Single-Cell Transcriptomics Reveals Evidence of Endothelial Dysfunction in the Brains of COVID-19 Patients with Implications for Glioblastoma Progression
Abstract
:1. Introduction
2. Materials and Methods
2.1. Single Cell RNA-Seq Dataset
2.2. Computational Analysis, Statistics, and Schematics
3. Results
3.1. Annotation of Major Brain Cell Types from Healthy and COVID-19 Patients’ Brain
3.2. Aberrant Expression of CLDN5 in Endothelial Cells during COVID-19
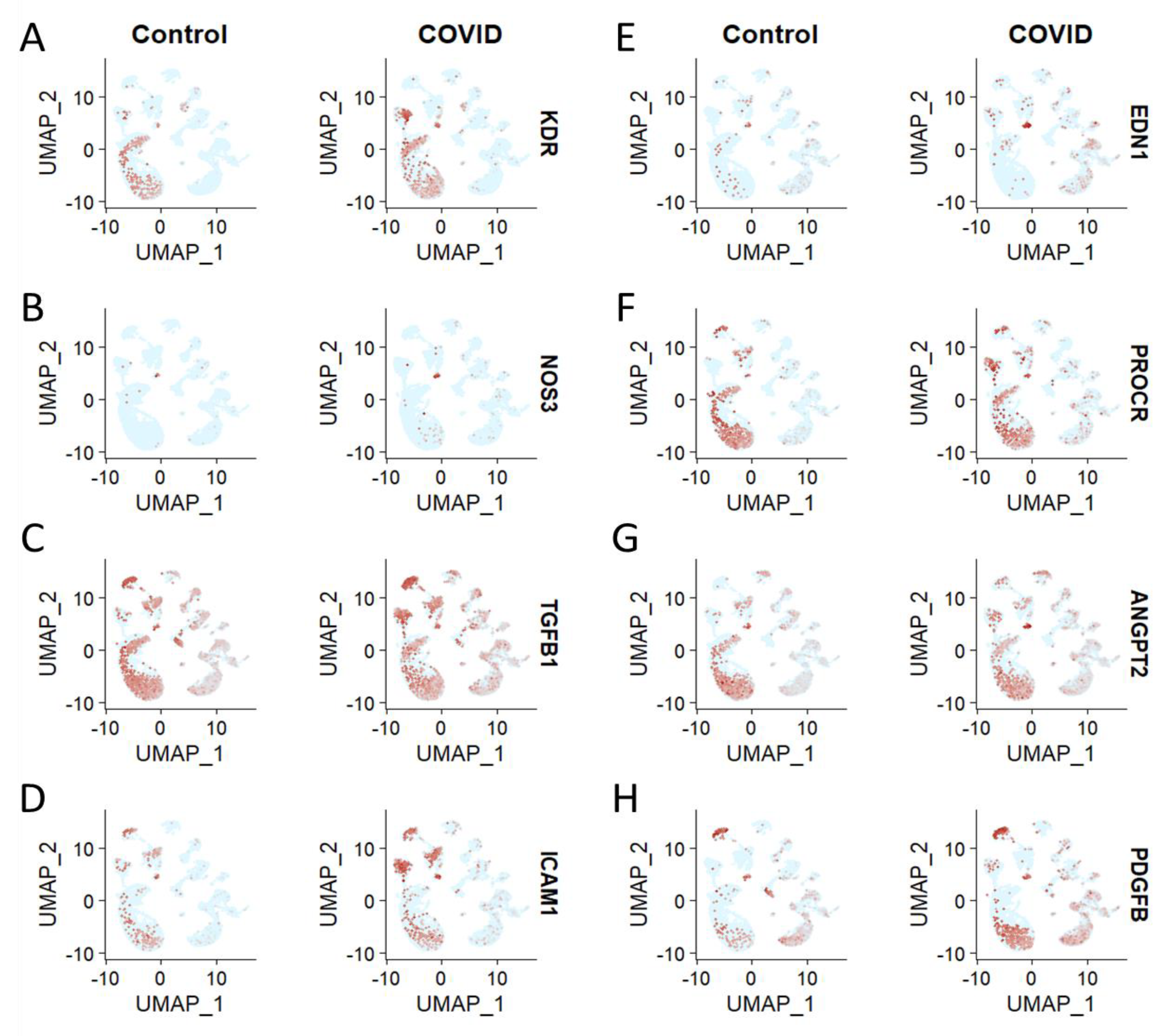
3.3. Brain Cell Type-Specific Altered Expression of Major Genes Involved in Endothelial Dysfunction in COVID-19
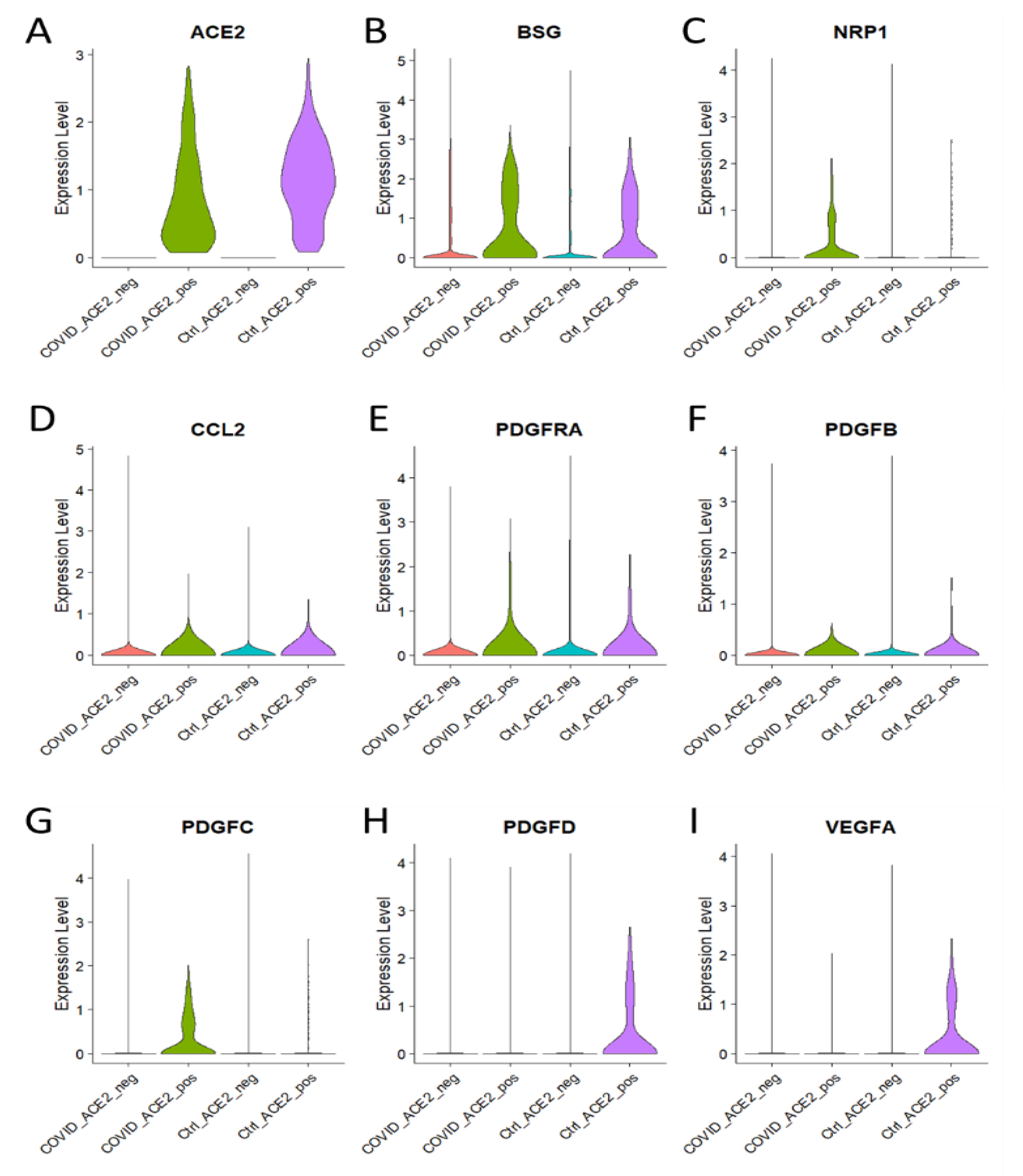
3.4. Enhanced Expression of Major Genes Involved in Endothelial Dysfunction in ACE2 Positive Brain Cells of COVID-19 Patients
3.5. C1qRs and MAC Are Overexpressed in KEGG Pathway in COVID-19
3.6. Brain Cell Type-Specific Altered Expression of CLDN5, c1qRs, and C5 in GBM Patient
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Thakur, A. Shedding Lights on the Extracellular Vesicles as Functional Mediator and Therapeutic Decoy for COVID-19. Life 2023, 13, 840. [Google Scholar] [CrossRef]
- Boni, M.F.; Lemey, P.; Jiang, X.; Lam, T.T.-Y.; Perry, B.W.; Castoe, T.A.; Rambaut, A.; Robertson, D.L. Evolutionary origins of the SARS-CoV-2 sarbecovirus lineage responsible for the COVID-19 pandemic. Nat. Microbiol. 2020, 5, 1408–1417. [Google Scholar] [CrossRef] [PubMed]
- Yang, R.-C.; Huang, K.; Zhang, H.-P.; Li, L.; Zhang, Y.-F.; Tan, C.; Chen, H.-C.; Jin, M.-L.; Wang, X.-R. SARS-CoV-2 productively infects human brain microvascular endothelial cells. J. Neuroinflamm. 2022, 19, 149. [Google Scholar] [CrossRef] [PubMed]
- Vassiliou, A.G.; Kotanidou, A.; Dimopoulou, I.; Orfanos, S.E. Endothelial Damage in Acute Respiratory Distress Syndrome. Int. J. Mol. Sci. 2020, 21, 8793. [Google Scholar] [CrossRef]
- Masaki, T.; Sawamura, T. Endothelin and endothelial dysfunction. Proc. Jpn. Acad. Ser. B 2006, 82, 17–24. [Google Scholar] [CrossRef]
- Xu, S.; Ilyas, I.; Weng, J. Endothelial dysfunction in COVID-19: An overview of evidence, biomarkers, mechanisms and potential therapies. Acta Pharmacol. Sin. 2023, 44, 695–709. [Google Scholar] [CrossRef]
- Wang, X.; Bove, A.M.; Simone, G.; Ma, B. Molecular Bases of VEGFR-2-Mediated Physiological Function and Pathological Role. Front. Cell Dev. Biol. 2020, 8, 599281. [Google Scholar] [CrossRef]
- Sandoo, A.; Veldhuijzen van Zanten, J.J.C.; Metsios, G.S.; Carroll, D.; Kitas, G.D. The Endothelium and Its Role in Regulating Vascular Tone. Open Cardiovasc. Med. J. 2010, 4, 302–312. [Google Scholar] [CrossRef]
- Gheblawi, M.; Wang, K.; Viveiros, A.; Nguyen, Q.; Zhong, J.-C.; Turner, A.J.; Raizada, M.K.; Grant, M.B.; Oudit, G.Y. Angiotensin-Converting Enzyme 2: SARS-CoV-2 Receptor and Regulator of the Renin-Angiotensin System. Circ. Res. 2020, 126, 1456–1474. [Google Scholar] [CrossRef]
