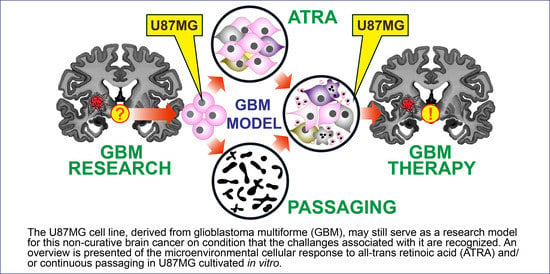
All-Trans Retinoic Acid Fosters the Multifarious U87MG Cell Line as a Model of Glioblastoma
Abstract
:1. Introduction
2. Materials and Methods
2.1. Propagation and Maintenance of U87MG
2.2. All Trans-Retinoic Acid (ATRA) Preparation
2.3. Wound Healing Assay
2.4. Determination of Proliferation with EdU Flow Cytometry
2.5. Total RNA Isolation and cDNA Synthesis
2.6. Real Time PCR Quantification of Long Non-Coding RNA and Endogenous Control Genes
2.7. Protein Extraction
2.8. Electrophoresis and Western Blotting
2.9. Cytogenetic Harvesting of U87MG
2.10. Wright Staining and Banding of U87MG Chromosomes
2.11. Giemsa Staining for Structural Chromosomal Abberations
2.12. Immunofluorescence
2.13. Statistical Analysis
2.14. Data Availability
3. Results
3.1. Reduced Expression of the Stem Cell Marker Prominin-1 (CD133) in U87MG in Response to ATRA
3.2. The Intercellular Adhesion Molecule (ICAM-1; CD54) Is Distributed throughout U87MG with Increased Expression in Response to ATRA
3.3. ATRA Increases Proliferation of U87MG in the Leading Edge in Comparison to the Established Cellular Monolayer
3.4. Long Non-Coding RNAs (lncRNA) at the Leading Edge Respond to ATRA Treatment in Early Passaged U87MG
3.5. Increased Prevalence of Chromosomal Aberrations with Passaging in U87MG Exposed to ATRA
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Lapointe, S.; Perry, A.; Butowski, N.A. Primary brain tumours in adults. Lancet 2018, 392, 432–446. [Google Scholar] [CrossRef]
- Qian, B.Z.; Pollard, J.W. Macrophage diversity enhances tumor progression and metastasis. Cell 2010, 141, 39–51. [Google Scholar] [CrossRef] [Green Version]
- Giakoumettis, D.; Kritis, A.; Foroglou, N. C6 cell line: The gold standard in glioma research. Hippokratia 2018, 22, 105–112. [Google Scholar]
- Furnari, F.B.; Fenton, T.; Bachoo, R.M.; Mukasa, A.; Stommel, J.M.; Stegh, A.; Hahn, W.C.; Ligon, K.L.; Louis, D.N.; Brennan, C.; et al. Malignant astrocytic glioma: Genetics, biology, and paths to treatment. Genes Dev. 2007, 21, 2683–2710. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Irtenkauf, S.M.; Sobiechowski, S.; Hasselbach, L.A.; Nelson, K.K.; Transou, A.D.; Carlton, E.T.; Mikkelsen, T.; Decarvalho, A.C. Optimization of Glioblastoma Mouse Orthotopic Xenograft Models for Translational Research. Comp. Med. 2017, 67, 300–314. [Google Scholar] [PubMed]
- Kutna, V.; O’Leary, V.B.; Newman, E.; Hoschl, C.; Ovsepian, S.V. Revisiting Brain Tuberous Sclerosis Complex in Rat and Human: Shared Molecular and Cellular Pathology Leads to Distinct Neurophysiological and Behavioral Phenotypes. Neurotherapeutics 2021. [Google Scholar] [CrossRef]
- Mawson, A.R. Retinoids in the treatment of glioma: A new perspective. Cancer Manag. Res. 2012, 4, 233–241. [Google Scholar] [CrossRef] [Green Version]
- Jones, T.; Zhang, B.; Major, S.; Webb, A. All-trans retinoic acid eluting poly(diol citrate) wafers for treatment of glioblastoma. J. Biomed. Mater. Res. B 2020, 108, 619–628. [Google Scholar] [CrossRef]
- Rhinn, M.; Dolle, P. Retinoic acid signalling during development. Development 2012, 139, 843–858. [Google Scholar] [CrossRef] [Green Version]
