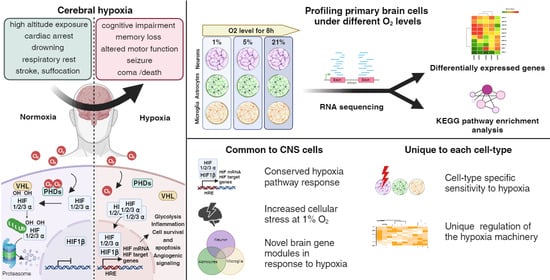
Transcriptional Responses of Different Brain Cell Types to Oxygen Decline
Abstract
:1. Introduction
2. Methods
2.1. Animals
2.2. Primary Mouse Brain Cell Cultures
2.3. Hypoxia Treatments
2.4. Library Preparation and Transcriptome Sequencing
2.5. RNA-Seq Analysis
2.6. Principal Component Analysis
2.7. Pathway Analysis
2.8. Statistics
3. Results
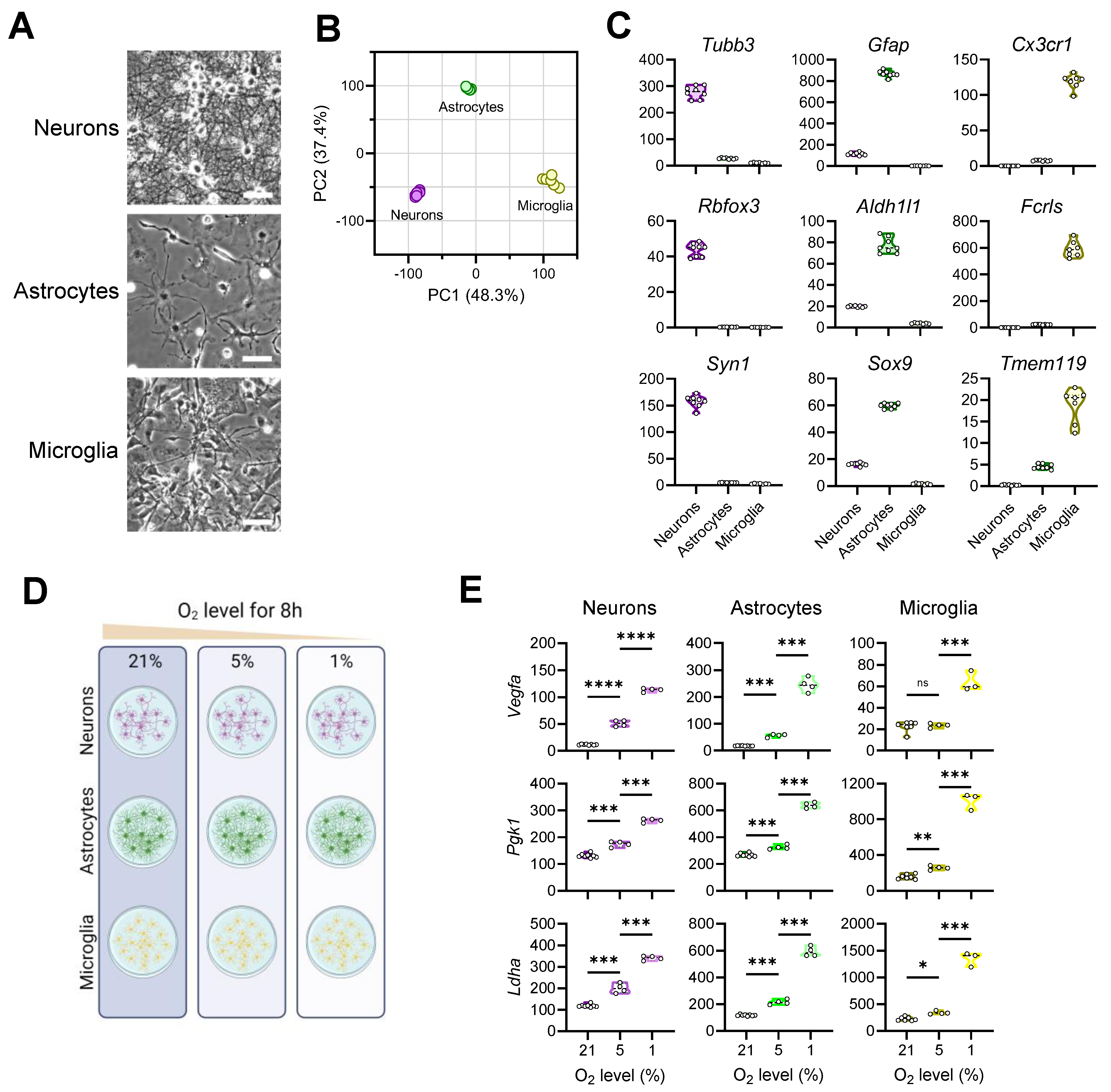