- Weiler, H. Endothelial protein C receptor: Location…with a pool! J. Thromb. Haemost. 2007, 5, 1391–1393. [Google Scholar] [CrossRef]
- Smith, J.D.; Bryant, S.R.; Couper, L.L.; Vary, C.P.H.; Gotwals, P.J.; Koteliansky, V.E.; Lindner, V. Soluble Transforming Growth Factor-β Type II Receptor Inhibits Negative Remodeling, Fibroblast Transdifferentiation, and Intimal Lesion Formation but Not Endothelial Growth. Circ. Res. 1999, 84, 1212–1222. [Google Scholar] [CrossRef] [PubMed]
- Jang, D.; Lee, A.-H.; Shin, H.-Y.; Song, H.-R.; Park, J.-H.; Kang, T.-B.; Lee, S.-R.; Yang, S.-H. The Role of Tumor Necrosis Factor Alpha (TNF-α) in Autoimmune Disease and Current TNF-α Inhibitors in Therapeutics. Int. J. Mol. Sci. 2021, 22, 2719. [Google Scholar] [CrossRef]
- Bui, T.M.; Wiesolek, H.L.; Sumagin, R. ICAM-1: A master regulator of cellular responses in inflammation, injury resolution, and tumorigenesis. J. Leukoc. Biol. 2020, 108, 787–799. [Google Scholar] [CrossRef] [PubMed]
- Zamorano Cuervo, N.; Grandvaux, N. ACE2: Evidence of role as entry receptor for SARS-CoV-2 and implications in comorbidities. Elife 2020, 9, e61390. [Google Scholar] [CrossRef] [PubMed]
- Ni, W.; Yang, X.; Yang, D.; Bao, J.; Li, R.; Xiao, Y.; Hou, C.; Wang, H.; Liu, J.; Yang, D.; et al. Role of angiotensin-converting enzyme 2 (ACE2) in COVID-19. Crit. Care 2020, 24, 422. [Google Scholar] [CrossRef] [PubMed]
- Wan, D.; Du, T.; Hong, W.; Chen, L.; Que, H.; Lu, S.; Peng, X. Neurological complications and infection mechanism of SARS-CoV-2. Signal Transduct. Target. Ther. 2021, 6, 406. [Google Scholar] [CrossRef]
- Jiao, L.; Yang, Y.; Yu, W.; Zhao, Y.; Long, H.; Gao, J.; Ding, K.; Ma, C.; Li, J.; Zhao, S.; et al. The olfactory route is a potential way for SARS-CoV-2 to invade the central nervous system of rhesus monkeys. Signal Transduct. Target. Ther. 2021, 6, 169. [Google Scholar] [CrossRef]
- Wenzel, J.; Lampe, J.; Müller-Fielitz, H.; Schuster, R.; Zille, M.; Müller, K.; Krohn, M.; Körbelin, J.; Zhang, L.; Özorhan, Ü.; et al. The SARS-CoV-2 main protease Mpro causes microvascular brain pathology by cleaving NEMO in brain endothelial cells. Nat. Neurosci. 2021, 24, 1522–1533. [Google Scholar] [CrossRef]
- Rhea, E.M.; Logsdon, A.F.; Hansen, K.M.; Williams, L.M.; Reed, M.J.; Baumann, K.K.; Holden, S.J.; Raber, J.; Banks, W.A.; Erickson, M.A. The S1 protein of SARS-CoV-2 crosses the blood–brain barrier in mice. Nat. Neurosci. 2021, 24, 368–378. [Google Scholar] [CrossRef]
- Buzhdygan, T.P.; DeOre, B.J.; Baldwin-Leclair, A.; Bullock, T.A.; McGary, H.M.; Khan, J.A.; Razmpour, R.; Hale, J.F.; Galie, P.A.; Potula, R.; et al. The SARS-CoV-2 spike protein alters barrier function in 2D static and 3D microfluidic in-vitro models of the human blood–brain barrier. Neurobiol. Dis. 2020, 146, 105131. [Google Scholar] [CrossRef]
- Constant, O.; Barthelemy, J.; Bolloré, K.; Tuaillon, E.; Gosselet, F.; Chable-Bessia, C.; Merida, P.; Muriaux, D.; Van de Perre, P.; Salinas, S.; et al. SARS-CoV-2 Poorly Replicates in Cells of the Human Blood-Brain Barrier Without Associated Deleterious Effects. Front. Immunol. 2021, 12, 697329. [Google Scholar] [CrossRef]
