Microfluidics-Driven Fabrication of a Low Cost and Ultrasensitive SERS-Based Paper Biosensor
Abstract
:Featured Application
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Synthesis of Gold Nanostars
2.3. Synthesis of Au@Ag Nanorods
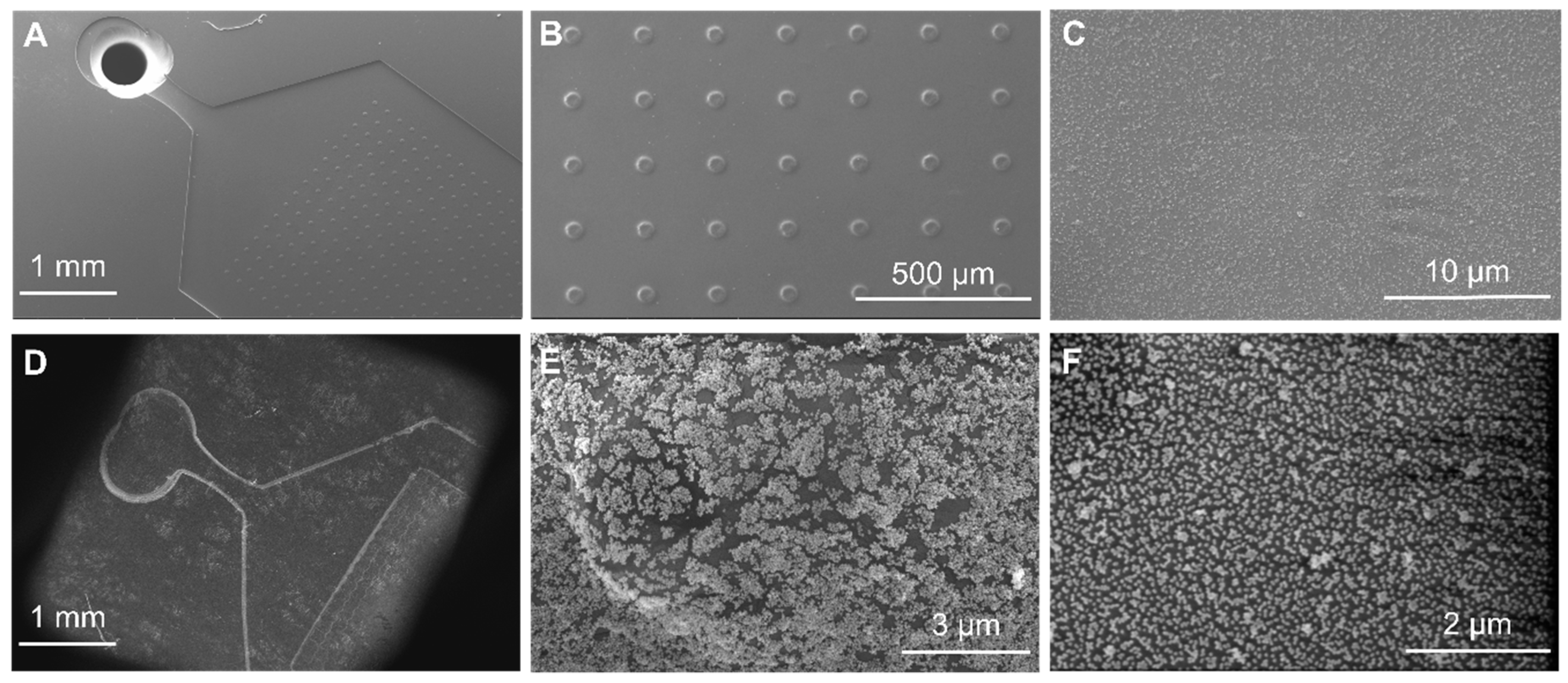
2.4. Optical and Morphological Characterization of the NPs
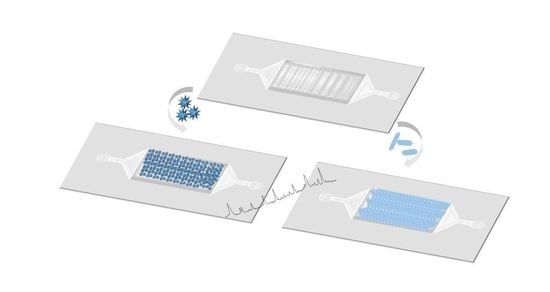
2.5. PDMS/Paper Device Fabrication
2.6. Assembly of NPs on PDMS-Paper Substrates
2.7. SERS Characterization
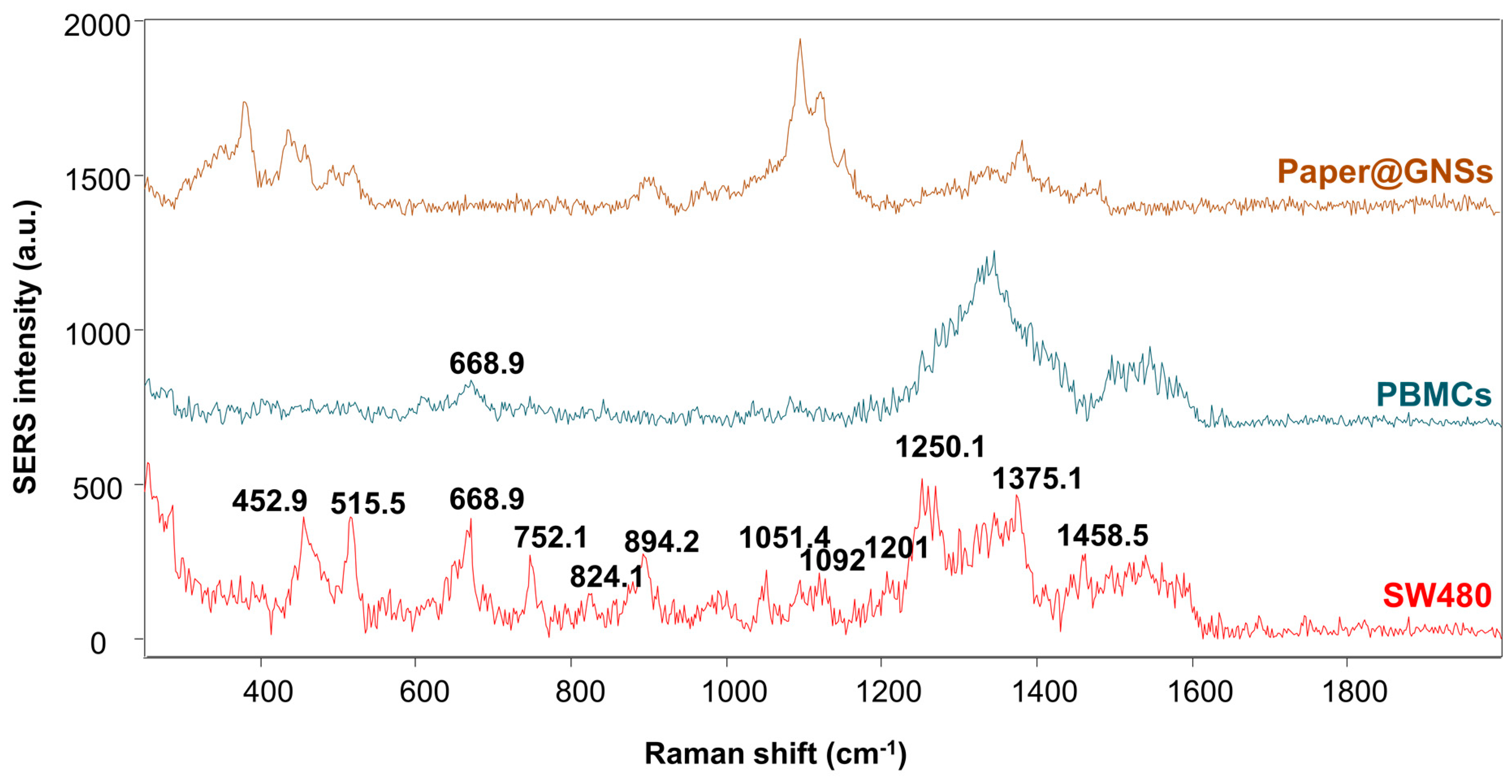
2.8. Cell Culture, Sample Preparation, and SERS Analysis
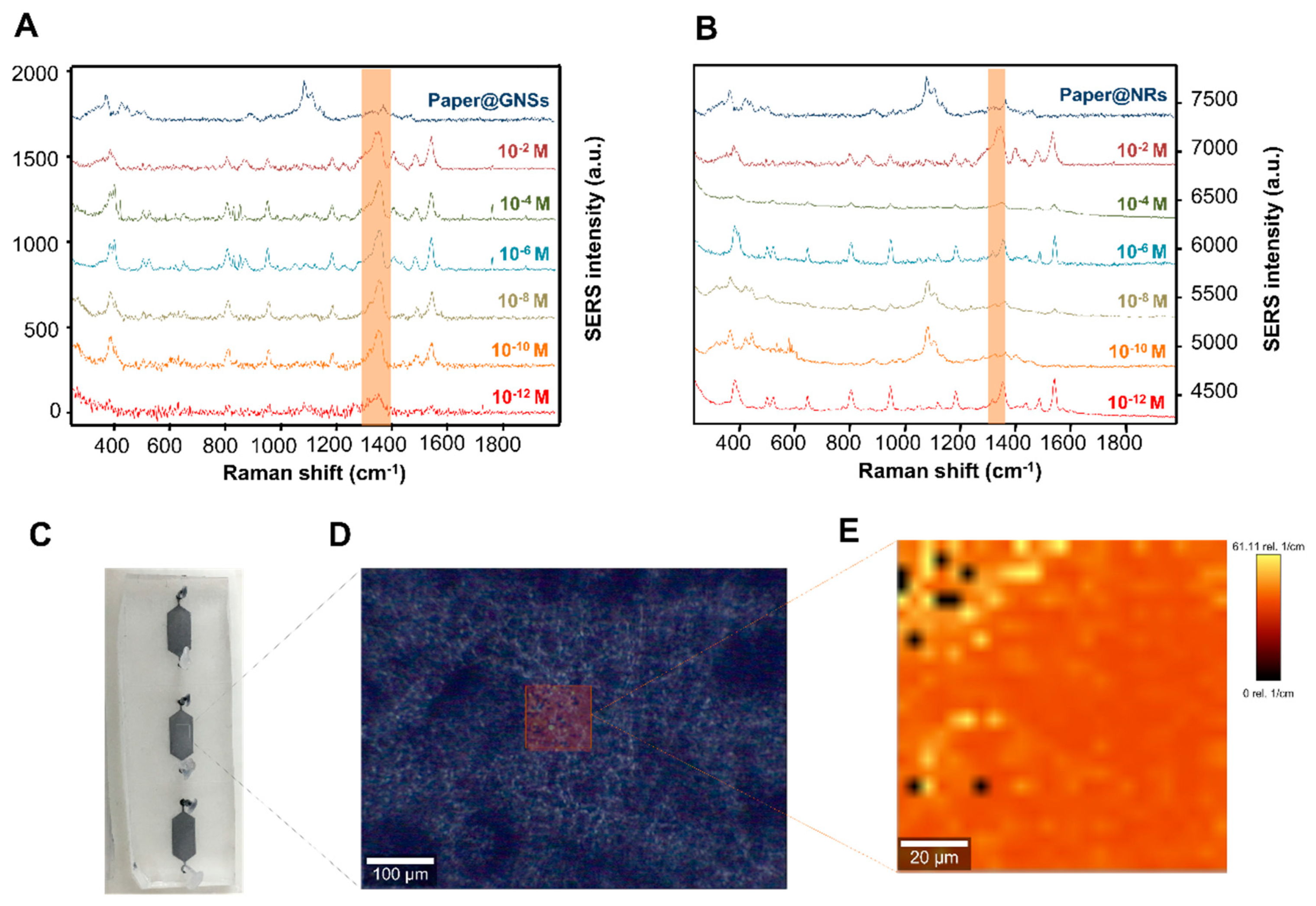
3. Results and Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Kant, K.; Abalde-Cela, S. Surface-Enhanced Raman Scattering Spectroscopy and Microfluidics: Towards Ultrasensitive Label-Free Sensing. Biosensors 2018, 8, 62. [Google Scholar] [CrossRef] [PubMed]
- Aroca, R. Surface-Enhanced Vibrational Spectroscopy; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2006; ISBN 978-0-471-60731-1. [Google Scholar]
- Kneipp, K.; Wang, Y.; Kneipp, H.; Perelman, L.T.; Itzkan, I.; Dasari, R.R.; Feld, M.S. Single molecule detection using surface-enhanced raman scattering (SERS). Phys. Rev. Lett. 1997, 78, 1667–1670. [Google Scholar] [CrossRef]
- Li, A.; Tang, L.; Song, D.; Song, S.; Ma, W.; Xu, L.; Kuang, H.; Wu, X.; Liu, L.; Chen, X.; Xu, C. A SERS-active sensor based on heterogeneous gold nanostar core–silver nanoparticle satellite assemblies for ultrasensitive detection of aflatoxinB1. Nanoscale 2016, 8, 1873–1878. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Yin, H.; Ma, W.; Kuang, H.; Wang, L.; Xu, C. Ultrasensitive SERS detection of mercury based on the assembled gold nanochains. Biosens. Bioelectron. 2015, 67, 472–476. [Google Scholar] [CrossRef]
- Etchegoin, P.; Maher, R.; Cohen, L.; Hartigan, H.; Brown, R.J.; Milton, M.J.; Gallop, J. New limits in ultrasensitive trace detection by surface enhanced Raman scattering (SERS). Chem. Phys. Lett. 2003, 375, 84–90. [Google Scholar] [CrossRef]
- Li, W.; Zhao, X.; Yi, Z.; Glushenkov, A.M.; Kong, L. Plasmonic substrates for surface enhanced Raman scattering. Anal. Chim. Acta 2017, 984, 19–41. [Google Scholar] [CrossRef]
