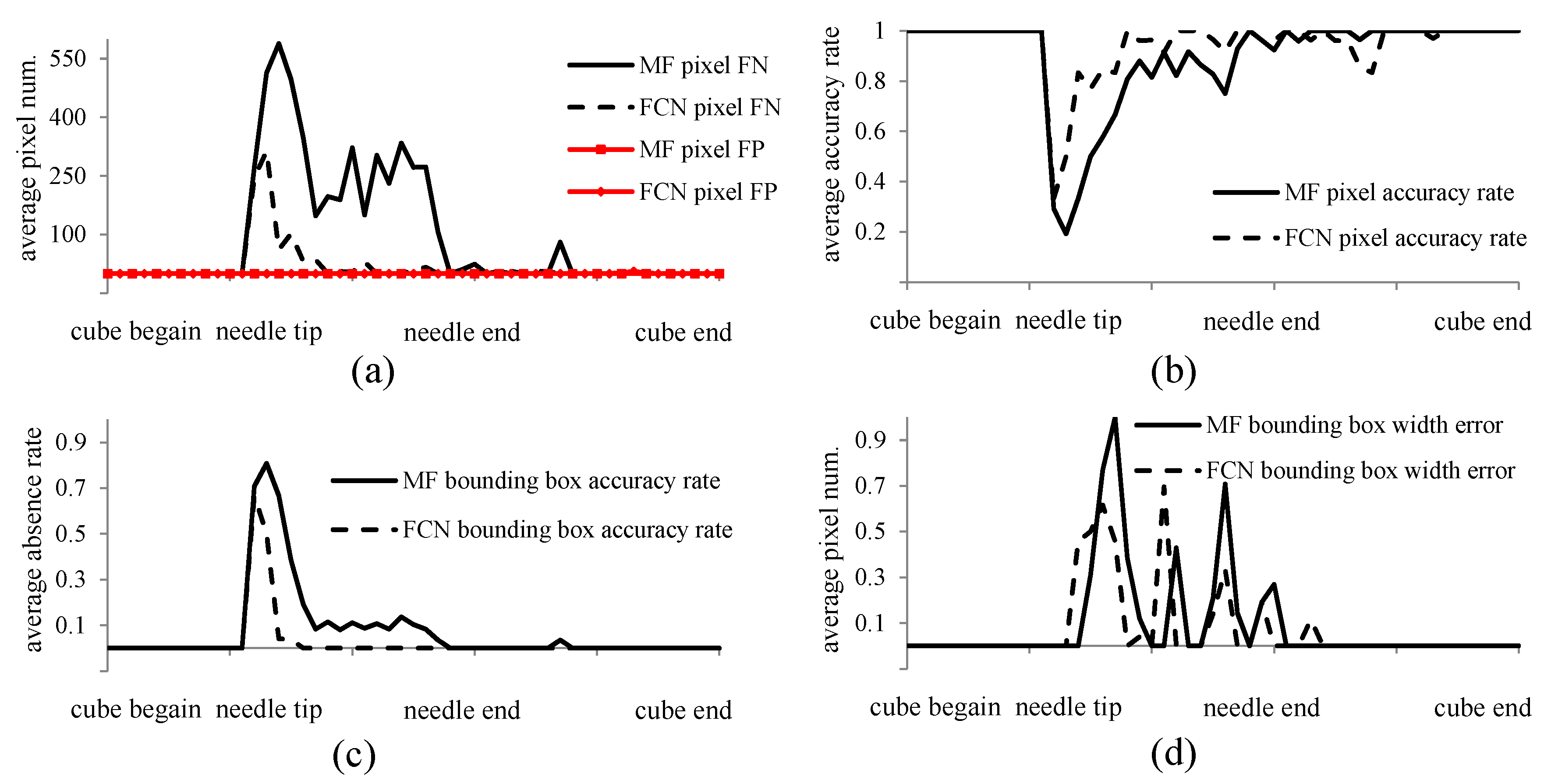
3.2. Results
4. Discussion
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- World Health Organization (WHO). Towards Universal Eye Health: A Global Action Plan 2014 to 2019 Report; WHO: Geneva, Switzerland, 2013. [Google Scholar]
- Rizzo, S.; Patelli, F.; Chow, D. Vitreo-Retinal Surgery: Progress III; Springer: New York, NY, USA, 2008. [Google Scholar]
- Nakano, T.; Sugita, N.; Ueta, T.; Tamaki, Y.; Mitsuishi, M. A parallel robot to assist vitreoretinal surgery. Int. J. Comput. Assist. Radiol. Surg. 2009, 4, 517–526. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.; Goldman, R.; Simaan, N.; Fine, H.; Chang, S. Design and theoretical evaluation of micro-surgical manipulators for orbital manipulation and intraocular dexterity. In Proceedings of the 2007 IEEE International Conference on Robotics and Automation, Roma, Italy, 10–14 April 2007; pp. 3389–3395. [Google Scholar]
- Bynoe, L.A.; Hutchins, R.K.; Lazarus, H.S.; Friedberg, M.A. Retinal endovascular surgery for central retinal vein occlusion: Initial experience of four surgeons. Retina 2005, 25, 625–632. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Chen, C.; Huang, X.; Huang, J. Instrument tracking via online learning in retinal microsurgery. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Boston, MA, USA, 14–18 September 2014; Springer: Cham, Switzerland, 2014; pp. 464–471. [Google Scholar]
- Rieke, N.; Tan, D.J.; Alsheakhali, M.; Tombari, F.; di San Filippo, C.A.; Belagiannis, V.; Eslami, A.; Navab, N. Surgical tool tracking and pose estimation in retinal microsurgery. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Munich, Germany, 5–9 October 2015; Springer: Cham, Switzerland, 2015; pp. 266–273. [Google Scholar]
- Sznitman, R.; Ali, K.; Richa, R.; Taylor, R.H.; Hager, G.D.; Fua, P. Data-driven visual tracking in retinal microsurgery. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Nice, France, 1–5 October 2012; Springer: Berlin, Germany; New York, NY, USA, 2012; pp. 568–575. [Google Scholar]
- Kwoh, Y.S.; Hou, J.; Jonckheere, E.A.; Hayati, S. A robot with improved absolute positioning accuracy for CT guided stereotactic brain surgery. IEEE Trans. Biomed. Eng. 1988, 35, 153–160. [Google Scholar] [CrossRef] [PubMed]
- McDannold, N.; Clement, G.; Black, P.; Jolesz, F.; Hynynen, K. Transcranial MRI-guided focused ultrasound surgery of brain tumors: Initial findings in three patients. Neurosurgery 2010, 66, 323. [Google Scholar] [CrossRef] [PubMed]