- Varga, Z.; Flammer, A.J.; Steiger, P.; Haberecker, M.; Andermatt, R.; Zinkernagel, A.S.; Mehra, M.R.; Schuepbach, R.A.; Ruschitzka, F.; Moch, H. Endothelial cell infection and endotheliitis in COVID-19. Lancet 2020, 395, 1417–1418. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Narayan, R.K.; Kumari, C.; Faiq, M.A.; Kulandhasamy, M.; Kant, K.; Pareek, V. SARS-CoV-2 cell entry receptor ACE2 mediated endothelial dysfunction leads to vascular thrombosis in COVID-19 patients. Med. Hypotheses 2020, 145, 110320. [Google Scholar] [CrossRef] [PubMed]
- Douyère, M.; Chastagner, P.; Boura, C. Neuropilin-1: A Key Protein to Consider in the Progression of Pediatric Brain Tumors. Front. Oncol. 2021, 11, 665634. [Google Scholar] [CrossRef] [PubMed]
- Zalpoor, H.; Shapourian, H.; Akbari, A.; Shahveh, S.; Haghshenas, L. Increased neuropilin-1 expression by COVID-19: A possible cause of long-term neurological complications and progression of primary brain tumors. Hum. Cell 2022, 35, 1301–1303. [Google Scholar] [CrossRef]
- Zalpoor, H.; Akbari, A.; Nabi-Afjadi, M. Ephrin (Eph) receptor and downstream signaling pathways: A promising potential targeted therapy for COVID-19 and associated cancers and diseases. Hum. Cell 2022, 35, 952–954. [Google Scholar] [CrossRef]
- Ali, M.; Wani, S.U.D.; Masoodi, M.H.; Khan, N.A.; Shivakumar, H.G.; Osmani, R.M.A.; Khan, K.A. Global Effect of COVID-19 Pandemic on Cancer Patients and its Treatment: A Systematic Review. Clin. Complement. Med. Pharmacol. 2022, 2, 100041. [Google Scholar] [CrossRef]
- Xu, C.; Thakur, A.; Li, Z.; Yang, T.; Zhao, C.; Li, Y.; Lee, Y.; Wu, C.-M.L. Determination of glioma cells’ malignancy and their response to TMZ via detecting exosomal BIGH3 by a TiO2-CTFE-AuNIs plasmonic biosensor. Chem. Eng. J. 2021, 415, 128948. [Google Scholar] [CrossRef]
- Thakur, A.; Qiu, G.; Xu, C.; Han, X.; Yang, T.; NG, S.P.; Chan, K.W.Y.; Wu, C.M.L.; Lee, Y. Label-free sensing of exosomal MCT1 and CD147 for tracking metabolic reprogramming and malignant progression in glioma. Sci. Adv. 2020, 6, eaaz6119. [Google Scholar] [CrossRef]
- Qiu, G.; Thakur, A.; Xu, C.; Ng, S.; Lee, Y.; Wu, C.L. Detection of Glioma-Derived Exosomes with the Biotinylated Antibody-Functionalized Titanium Nitride Plasmonic Biosensor. Adv. Funct. Mater. 2019, 29, 1806761. [Google Scholar] [CrossRef]
- Lozano-Sanchez, F.; Ursu, R.; Di-Stefano, A.L.; Ducray, F.; Younan, N.; Touat, M.; Groh, M.; Agguini, H.; Belin, C.; Garnier, L.; et al. Descriptive and retrospective analysis of diffuse glioma patients with symptomatic SARS-CoV2 infection during the first wave of the pandemic. Neuro-Oncol. Adv. 2021, 3, vdab078. [Google Scholar] [CrossRef]