- Abalde-Cela, S.; Carregal-Romero, S.; Coelho, J.P.; Guerrero-Martínez, A. Recent progress on colloidal metal nanoparticles as signal enhancers in nanosensing. Adv. Colloid Interface Sci. 2016, 233, 255–270. [Google Scholar] [CrossRef] [PubMed]
- Hamon, C.; Liz-Marzán, L.M. Colloidal design of plasmonic sensors based on surface enhanced Raman scattering. J. Colloid Interface Sci. 2018, 512, 834–843. [Google Scholar] [CrossRef] [PubMed]
- LeRu, E.C.; Etchegoin, P. Principles of Surface-Enhanced Raman Spectroscopy; Elsevier: Amsterdam, The Netherlands, 2009; ISBN 9780444527790. [Google Scholar]
- Schlücker, S. Surface-Enhanced Raman Spectroscopy: Concepts and Chemical Applications. Angew. Chemie Int. Ed. 2014, 53, 4756–4795. [Google Scholar] [CrossRef] [PubMed]
- Kim, N.H.; Hwang, W.; Baek, K.; Rohman, M.R.; Kim, J.; Kim, H.W.; Mun, J.; Lee, S.Y.; Yun, G.; Murray, J.; et al. Smart SERS Hot Spots: Single Molecules Can Be Positioned in a Plasmonic Nanojunction Using Host–Guest Chemistry. J. Am. Chem. Soc. 2018, 140, 4705–4711. [Google Scholar] [CrossRef]
- Chen, A.; DePrince, A.E.; Demortière, A.; Joshi-Imre, A.; Shevchenko, E.V.; Gray, S.K.; Welp, U.; Vlasko-Vlasov, V.K. Self-Assembled Large Au Nanoparticle Arrays with Regular Hot Spots for SERS. Small 2011, 7, 2365–2371. [Google Scholar] [CrossRef]
- Sugawa, K.; Akiyama, T.; Tanoue, Y.; Harumoto, T.; Yanagida, S.; Yasumori, A.; Tomita, S.; Otsuki, J. Particle size dependence of the surface-enhanced Raman scattering properties of densely arranged two-dimensional assemblies of Au(core)–Ag(shell) nanospheres. Phys. Chem. Chem. Phys. 2015, 17, 21182–21189. [Google Scholar] [CrossRef]
- Wustholz, K.L.; Henry, A.-I.; McMahon, J.M.; Freeman, R.G.; Valley, N.; Piotti, M.E.; Natan, M.J.; Schatz, G.C.; Van Duyne, R.P. Structure−Activity Relationships in Gold Nanoparticle Dimers and Trimers for Surface-Enhanced Raman Spectroscopy. J. Am. Chem. Soc. 2010, 132, 10903–10910. [Google Scholar] [CrossRef] [PubMed]