- McVeigh, E.R.; Guttman, M.A.; Lederman, R.J.; Li, M.; Kocaturk, O.; Hunt, T.; Kozlov, S.; Horvath, K.A. Real-time interactive MRI-guided cardiac surgery: Aortic valve replacement using a direct apical approach. Magn. Reson. Med. 2006, 56, 958–964. [Google Scholar] [CrossRef] [PubMed]
- Vrooijink, G.J.; Abayazid, M.; Misra, S. Real-time three-dimensional flexible needle tracking using two-dimensional ultrasound. In Proceedings of the 2013 IEEE International Conference on Robotics and Automation (ICRA), Karlsruhe, Germany, 6–10 May 2013; pp. 1688–1693. [Google Scholar]
- Yasui, K.; Kanazawa, S.; Sano, Y.; Fujiwara, T.; Kagawa, S.; Mimura, H.; Dendo, S.; Mukai, T.; Fujiwara, H.; Iguchi, T.; et al. Thoracic Tumors Treated with CT-guided Radiofrequency Ablation: Initial Experience 1. Radiology 2004, 231, 850–857. [Google Scholar] [CrossRef] [PubMed]
- Lam, T.T.; Miller, P.; Howard, S.; Nork, T.M. Validation of a Rabbit Model of Choroidal Neovascularization Induced by a Subretinal Injection of FGF-LPS. Investig. Ophthalmol. Vis. Sci. 2014, 55, 1204. [Google Scholar]
- Huang, D.; Swanson, E.A.; Lin, C.P.; Schuman, J.S.; Stinson, W.G.; Chang, W.; Hee, M.R.; Flotte, T.; Gregory, K.; Puliafito, C.A.; et al. Optical coherence tomography. Science 1991, 254, 1178. [Google Scholar] [CrossRef]
- Ehlers, J.P.; Srivastava, S.K.; Feiler, D.; Noonan, A.I.; Rollins, A.M.; Tao, Y.K. Integrative advances for OCT-guided ophthalmic surgery and intraoperative OCT: Microscope integration, surgical instrumentation, and heads-up display surgeon feedback. PLoS One 2014, 9, e105224. [Google Scholar] [CrossRef] [PubMed]
- Ehlers, J.P.; Tao, Y.K.; Srivastava, S.K. The Value of Intraoperative OCT Imaging in Vitreoretinal Surgery. Curr. Opin. Ophthalmol. 2014, 25, 221. [Google Scholar] [CrossRef] [PubMed]
- Adebar, T.K.; Fletcher, A.E.; Okamura, A.M. 3-D ultrasound-guided robotic needle steering in biological tissue. IEEE Trans. Biomed. Eng. 2014, 61, 2899–2910. [Google Scholar] [CrossRef] [PubMed]
- Viehland, C.; Keller, B.; Carrasco-Zevallos, O.M.; Nankivil, D.; Shen, L.; Mangalesh, S.; Viet, D.T.; Kuo, A.N.; Toth, C.A.; Izatt, J.A. Enhanced volumetric visualization for real time 4D intraoperative ophthalmic swept-source OCT. Biomed. Opt. Express 2016, 7, 1815–1829. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Kang, J.U. Real-time 4D signal processing and visualization using graphics processing unit on a regular nonlinear-k Fourier-domain OCT system. Opt. Express 2010, 18, 11772–11784. [Google Scholar] [CrossRef] [PubMed]
- El-Haddad, M.T.; Ehlers, J.P.; Srivastava, S.K.; Tao, Y.K. Automated real-time instrument tracking for microscope-integrated intraoperative OCT imaging of ophthalmic surgical maneuvers. Proceedings of SPIE BiOS. International Society for Optics and Photonics, San Francisco, CA, USA, 7 February 2015; p. 930707. [Google Scholar]