- Takata, F.; Nakagawa, S.; Matsumoto, J.; Dohgu, S. Blood-Brain Barrier Dysfunction Amplifies the Development of Neuroinflammation: Understanding of Cellular Events in Brain Microvascular Endothelial Cells for Prevention and Treatment of BBB Dysfunction. Front. Cell. Neurosci. 2021, 15, 661838. [Google Scholar] [CrossRef] [PubMed]
- Kadry, H.; Noorani, B.; Cucullo, L. A blood–brain barrier overview on structure, function, impairment, and biomarkers of integrity. Fluids Barriers CNS 2020, 17, 69. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zhou, L.; Bao, L.; Liu, J.; Zhu, H.; Lv, Q.; Liu, R.; Chen, W.; Tong, W.; Wei, Q.; et al. SARS-CoV-2 crosses the blood–brain barrier accompanied with basement membrane disruption without tight junctions alteration. Signal Transduct. Target. Ther. 2021, 6, 337. [Google Scholar] [CrossRef] [PubMed]
- Galea, I. The blood–brain barrier in systemic infection and inflammation. Cell. Mol. Immunol. 2021, 18, 2489–2501. [Google Scholar] [CrossRef]
- Chen, Y.; Yang, W.; Chen, F.; Cui, L. COVID-19 and cognitive impairment: Neuroinvasive and blood—brain barrier dysfunction. J. Neuroinflamm. 2022, 19, 222. [Google Scholar] [CrossRef]
- Montiel, V.; Lobysheva, I.; Gérard, L.; Vermeersch, M.; Perez-Morga, D.; Castelein, T.; Mesland, J.-B.; Hantson, P.; Collienne, C.; Gruson, D.; et al. Oxidative stress-induced endothelial dysfunction and decreased vascular nitric oxide in COVID-19 patients. eBioMedicine 2022, 77, 103893. [Google Scholar] [CrossRef]
- Ma, S.-C.; Li, Q.; Peng, J.-Y.; Zhouwen, J.-L.; Diao, J.-F.; Niu, J.-X.; Wang, X.; Guan, X.-D.; Jia, W.; Jiang, W.-G. Claudin-5 regulates blood-brain barrier permeability by modifying brain microvascular endothelial cell proliferation, migration, and adhesion to prevent lung cancer metastasis. CNS Neurosci. Ther. 2017, 23, 947–960. [Google Scholar] [CrossRef]
- Jeon, H.; Kim, M.; Park, W.; Lim, J.S.; Lee, E.; Cha, H.; Ahn, J.S.; Kim, J.H.; Hong, S.H.; Park, J.E.; et al. Upregulation of AQP4 Improves Blood–Brain Barrier Integrity and Perihematomal Edema Following Intracerebral Hemorrhage. Neurotherapeutics 2021, 18, 2692–2706. [Google Scholar] [CrossRef]
- Zhao, F.; Deng, J.; Xu, X.; Cao, F.; Lu, K.; Li, D.; Cheng, X.; Wang, X.; Zhao, Y. Aquaporin-4 deletion ameliorates hypoglycemia-induced BBB permeability by inhibiting inflammatory responses. J. Neuroinflamm. 2018, 15, 157. [Google Scholar] [CrossRef]
- Podjaski, C.; Alvarez, J.I.; Bourbonniere, L.; Larouche, S.; Terouz, S.; Bin, J.M.; Lécuyer, M.-A.; Saint-Laurent, O.; Larochelle, C.; Darlington, P.J.; et al. Netrin 1 regulates blood–brain barrier function and neuroinflammation. Brain 2015, 138, 1598–1612. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.-Y.; Gao, F.; Yang, X.-M.; Qin, X.; Chen, G.-Z.; Li, D.; Dang, B.-Q.; Chen, G. Matrix metalloproteinase-9 regulates the blood brain barrier via the hedgehog pathway in a rat model of traumatic brain injury. Brain Res. 2020, 1727, 146553. [Google Scholar] [CrossRef] [PubMed]
- Goldeman, C.; Ozgür, B.; Brodin, B. Culture-induced changes in mRNA expression levels of efflux and SLC-transporters in brain endothelial cells. Fluids Barriers CNS 2020, 17, 32. [Google Scholar] [CrossRef] [PubMed]