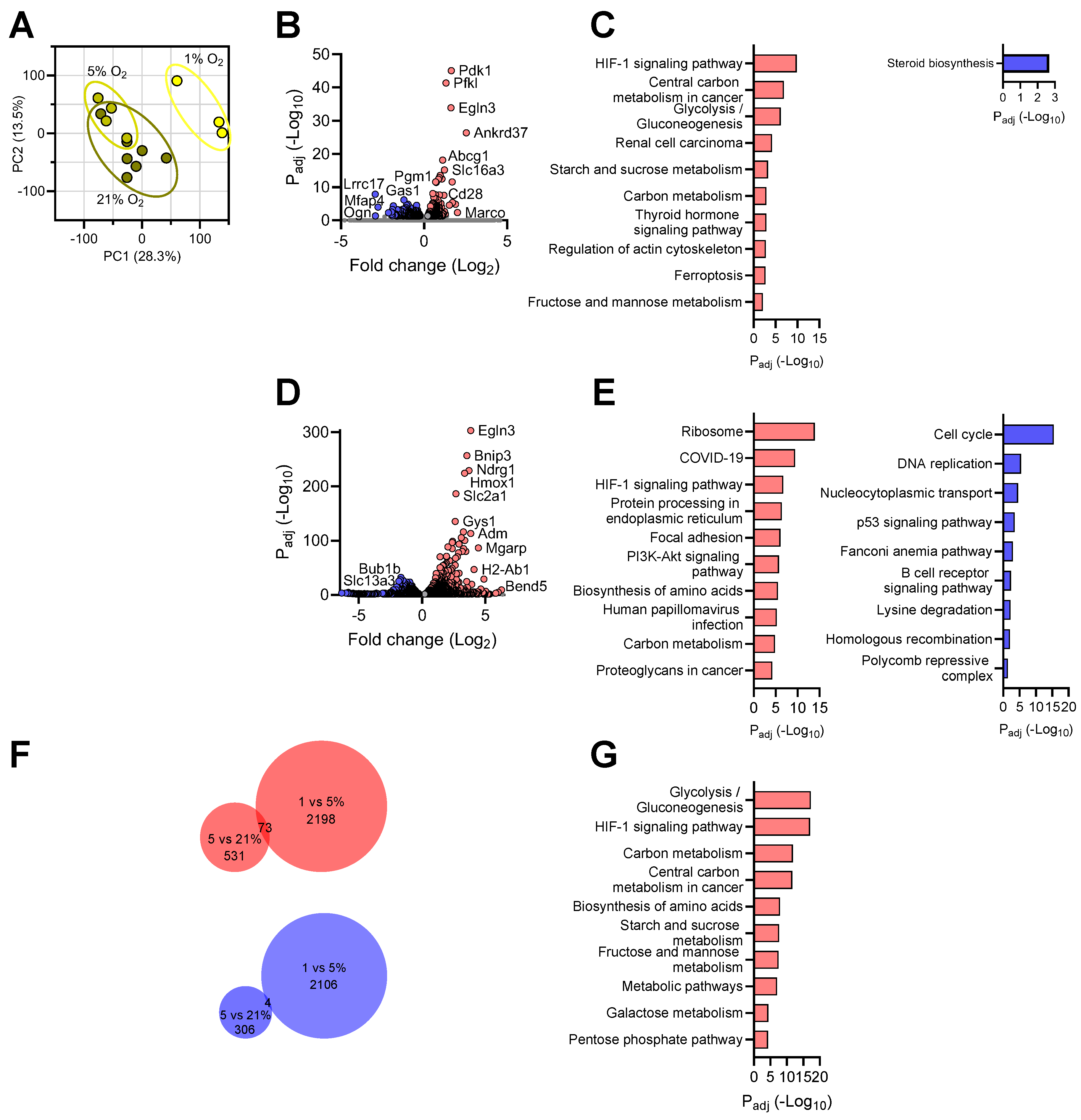
3.1. Exposing Primary Mouse Brain Cells to Different Oxygen Levels
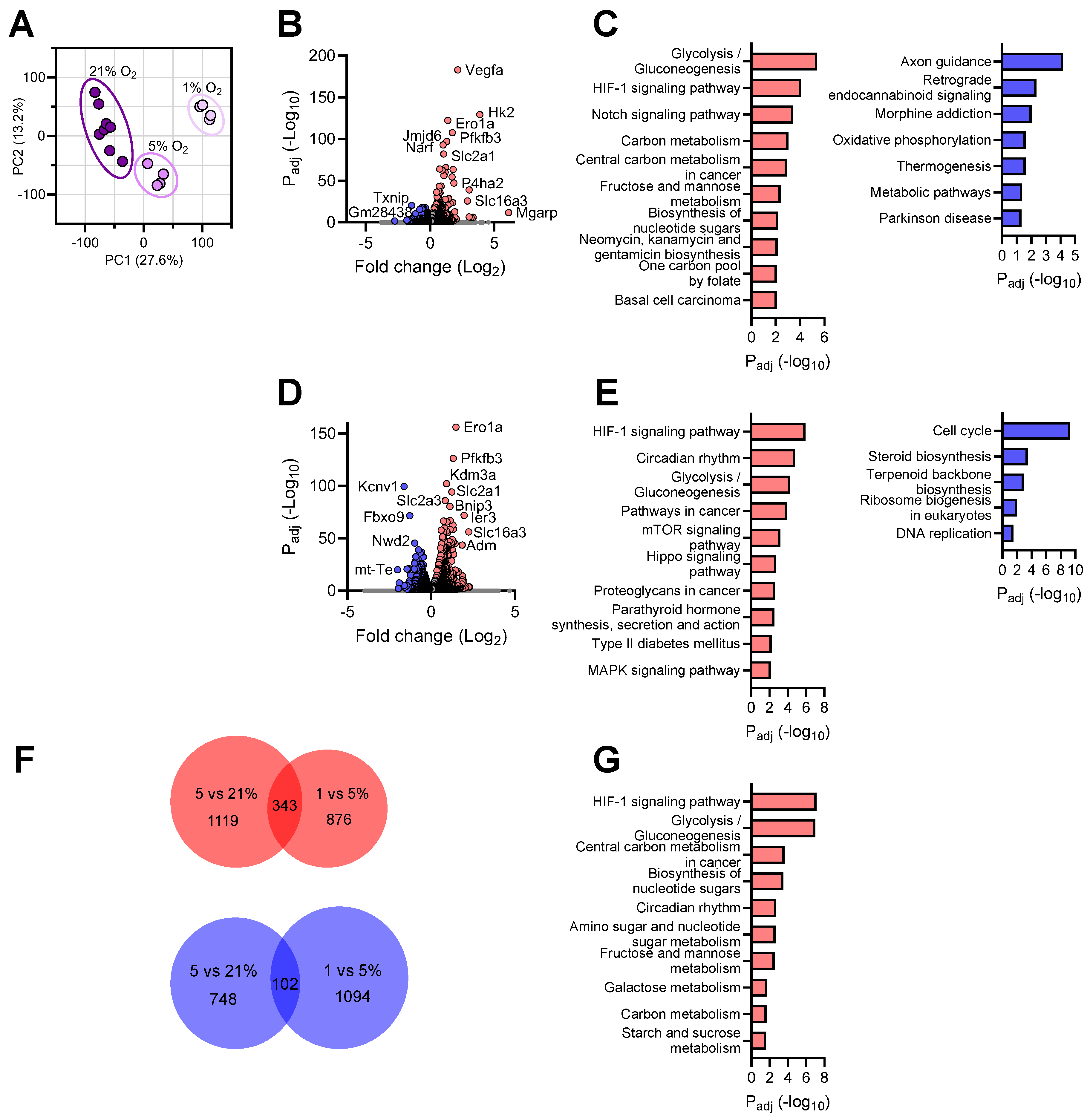
3.2. Transcriptional Responses of Neurons to Reduced Oxygen Availability