- Környei, Z.; Gócza, E.; Rühl, R.; Orsolits, B.; Vörös, E.; Szabó, B.; Vágovits, B.; Madarász, E. Astroglia-derived retinoic acid is a key factor in glia-induced neurogenesis. FASEB J. 2007, 21, 2496–2509. [Google Scholar] [CrossRef]
- Dolgin, E. Venerable brain-cancer cell line faces identity crisis. Nature 2016, 537, 149–150. [Google Scholar] [CrossRef] [Green Version]
- Allen, M.; Bjerke, M.; Edlund, H.; Nelander, S.; Westermark, B. Origin of the U87MG glioma cell line: Good news and bad news. Sci. Transl. Med. 2016, 8, 354re3. [Google Scholar] [CrossRef]
- O’Leary, V.B.; Hain, S.; Maugg, D.; Smida, J.; Azimzadeh, O.; Tapio, S.; Ovsepian, S.V.; Atkinson, M.J. Long non-coding RNA PARTICLE bridges histone and DNA methylation. Sci. Rep. 2017, 7, 1790. [Google Scholar] [CrossRef] [Green Version]
- O’Leary, V.B.; Maugg, D.; Smida, J.; Baumhoer, D.; Nathrath, M.; Ovsepian, S.V.; Atkinson, M.J. The long non-coding RNA PARTICLE is associated with WWOX and the absence of FRA16D breakage in osteosarcoma patients. Oncotarget 2017, 8, 87431–87441. [Google Scholar] [CrossRef] [Green Version]
- O’Leary, V.B.; Ovsepian, S.V.; Carrascosa, L.G.; Buske, F.A.; Radulovic, V.; Niyazi, M.; Moertl, S.; Trau, M.; Atkinson, M.J.; Anastasov, N. PARTICLE, a Triplex-Forming Long ncRNA, Regulates Locus-Specific Methylation in Response to Low-Dose Irradiation. Cell Rep. 2015, 11, 474–485. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Leary, V.B.; Ovsepian, S.V.; Smida, J.; Atkinson, M.J. PARTICLE—The RNA podium for genomic silencers. J. Cell. Physiol. 2019, 234, 19464–19470. [Google Scholar] [CrossRef]
- O’Leary, V.B.; Smida, J.; Buske, F.A.; Carrascosa, L.G.; Azimzadeh, O.; Maugg, D.; Hain, S.; Tapio, S.; Heidenreich, W.; Kerr, J.; et al. PARTICLE triplexes cluster in the tumor suppressor WWOX and may extend throughout the human genome. Sci. Rep. 2017, 7, 7163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, J.; Hodges, T.R.; Song, R.; Gong, Y.; Calin, G.A.; Heimberger, A.B.; Zhao, H. Serum HOTAIR and GAS5 levels as predictors of survival in patients with glioblastoma. Mol. Carcinog. 2018, 57, 137–141. [Google Scholar] [CrossRef] [PubMed]
- Toraih, E.A.; Alghamdi, S.A.; El-Wazir, A.; Hosny, M.M.; Hussein, M.H.; Khashana, M.S.; Fawzy, M.S. Dual biomarkers long non-coding RNA GAS5 and microRNA-34a co-expression signature in common solid tumors. PLoS ONE 2018, 13, e0198231. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Xin, S.; Zhang, K.; Shi, R.; Bao, X. Low GAS5 Levels as a Predictor of Poor Survival in Patients with Lower-Grade Gliomas. J. Oncol. 2019, 2019, 1785042. [Google Scholar] [CrossRef] [Green Version]
- Gadji, M.; Crous-Tsanaclis, A.M.; Mathieu, D.; Mai, S.; Fortin, D.; Drouin, R. A new der(1;7)(q10;p10) leading to a singular 1p loss in a case of glioblastoma with oligodendroglioma component. Neuropathology 2014, 34, 170–178. [Google Scholar] [CrossRef]
- Kútna, V.; Uttl, L.; Waltereit, R.; Krištofiková, Z.; Kaping, D.; Petrásek, T.; Hoschl, C.; Ovsepian, S.V. Tuberous Sclerosis (tsc2+/−) Model Eker Rats Reveals Extensive Neuronal Loss with Microglial Invasion and Vascular Remodeling Related to Brain Neoplasia. Neurotherapeutics 2020, 17, 329–339. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Guo, Y.; Li, Y.; Li, W.; Zheng, X.; Xia, H.; Mao, Q. Detection of CD133 expression in U87 glioblastoma cells using a novel anti-CD133 monoclonal antibody. Oncol. Lett. 2015, 9, 2603–2608. [Google Scholar] [CrossRef] [PubMed]