- Jovic, D.; Liang, X.; Zeng, H.; Lin, L.; Xu, F.; Luo, Y. Single-cell RNA sequencing technologies and applications: A brief overview. Clin. Transl. Med. 2022, 12, e694. [Google Scholar] [CrossRef]
- Lei, Y.; Tang, R.; Xu, J.; Wang, W.; Zhang, B.; Liu, J.; Yu, X.; Shi, S. Applications of single-cell sequencing in cancer research: Progress and perspectives. J. Hematol. Oncol. 2021, 14, 91. [Google Scholar] [CrossRef]
- Kotliar, D.; Veres, A.; Nagy, M.A.; Tabrizi, S.; Hodis, E.; Melton, D.A.; Sabeti, P.C. Identifying gene expression programs of cell-type identity and cellular activity with single-cell RNA-Seq. Elife 2019, 8, e43803. [Google Scholar] [CrossRef]
- Kadiyska, T.; Tourtourikov, I.; Dabchev, K.; Cherneva, R.; Stoynev, N.; Hadjiolova, R.; Mitev, V.; Spandidos, D.; Adamaki, M.; Zoumpourlis, V. Role of endothelial dysfunction in the severity of COVID-19 infection (Review). Mol. Med. Rep. 2022, 26, 351. [Google Scholar] [CrossRef]
- Ruhl, L.; Pink, I.; Kühne, J.F.; Beushausen, K.; Keil, J.; Christoph, S.; Sauer, A.; Boblitz, L.; Schmidt, J.; David, S.; et al. Endothelial dysfunction contributes to severe COVID-19 in combination with dysregulated lymphocyte responses and cytokine networks. Signal Transduct. Target. Ther. 2021, 6, 418. [Google Scholar] [CrossRef]
- Wang, S.; Yao, X.; Ma, S.; Ping, Y.; Fan, Y.; Sun, S.; He, Z.; Shi, Y.; Sun, L.; Xiao, S.; et al. A single-cell transcriptomic landscape of the lungs of patients with COVID-19. Nat. Cell Biol. 2021, 23, 1314–1328. [Google Scholar] [CrossRef]
- Delorey, T.M.; Ziegler, C.G.K.; Heimberg, G.; Normand, R.; Yang, Y.; Segerstolpe, A.; Abbondanza, D.; Fleming, S.J.; Subramanian, A.; Montoro, D.T.; et al. A single-cell and spatial atlas of autopsy tissues reveals pathology and cellular targets of SARS-CoV-2. bioRxiv 2021. [Google Scholar] [CrossRef]
- Ahern, D.J.; Ai, Z.; Ainsworth, M.; Allan, C.; Allcock, A.; Angus, B.; Ansari, M.A.; Arancibia-Cárcamo, C.V.; Aschenbrenner, D.; Attar, M.; et al. A blood atlas of COVID-19 defines hallmarks of disease severity and specificity. Cell 2022, 185, 916–938.e58. [Google Scholar] [CrossRef]
- Chen, J.; Tan, R.; Mo, Y.; Zhang, J. The blood-brain barrier in health, neurological diseases, and COVID-19. Fundam. Res. 2022, 2, 817–826. [Google Scholar] [CrossRef]
- Hashimoto, R.; Takahashi, J.; Shirakura, K.; Funatsu, R.; Kosugi, K.; Deguchi, S.; Yamamoto, M.; Tsunoda, Y.; Morita, M.; Muraoka, K.; et al. SARS-CoV-2 disrupts respiratory vascular barriers by suppressing Claudin-5 expression. Sci. Adv. 2022, 8, eabo6783. [Google Scholar] [CrossRef] [PubMed]
- Maldonado, F.; Morales, D.; Díaz-Papapietro, C.; Valdés, C.; Fernandez, C.; Valls, N.; Lazo, M.; Espinoza, C.; González, R.; Gutiérrez, R.; et al. Relationship Between Endothelial and Angiogenesis Biomarkers Envisage Mortality in a Prospective Cohort of COVID-19 Patients Requiring Respiratory Support. Front. Med. 2022, 9, 654. [Google Scholar] [CrossRef] [PubMed]