- Tanoue, Y.; Sugawa, K.; Yamamuro, T.; Akiyama, T. Densely arranged two-dimensional silver nanoparticle assemblies with optical uniformity over vast areas as excellent surface-enhanced Raman scattering substrates. Phys. Chem. Chem. Phys. 2013, 15, 15802–15805. [Google Scholar] [CrossRef] [PubMed]
- Reguera, J.; Langer, J.; Jimé Nez De Aberasturi, D.; Liz-Marzá, L.M. Anisotropic metal nanoparticles for surface enhanced Raman scattering. This J. is Cite this Chem. Soc. Rev. 2017, 3866, 3866–3885. [Google Scholar] [CrossRef] [PubMed]
- Nery, E.W.; Kubota, L.T. Sensing approaches on paper-based devices: a review. Anal. Bioanal. Chem. 2013, 405, 7573–7595. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.H.; Hankus, M.E.; Tian, L.; Pellegrino, P.M.; Singamaneni, S. Highly Sensitive Surface Enhanced Raman Scattering Substrates Based on Filter Paper Loaded with Plasmonic Nanostructures. Anal. Chem. 2011, 83, 8953–8958. [Google Scholar] [CrossRef] [PubMed]
- Betz, J.F.; Yu, W.W.; Cheng, Y.; White, I.M.; Rubloff, G.W. Simple SERS substrates: Powerful, portable, and full of potential. Phys. Chem. Chem. Phys. 2014, 16, 2224–2239. [Google Scholar] [CrossRef]
- Zhang, R.; Xu, B.-B.; Liu, X.-Q.; Zhang, Y.-L.; Xu, Y.; Chen, Q.-D.; Sun, H.-B. Highly efficient SERS test strips. Chem. Commun. 2012, 48, 5913–5915. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.-C.; Chou, S.-Y.; Tseng, Y.-C.; Tseng, S.-C.; Yen, Y.-T.; Chen, H.-L. Single-shot laser treatment provides quasi-three-dimensional paper-based substrates for SERS with attomolar sensitivity. Nanoscale 2015, 7, 1667–1677. [Google Scholar] [CrossRef] [PubMed]
- Cheng, M.-L.; Tsai, B.-C.; Yang, J. Silver nanoparticle-treated filter paper as a highly sensitive surface-enhanced Raman scattering (SERS) substrate for detection of tyrosine in aqueous solution. Anal. Chim. Acta 2011, 708, 89–96. [Google Scholar] [CrossRef] [PubMed]
- Ngo, Y.H.; Li, D.; Simon, G.P.; Garnier, G. Gold Nanoparticle–Paper as a Three-Dimensional Surface Enhanced Raman Scattering Substrate. Langmuir 2012, 28, 8782–8790. [Google Scholar] [CrossRef] [PubMed]