- Mei, D.; Lv, B.; Chen, B.; Xiao, S.; Jiang, J.; Xie, Y.; Jiang, L. All-trans retinoic acid suppresses malignant characteristics of CD133-positive thyroid cancer stem cells and induces apoptosis. PLoS ONE 2017, 12, e0182835. [Google Scholar] [CrossRef] [PubMed]
- Piao, Y.; Henry, V.; Tiao, N.; Park, S.Y.; Martinez-Ledesma, J.; Dong, J.W. Targeting intercellular adhesion molecule-1 prolongs survival in mice bearing bevacizumab-resistant glioblastoma. Oncotarget 2017, 8, 96970–96983. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, L.; Li, H.; Zhan, Y. All-trans retinoic acid enhances temozolomide-induced autophagy in human glioma cells U251 via targeting Keap1/Nrf2/ARE signaling pathway. Oncol. Lett. 2017, 14, 2709–2714. [Google Scholar] [CrossRef]
- Jia, P.F.; Gu, W.T.; Zhang, W.F.; Li, F. Treatment of recurrent malignant gliomas with 13-cis-retinoic acid naphthalene triazole. Neurol. Sci. 2015, 36, 717–721. [Google Scholar] [CrossRef]
- Chen, P.H.; Shih, C.M.; Chang, W.C.; Cheng, C.H.; Lin, C.W.; Ho, K.H.; Su, P.C.; Chen, K.C. MicroRNA-302b-inhibited E2F3 transcription factor is related to all trans retinoic acid-induced glioma cell apoptosis. J. Neurochem. 2014, 131, 731–742. [Google Scholar] [CrossRef] [Green Version]
- Das, A.; Banik, N.L.; Ray, S.K. Molecular mechanisms of the combination of retinoid and interferon-gamma for inducing differentiation and increasing apoptosis in human glioblastoma T98G and U87MG cells. Neurochem. Res. 2009, 34, 87–101. [Google Scholar] [CrossRef] [PubMed]
- Liang, C.; Yang, L.; Guo, S. All-trans retinoic acid inhibits migration, invasion and proliferation, and promotes apoptosis in glioma cells in vitro. Oncol. Lett. 2015, 9, 2833–2838. [Google Scholar] [CrossRef] [Green Version]
- Johnston, A.L.; Lun, X.; Rahn, J.J.; Liacini, A.; Wang, L.; Hamilton, M.G.; Parney, I.F.; Hempstead, B.L.; Robbins, S.M.; Forsyth, P.A.; et al. The p75 neurotrophin receptor is a central regulator of glioma invasion. PLoS Biol. 2007, 5, e212. [Google Scholar] [CrossRef]
- Guo, X.; Deng, K.; Wang, H.; Xia, J.; Shan, T.; Liang, Z.; Yao, L.; Jin, S. GAS5 Inhibits Gastric Cancer Cell Proliferation Partly by Modulating CDK6. Oncol. Res. Treat. 2015, 38, 362–366. [Google Scholar] [CrossRef]
- Yan, Z.; Ruoyu, L.; Xing, L.; Hua, L.; Jun, Z.; Yaqin, P.; Lu, W.; Aili, T.; Yuzi, Z.; Lin, M.; et al. Long non-coding RNA GAS5 regulates the growth and metastasis of human cervical cancer cells via induction of apoptosis and cell cycle arrest. Arch. Biochem. Biophys. 2020, 684, 108320. [Google Scholar] [CrossRef]
- Wick, W.; Platten, M. Understanding and Treating Glioblastoma. Neurol. Clin. 2018, 36, 485–499. [Google Scholar] [CrossRef] [PubMed]
- Bhatia, M. AC133 expression in human stem cells. Leukemia 2001, 15, 1685–1688. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Motegi, H.; Kamoshima, Y.; Terasaka, S.; Kobayashi, H.; Houkin, K. A novel adherent culture method of glioblastoma cells expressing CD133 using collagen-1-coated plates. Hokkaido Igaku Zasshi Hokkaido J. Med. Sci. 2012, 87, 147–151. [Google Scholar]