- Fabiś, M.; Gorzelak-Pabiś, P.; Satała, J.; Pawlos, A.; Fabiś, J.; Broncel, M. The relationship between COVID-19 severity, markers of endothelial impairment and Simple Covid Risk Index. Pol. Arch. Intern. Med. 2022, 132, 16348. [Google Scholar] [CrossRef] [PubMed]
- Ramírez-Martínez, G.; Jiménez-Álvarez, L.A.; Cruz-Lagunas, A.; Ignacio-Cortés, S.; Gómez-García, I.A.; Rodríguez-Reyna, T.S.; Choreño-Parra, J.A.; Zúñiga, J. Possible Role of Matrix Metalloproteinases and TGF-β in COVID-19 Severity and Sequelae. J. Interf. Cytokine Res. 2022, 42, 352–368. [Google Scholar] [CrossRef]
- Nagashima, S.; Mendes, M.C.; Camargo Martins, A.P.; Borges, N.H.; Godoy, T.M.; Miggiolaro, A.F.R.D.S.; da Silva Dezidério, F.; Machado-Souza, C.; de Noronha, L. Endothelial Dysfunction and Thrombosis in Patients with COVID-19—Brief Report. Arterioscler. Thromb. Vasc. Biol. 2020, 40, 2404–2407. [Google Scholar] [CrossRef]
- Willems, L.H.; Nagy, M.; ten Cate, H.; Spronk, H.M.H.; Groh, L.A.; Leentjens, J.; Janssen, N.A.F.; Netea, M.G.; Thijssen, D.H.J.; Hannink, G.; et al. Sustained inflammation, coagulation activation and elevated endothelin-1 levels without macrovascular dysfunction at 3 months after COVID-19. Thromb. Res. 2022, 209, 106–114. [Google Scholar] [CrossRef]
- Bayrakci, N.; Ozkan, G.; Mutlu, L.C.; Erdem, L.; Yildirim, I.; Gulen, D.; Celikkol, A. Relationship between serum soluble endothelial protein C receptor level and COVID-19 findings. Blood Coagul. Fibrinol. 2021, 32, 550–555. [Google Scholar] [CrossRef]
- Flaumenhaft, R.; Enjyoji, K.; Schmaier, A.A. Vasculopathy in COVID-19. Blood 2022, 140, 222–235. [Google Scholar] [CrossRef]
- Price, D.R.; Benedetti, E.; Hoffman, K.L.; Gomez-Escobar, L.; Alvarez-Mulett, S.; Capili, A.; Sarwath, H.; Parkhurst, C.N.; Lafond, E.; Weidman, K.; et al. Angiopoietin 2 Is Associated with Vascular Necroptosis Induction in Coronavirus Disease 2019 Acute Respiratory Distress Syndrome. Am. J. Pathol. 2022, 192, 1001–1015. [Google Scholar] [CrossRef] [PubMed]
- Raica, M.; Cimpean, A.M. Platelet-Derived Growth Factor (PDGF)/PDGF Receptors (PDGFR) Axis as Target for Antitumor and Antiangiogenic Therapy. Pharmaceuticals 2010, 3, 572–599. [Google Scholar] [CrossRef]
- Canale, M.P.; Menghini, R.; Martelli, E.; Federici, M. COVID-19–Associated Endothelial Dysfunction and Microvascular Injury. Card. Electrophysiol. Clin. 2022, 14, 21–28. [Google Scholar] [CrossRef] [PubMed]
- Kalejaiye, T.D.; Bhattacharya, R.; Burt, M.A.; Travieso, T.; Okafor, A.E.; Mou, X.; Blasi, M.; Musah, S. SARS-CoV-2 Employ BSG/CD147 and ACE2 Receptors to Directly Infect Human Induced Pluripotent Stem Cell-Derived Kidney Podocytes. Front. Cell Dev. Biol. 2022, 10, 855340. [Google Scholar] [CrossRef]
- Cantuti-Castelvetri, L.; Ojha, R.; Pedro, L.D.; Djannatian, M.; Franz, J.; Kuivanen, S.; van der Meer, F.; Kallio, K.; Kaya, T.; Anastasina, M.; et al. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science 2020, 370, 856–860. [Google Scholar] [CrossRef]