- Polavarapu, L.; La Porta, A.; Novikov, S.M.; Coronado-Puchau, M.; Liz-Marzán, L.M. Pen-on-paper approach toward the design of universal surface enhanced Raman scattering substrates. Small 2014, 10, 3065–3071. [Google Scholar] [CrossRef] [PubMed]
- Yu, W.W.; White, I.M. Inkjet-printed paper-based SERS dipsticks and swabs for trace chemical detection. Analyst 2013, 138, 1020–1025. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Li, B.; Chen, L.; Wang, Y.; Gao, D.; Ma, X.; Wu, A. Brushing, a simple way to fabricate SERS active paper substrates. Anal. Methods 2014, 6, 2066–2071. [Google Scholar] [CrossRef] [Green Version]
- Huang, Z.; Nagpal, A.; Siddhanta, S.; Barman, I. Leveraging coffee-ring effect on plasmonic paper substrate for sensitive analyte detection using Raman spectroscopy. J. Raman Spectrosc. 2018, 49, 1552–1558. [Google Scholar] [CrossRef]
- Abalde-Cela, S.; Taladriz-Blanco, P.; De Oliveira, M.G.; Abell, C. Droplet microfluidics for the highly controlled synthesis of branched gold nanoparticles. Sci. Rep. 2018, 8. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Graña, S.; Fernández-López, C.; Polavarapu, L.; Salmon, J.-B.; Leng, J.; Pastoriza-Santos, I.; Pérez-Juste, J. Gold Nanooctahedra with Tunable Size and Microfluidic-Induced 3D Assembly for Highly Uniform SERS-Active Supercrystals. Chem. Mater. 2015, 27, 8310–8317. [Google Scholar] [CrossRef]
- Senthil Kumar, P.; Pastoriza-Santos, I.; Rodríguez-González, B.; Javier García de Abajo, F.; Liz-Marzán, L.M. High-yield synthesis and optical response of gold nanostars. Nanotechnology 2008, 19, 015606. [Google Scholar] [CrossRef]
- Turkevich, J.; Stevenson, P.C.; Hillier, J. A study of the nucleation and growth processes in the synthesis of colloidal gold. Discuss. Faraday Soc. 1951, 11, 55–75. [Google Scholar] [CrossRef]
- Nikoobakht, B.; El-Sayed, M.A. Preparation and Growth Mechanism of Gold Nanorods (NRs) Using Seed-Mediated Growth Method. Chem. Mater. 2003, 15, 1957–1962. [Google Scholar] [CrossRef]
- Whitesides, G.M.; Stroock, A.D. Flexible Methods for Microfluidics. Phys. Today 2001, 54, 42–48. [Google Scholar] [CrossRef] [Green Version]