- Piao, Y.; Liang, J.; Holmes, L.; Zurita, A.J.; Henry, V.; Heymach, J.V.; De Groot, J.F. Glioblastoma resistance to anti-VEGF therapy is associated with myeloid cell infiltration, stem cell accumulation, and a mesenchymal phenotype. Neuro-Oncology 2012, 14, 1379–1392. [Google Scholar] [CrossRef] [Green Version]
- Tang, X.H.; Gudas, L.J. Retinoids, retinoic acid receptors, and cancer. Annu. Rev. Pathol. 2011, 6, 345–364. [Google Scholar] [CrossRef]
- Lu, J.; Zhang, F.; Zhao, D.; Hong, L.; Min, J.; Zhang, L.; Li, F.; Yan, Y.; Li, H.; Ma, Y.; et al. ATRA-inhibited proliferation in glioma cells is associated with subcellular redistribution of beta-catenin via up-regulation of Axin. J. Neurooncol. 2008, 87, 271–277. [Google Scholar] [CrossRef]
- Zhao, X.; Wang, P.; Liu, J.; Zheng, J.; Liu, Y.; Chen, J.; Xue, Y. GAS5 Exerts Tumor-suppressive Functions in Human Glioma Cells by Targeting miR-222. Mol. Ther. 2015, 23, 1899–1911. [Google Scholar] [CrossRef] [Green Version]
- Zeng, T.; Li, L.; Zhou, Y.; Gao, L. Exploring Long Noncoding RNAs in Glioblastoma: Regulatory Mechanisms and Clinical Potentials. Int. J. Genom. 2018, 2018, 2895958. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salta, E.; De Strooper, B. Non-coding RNAs with essential roles in neurodegenerative disorders. Lancet Neurol. 2012, 11, 189–200. [Google Scholar] [CrossRef]
- Ma, X.L.; Zhu, W.D.; Tian, L.X.; Sun, W.D.; Shang, F.; Lin, Q.T.; Zhang, H.Q. Long non-coding RNA TUSC7 expression is independently predictive of outcome in glioma. Eur. Rev. Med. Pharmacol. Sci. 2017, 21, 3605–3610. [Google Scholar] [PubMed]
- Zhang, X.; Sun, S.; Pu, J.K.S.; Tsang, A.C.O.; Lee, D.; Man, V.O.Y.; Lui, W.M.; Wong, S.T.S.; Leung, G.K.K. Long non-coding RNA expression profiles predict clinical phenotypes in glioma. Neurobiol. Dis. 2012, 48, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Clark, M.J.; Homer, N.; O’Connor, B.D.; Chen, Z.; Eskin, A.; Lee, H.; Merriman, B.; Nelson, S.F. U87MG decoded: The genomic sequence of a cytogenetically aberrant human cancer cell line. PLoS Genet. 2010, 6, e1000832. [Google Scholar] [CrossRef] [PubMed]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pokorná, M.; Hudec, M.; Juříčková, I.; Vácha, M.; Polívková, Z.; Kútna, V.; Pala, J.; Ovsepian, S.V.; Černá, M.; O’Leary, V.B. All-Trans Retinoic Acid Fosters the Multifarious U87MG Cell Line as a Model of Glioblastoma. Brain Sci. 2021, 11, 812. https://doi.org/10.3390/brainsci11060812
Pokorná M, Hudec M, Juříčková I, Vácha M, Polívková Z, Kútna V, Pala J, Ovsepian SV, Černá M, O’Leary VB. All-Trans Retinoic Acid Fosters the Multifarious U87MG Cell Line as a Model of Glioblastoma. Brain Sciences. 2021; 11(6):812. https://doi.org/10.3390/brainsci11060812
Chicago/Turabian StylePokorná, Markéta, Michael Hudec, Iva Juříčková, Michael Vácha, Zdeňka Polívková, Viera Kútna, Jan Pala, Saak V. Ovsepian, Marie Černá, and Valerie Bríd O’Leary. 2021. "All-Trans Retinoic Acid Fosters the Multifarious U87MG Cell Line as a Model of Glioblastoma" Brain Sciences 11, no. 6: 812. https://doi.org/10.3390/brainsci11060812