- Roodaki, H.; Filippatos, K.; Eslami, A.; Navab, N. Introducing augmented reality to optical coherence tomography in ophthalmic microsurgery. In Proceedings of the 2015 IEEE International Symposium on Mixed and Augmented Reality (ISMAR), Fukuoka, Japan, 29 September–3 October 2015; pp. 1–6. [Google Scholar]
- Long, J.; Shelhamer, E.; Darrell, T. Fully convolutional networks for semantic segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 3431–3440. [Google Scholar]
- Ahlers, C.; Simader, C.; Geitzenauer, W.; Stock, G.; Stetson, P.; Dastmalchi, S.; Schmidt-Erfurth, U. Automatic segmentation in three-dimensional analysis of fibrovascular pigmentepithelial detachment using high-definition optical coherence tomography. Br. J. Ophthalmol. 2008, 92, 197–203. [Google Scholar] [CrossRef] [PubMed]
- Milletari, F.; Navab, N.; Ahmadi, S.A. V-net: Fully convolutional neural networks for volumetric medical image segmentation. In Proceedings of the 2016 Fourth International Conference on 3D Vision (3DV), Stanford, CA, USA, 25–28 October 2016; pp. 565–571. [Google Scholar]
- Jia, Y.; Shelhamer, E.; Donahue, J.; Karayev, S.; Long, J.; Girshick, R.; Guadarrama, S.; Darrell, T. Caffe: Convolutional Architecture for Fast Feature Embedding. arXiv, 2014; arXiv:1408.5093. [Google Scholar]
| Stage | Mean (ms) | Variance (ms) |
|---|---|---|
| Loading | 0.71 | 0.10 |
| Filtering | 7.87 | 2.22 |
| Detection | 17.02 | 20.55 |
| Total | 25.6 | 22.6 |
| Stage | Mean (ms) | Variance (ms) |
|---|---|---|
| Loading | 0.72 | 0.11 |
| CNN | 97.46 | 4.67 |
| Fusion | 6.63 | 0.38 |
| Total | 121.83 | 4.91 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, M.; Roodaki, H.; Eslami, A.; Chen, G.; Huang, K.; Maier, M.; Lohmann, C.P.; Knoll, A.; Nasseri, M.A. Needle Segmentation in Volumetric Optical Coherence Tomography Images for Ophthalmic Microsurgery. Appl. Sci. 2017, 7, 748. https://doi.org/10.3390/app7080748
Zhou M, Roodaki H, Eslami A, Chen G, Huang K, Maier M, Lohmann CP, Knoll A, Nasseri MA. Needle Segmentation in Volumetric Optical Coherence Tomography Images for Ophthalmic Microsurgery. Applied Sciences. 2017; 7(8):748. https://doi.org/10.3390/app7080748
Chicago/Turabian StyleZhou, Mingchuan, Hessam Roodaki, Abouzar Eslami, Guang Chen, Kai Huang, Mathias Maier, Chris P. Lohmann, Alois Knoll, and Mohammad Ali Nasseri. 2017. "Needle Segmentation in Volumetric Optical Coherence Tomography Images for Ophthalmic Microsurgery" Applied Sciences 7, no. 8: 748. https://doi.org/10.3390/app7080748