- Duffy, D.C.; McDonald, J.C.; Schueller, O.J.A.; Whitesides, G.M. Rapid Prototyping of Microfluidic Systems in Poly(dimethylsiloxane). Anal. Chem. 1998, 70, 4974–4984. [Google Scholar] [CrossRef]
- Abalde-Cela, S.; Gould, A.; Liu, X.; Kazamia, E.; Smith, A.G.; Abell, C. High-throughput detection of ethanol-producing cyanobacteria in a microdroplet platform. J. R. Soc. Interface 2015, 12. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.; Li, P.; Zhu, Q.; Wu, M.; Li, H.; Lu, F. Functional paper-based SERS substrate for rapid and sensitive detection of Sudan dyes in herbal medicine. Spectrochim. Acta Part. A Mol. Biomol. Spectrosc. 2018, 196, 110–116. [Google Scholar] [CrossRef]
- Meng, Y.; Lai, Y.; Jiang, X.; Zhao, Q.; Zhan, J. Silver nanoparticles decorated filter paper via self-sacrificing reduction for membrane extraction surface-enhanced Raman spectroscopy detection. Analyst 2013, 138, 2090–2095. [Google Scholar] [CrossRef]
- Zhu, Y.; Li, M.; Yu, D.; Yang, L. A novel paper rag as ‘D-SERS’ substrate for detection of pesticide residues at various peels. Talanta 2014, 128, 117–124. [Google Scholar] [CrossRef]
- Villa, J.E.L.; dos Santos, D.P.; Poppi, R.J. Fabrication of gold nanoparticle-coated paper and its use as a sensitive substrate for quantitative SERS analysis. Microchim. Acta 2016, 183, 2745–2752. [Google Scholar] [CrossRef]
- Enustun, B.V.; Turkevich, J. Coagulation of Colloidal Gold. J. Am. Chem. Soc. 1963, 85, 3317–3328. [Google Scholar] [CrossRef]
- Wiley, J.H.; Atalla, R.H. Band assignments in the raman spectra of celluloses. Carbohydr. Res. 1987, 160, 113–129. [Google Scholar] [CrossRef]
- Wu, X.; Gao, S.; Wang, J.-S.; Wang, H.; Huang, Y.-W.; Zhao, Y. The surface-enhanced Raman spectra of aflatoxins: Spectral analysis, density functional theory calculation, detection and differentiation. Analyst 2012, 137, 4226–4234. [Google Scholar] [CrossRef] [PubMed]
- Armbruster, D.A.; Pry, T. Limit of blank, limit of detection and limit of quantitation. Clin. Biochem. Rev. 2008, 29 (Suppl. 1), S49–S52. [Google Scholar]
- Shrivastava, A.; Gupta, V. Methods for the determination of limit of detection and limit of quantitation of the analytical methods. Chronicles Young Sci. 2011, 2, 21–25. [Google Scholar] [CrossRef]