- Li, Z.; Zhang, Y.; Fang, J.; Xu, Z.; Zhang, H.; Mao, M.; Chen, Y.; Zhang, L.; Pian, C. NcPath: A novel platform for visualization and enrichment analysis of human non-coding RNA and KEGG signaling pathways. Bioinformatics 2023, 39, btac812. [Google Scholar] [CrossRef]
- Lu, L.; Liu, L.-P.; Gui, R.; Dong, H.; Su, Y.-R.; Zhou, X.-H.; Liu, F.-X. Discovering common pathogenetic processes between COVID-19 and sepsis by bioinformatics and system biology approach. Front. Immunol. 2022, 13, 975848. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S.; Furumichi, M.; Tanabe, M.; Hirakawa, M. KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res. 2010, 38, D355–D360. [Google Scholar] [CrossRef]
- Taheri, G.; Habibi, M. Comprehensive analysis of pathways in Coronavirus 2019 (COVID-19) using an unsupervised machine learning method. Appl. Soft Comput. 2022, 128, 109510. [Google Scholar] [CrossRef]
- Shagdarsuren, E.; Bidzhekov, K.; Djalali-Talab, Y.; Liehn, E.A.; Hristov, M.; Matthijsen, R.A.; Buurman, W.A.; Zernecke, A.; Weber, C. C1-Esterase Inhibitor Protects Against Neointima Formation After Arterial Injury in Atherosclerosis-Prone Mice. Circulation 2008, 117, 70–78. [Google Scholar] [CrossRef]
- Kilgore, K.S.; Flory, C.M.; Miller, B.F.; Evans, V.M.; Warren, J.S. The membrane attack complex of complement induces interleukin-8 and monocyte chemoattractant protein-1 secretion from human umbilical vein endothelial cells. Am. J. Pathol. 1996, 149, 953–961. [Google Scholar] [PubMed]
- Neftel, C.; Laffy, J.; Filbin, M.G.; Hara, T.; Shore, M.E.; Rahme, G.J.; Richman, A.R.; Silverbush, D.; Shaw, M.L.; Hebert, C.M.; et al. An Integrative Model of Cellular States, Plasticity, and Genetics for Glioblastoma. Cell 2019, 178, 835–849.e21. [Google Scholar] [CrossRef] [PubMed]
- Ma, S.; Li, Q.; Peng, J.; Zhouwen, J.; Zhang, D.; Zhang, C.; Jiang, W.; Jia, W. CLDN5 affects lncRNAs acting as ceRNA dynamics contributing to regulating blood-brain barrier permeability in tumor brain metastasis. Oncol. Rep. 2018, 39, 1441–1453. [Google Scholar] [CrossRef]
- Karnati, H.; Panigrahi, M.; Shaik, N.; Greig, N.; Bagadi, S.; Kamal, M.; Kapalavayi, N. Down Regulated Expression of Claudin-1 and Claudin-5 and Up Regulation of β-Catenin: Association with Human Glioma Progression. CNS Neurol. Disord.—Drug Targets 2014, 13, 1413–1426. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Xie, Y.; Tang, J.; Liu, B.; Luo, Y.; He, Q.; Zhang, L.; Xin, L.; Wang, J.; Wang, S.; et al. Uncovering a Distinct Gene Signature in Endothelial Cells Associated With Contrast Enhancement in Glioblastoma. Front. Oncol. 2021, 11, 683367. [Google Scholar] [CrossRef]
- Wang, X.; Zhou, R.; Xiong, Y.; Zhou, L.; Yan, X.; Wang, M.; Li, F.; Xie, C.; Zhang, Y.; Huang, Z.; et al. Sequential fate-switches in stem-like cells drive the tumorigenic trajectory from human neural stem cells to malignant glioma. Cell Res. 2021, 31, 684–702. [Google Scholar] [CrossRef]