- Abalde-Cela, S.; Ho, S.; Rodríguez-González, B.; Correa-Duarte, M.A.; Álvarez-Puebla, R.A.; Liz-Marzán, L.M.; Kotov, N.A. Loading of exponentially grown LBL films with silver nanoparticles and their application to generalized SERS detection. Angew. Chemie-Int. Ed. 2009, 48, 5326–5329. [Google Scholar] [CrossRef]
- Šubr, M.; Petr, M.; Kylián, O.; Štěpánek, J.; Veis, M.; Procházka, M. Anisotropic Optical Response of Silver Nanorod Arrays: Surface Enhanced Raman Scattering Polarization and Angular Dependences Confronted with Ellipsometric Parameters. Sci. Rep. 2017, 7, 4293. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Puebla, R.A.; Agarwal, A.; Manna, P.; Khanal, B.P.; Aldeanueva-Potel, P.; Carbó-Argibay, E.; Pazos-Pérez, N.; Vigderman, L.; Zubarev, E.R.; Kotov, N.A.; Liz-Marzán, L.M. Gold nanorods 3D-supercrystals as surface enhanced Raman scattering spectroscopy substrates for the rapid detection of scrambled prions. Proc. Natl. Acad. Sci. 2011, 108, 8157–8161. [Google Scholar] [CrossRef] [Green Version]
- Kuppusamy, P.; Govindan, N.; Yusoff, M.M.; Ichwan, S.J.A. Proteins are potent biomarkers to detect colon cancer progression. Saudi J. Biol. Sci. 2017. [Google Scholar] [CrossRef] [PubMed]
- Alix-Panabières, C.; Pantel, K. Circulating Tumor Cells: Liquid Biopsy of Cancer. Clin. Chem. 2013, 59, 110–118. [Google Scholar] [CrossRef]
- Abalde-Cela, S.; Piairo, P.; Diéguez, L. The Significance of Circulating Tumour Cells in the Clinic. Acta Cytol. 2019, 1–13. [Google Scholar] [CrossRef]
- Li, S.; Zhang, Y.; Xu, J.; Li, L.; Zeng, Q.; Lin, L.; Guo, Z.; Liu, Z.; Xiong, H.; Liu, S. Noninvasive prostate cancer screening based on serum surface-enhanced Raman spectroscopy and support vector machine. Appl. Phys. Lett. 2014. [Google Scholar] [CrossRef]
- Cai, C.; Chen, R.; Lin, J.; Li, Y.; Feng, S. Micro-Raman spectroscopy of single leukemic cells. Chin. Opt. Lett. 2008, 6, 938–940. [Google Scholar]
- Lin, D.; Pan, J.; Huang, H.; Chen, G.; Qiu, S.; Shi, H.; Chen, W.; Yu, Y.; Feng, S.; Chen, R. Label-free blood plasma test based on surface-enhanced Raman scattering for tumor stages detection in nasopharyngeal cancer. Sci. Rep. 2014. [Google Scholar] [CrossRef] [PubMed]
- Feng, S.; Lin, J.; Cheng, M.; Li, Y.-Z.; Chen, G.; Huang, Z.; Yu, Y.; Chen, R.; Zeng, H. Gold Nanoparticle Based Surface-Enhanced Raman Scattering Spectroscopy of Cancerous and Normal Nasopharyngeal Tissues under Near-Infrared Laser Excitation. Appl. Spectrosc. 2009, 63, 1089–1094. [Google Scholar] [CrossRef] [PubMed]
- Lin, D.; Feng, S.; Pan, J.; Chen, Y.; Lin, J.; Chen, G.; Xie, S.; Zeng, H.; Chen, R. Colorectal Cancer Detection by Gold Nanoparticle Based Surface-Enhanced Raman Spectroscopy of Blood Serum and Statistical Analysis. Opt. Express 2011, 19, 13565–13577. [Google Scholar] [CrossRef]
- Feng, S.; Chen, R.; Lin, J.; Pan, J.; Chen, G.; Li, Y.; Cheng, M.; Huang, Z.; Chen, J.; Zeng Haishan, H. Nasopharyngeal cancer detection based on blood plasma surface-enhanced Raman spectroscopy and multivariate analysis. Biosens. Bioelectron. 2010, 25, 2414–2419. [Google Scholar] [CrossRef] [PubMed]
- Duraipandian, S.; Zheng, W.; Ng, J.; Low, J.J.H.; Ilancheran, A.; Huang, Z. In vivo diagnosis of cervical precancer using Raman spectroscopy and genetic algorithm techniques. Analyst 2011, 136, 4328–4336. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Chen, R.; Feng, S.; Pan, J.; Li, Y.; Chen, G.; Cheng, M.; Huang, Z.; Yu, Y.; Zeng, H. A novel blood plasma analysis technique combining membrane electrophoresis with silver nanoparticle-based SERS spectroscopy for potential applications in noninvasive cancer detection. Nanomed. Nanotechnol. Biol. Med. 2011, 7, 655–663. [Google Scholar] [CrossRef] [PubMed]
- Chen Yong Kah, J.; Wei Kho, K.; Guat Leng Lee, C.; James Richard Sheppard, C.; Xiang Shen, Z.; Chee Soo, K.; Carolene Olivo, M. Early diagnosis of oral cancer based on the surface plasmon resonance of gold nanoparticles. Int. J. Nanomed. 2007, 2, 785–798. [Google Scholar]



| Raman Shift (cm−1) | Vibrational Mode | Peak Assignment | Reference |
|---|---|---|---|
| 516 | S–S Disulfide stretching | Proteins | [52] |
| 638–642 | C–S Stretch C–C Twist | Tyrosine | [53,54] |
| 649 | C–C twisting mode | D-glucose, lactose | [55] |