- Mangogna, A.; Belmonte, B.; Agostinis, C.; Zacchi, P.; Iacopino, D.G.; Martorana, A.; Rodolico, V.; Bonazza, D.; Zanconati, F.; Kishore, U.; et al. Prognostic Implications of the Complement Protein C1q in Gliomas. Front. Immunol. 2019, 10, 2366. [Google Scholar] [CrossRef]
- Zhu, H.; Yu, X.; Zhang, S.; Shu, K. Targeting the Complement Pathway in Malignant Glioma Microenvironments. Front. Cell Dev. Biol. 2021, 9, 657472. [Google Scholar] [CrossRef]
- Bouwens van der Vlis, T.A.M.; Kros, J.M.; Mustafa, D.A.M.; van Wijck, R.T.A.; Ackermans, L.; van Hagen, P.M.; van der Spek, P.J. The complement system in glioblastoma multiforme. Acta Neuropathol. Commun. 2018, 6, 91. [Google Scholar] [CrossRef]
- Prasad, M.; Leon, M.; Lerman, L.O.; Lerman, A. Viral Endothelial Dysfunction: A Unifying Mechanism for COVID-19. Mayo Clin. Proc. 2021, 96, 3099–3108. [Google Scholar] [CrossRef]
- Sinha, S.; Kundu, C.N. Cancer and COVID-19: Why are cancer patients more susceptible to COVID-19? Med. Oncol. 2021, 38, 101. [Google Scholar] [CrossRef] [PubMed]
- Otifi, H.M.; Adiga, B.K. Endothelial Dysfunction in COVID-19 Infection. Am. J. Med. Sci. 2022, 363, 281–287. [Google Scholar] [CrossRef] [PubMed]
- Khan, I.; Hatiboglu, M.A. Can COVID-19 induce glioma tumorogenesis through binding cell receptors? Med. Hypotheses 2020, 144, 110009. [Google Scholar] [CrossRef] [PubMed]
- Lv, D.; Gimple, R.C.; Zhong, C.; Wu, Q.; Yang, K.; Prager, B.C.; Godugu, B.; Qiu, Z.; Zhao, L.; Zhang, G.; et al. PDGF signaling inhibits mitophagy in glioblastoma stem cells through N6-methyladenosine. Dev. Cell 2022, 57, 1466–1481.e6. [Google Scholar] [CrossRef]
- Gangoso, E.; Southgate, B.; Bradley, L.; Rus, S.; Galvez-Cancino, F.; McGivern, N.; Güç, E.; Kapourani, C.-A.; Byron, A.; Ferguson, K.M.; et al. Glioblastomas acquire myeloid-affiliated transcriptional programs via epigenetic immunoediting to elicit immune evasion. Cell 2021, 184, 2454–2470.e26. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Thakur, A.; Liang, L.; Banerjee, S.; Zhang, K. Single-Cell Transcriptomics Reveals Evidence of Endothelial Dysfunction in the Brains of COVID-19 Patients with Implications for Glioblastoma Progression. Brain Sci. 2023, 13, 762. https://doi.org/10.3390/brainsci13050762
Thakur A, Liang L, Banerjee S, Zhang K. Single-Cell Transcriptomics Reveals Evidence of Endothelial Dysfunction in the Brains of COVID-19 Patients with Implications for Glioblastoma Progression. Brain Sciences. 2023; 13(5):762. https://doi.org/10.3390/brainsci13050762
Chicago/Turabian StyleThakur, Abhimanyu, Lifan Liang, Sourav Banerjee, and Kui Zhang. 2023. "Single-Cell Transcriptomics Reveals Evidence of Endothelial Dysfunction in the Brains of COVID-19 Patients with Implications for Glioblastoma Progression" Brain Sciences 13, no. 5: 762. https://doi.org/10.3390/brainsci13050762