| 725–728 | δ(C–H) | Adenine, coenzyme | [52,56] |
| 758 | Tryptophan | [53] | |
| 823 | Ring Breathing | Tyrosine | [56], |
| 813, 886 | v (C–C–O) v (C–O–H) | L-Serine, glutathione, Glutathione, d-(C)-galactosamine | [57] |
| 925–935 | C–C stretching mode, C–C α helix | Proteins and collagen | [58] |
| 979–999 | Phospholipids, glucose-I′-phosphate, DNA (around 980 cm−1) | [53,58] | |
| 1046, 1048 | C–H bedding | Glycogen: C–O, C–C | [59], [53] |
| 1080–1099 | C–C stretching, v(PO2−), v(CC), v(COC), | Nucleic acids, phospholipids | [58] |
| 1094–1134 | C–N stretch, C–C stretching | Lipids, D-mannos | [53], [54] |
| 1206 | Ring vibration | Tyrosine | [56] |
| 1240–1260 | C–N, N–H | Amide III | [58] |
| 1269 | δ (=CH) | Phospholipids | [60] |
| 1316–1329 | CH vibration, CH2 twisting | DNA/RNA, lipids | [55], [52] |
| 1331 | v(C–H) | Nucleic acid bases | [57] |
| 1347 | CH3CH2 wagging | Tryptophan, adenine, guanine | [52] |
| 1400–1420 | CH3 | Proteins, amino acids (around 1414 cm−1) | [58] |
| 1441–1445 | δ(CH2), δ(CH3) | Proteins, Collagen and phospholipids | [52], [57] |
| 1607 | Phenylalanine or Tyrosine | [59] | |
| 1625–1699 | ν(C=O) | Amide I (α-helix), collagen, Amide I | [56,57,58] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Teixeira, A.; Hernández-Rodríguez, J.F.; Wu, L.; Oliveira, K.; Kant, K.; Piairo, P.; Diéguez, L.; Abalde-Cela, S. Microfluidics-Driven Fabrication of a Low Cost and Ultrasensitive SERS-Based Paper Biosensor. Appl. Sci. 2019, 9, 1387. https://doi.org/10.3390/app9071387
Teixeira A, Hernández-Rodríguez JF, Wu L, Oliveira K, Kant K, Piairo P, Diéguez L, Abalde-Cela S. Microfluidics-Driven Fabrication of a Low Cost and Ultrasensitive SERS-Based Paper Biosensor. Applied Sciences. 2019; 9(7):1387. https://doi.org/10.3390/app9071387
Chicago/Turabian StyleTeixeira, Alexandra, Juan F. Hernández-Rodríguez, Lei Wu, Kevin Oliveira, Krishna Kant, Paulina Piairo, Lorena Diéguez, and Sara Abalde-Cela. 2019. "Microfluidics-Driven Fabrication of a Low Cost and Ultrasensitive SERS-Based Paper Biosensor" Applied Sciences 9, no. 7: 1387. https://doi.org/10.3390/app9071387