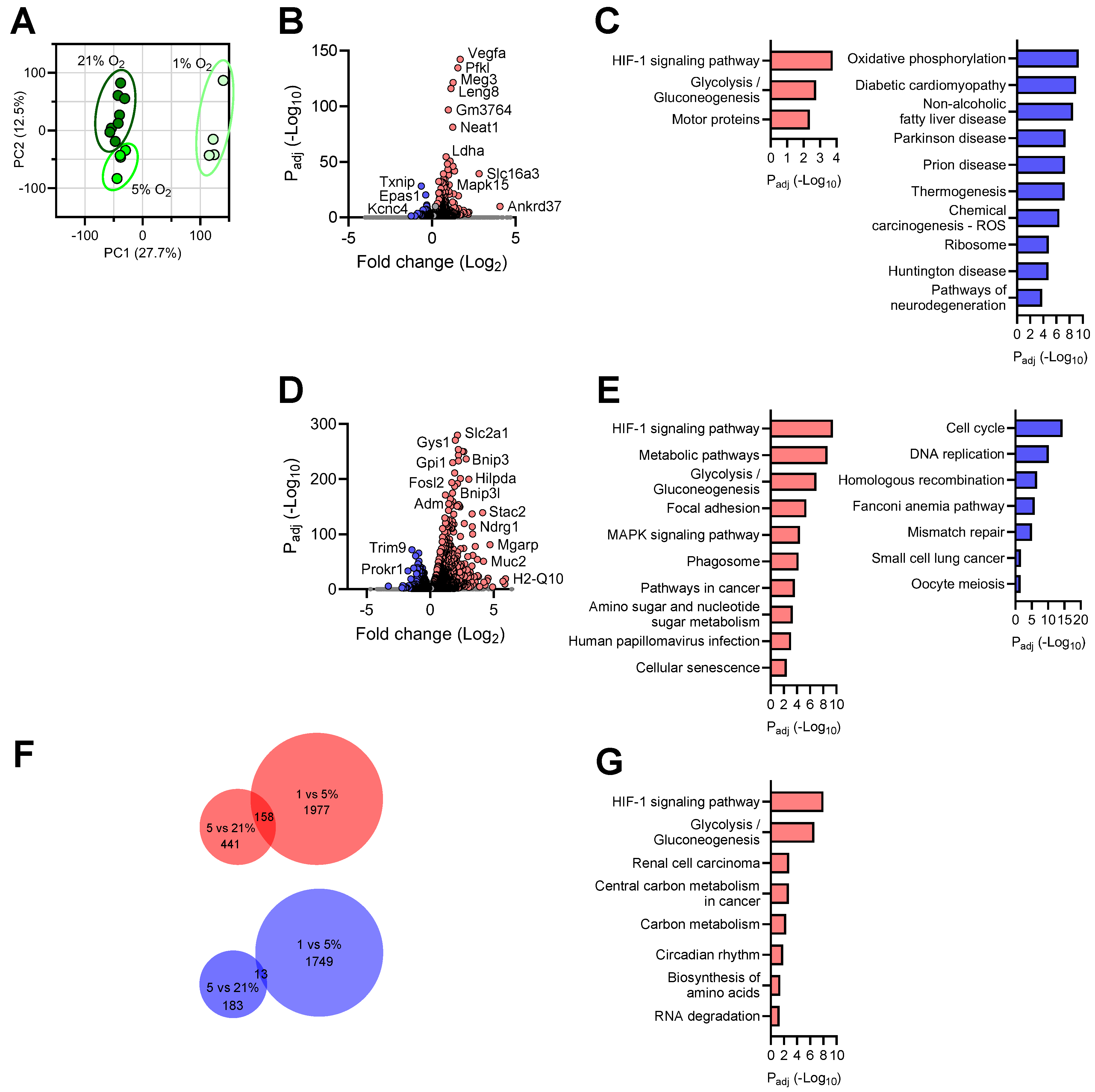
3.3. Transcriptional Responses of Astrocytes to Reduced Oxygen Availability
3.4. Transcriptional Responses of Microglia to Reduced Oxygen Availability
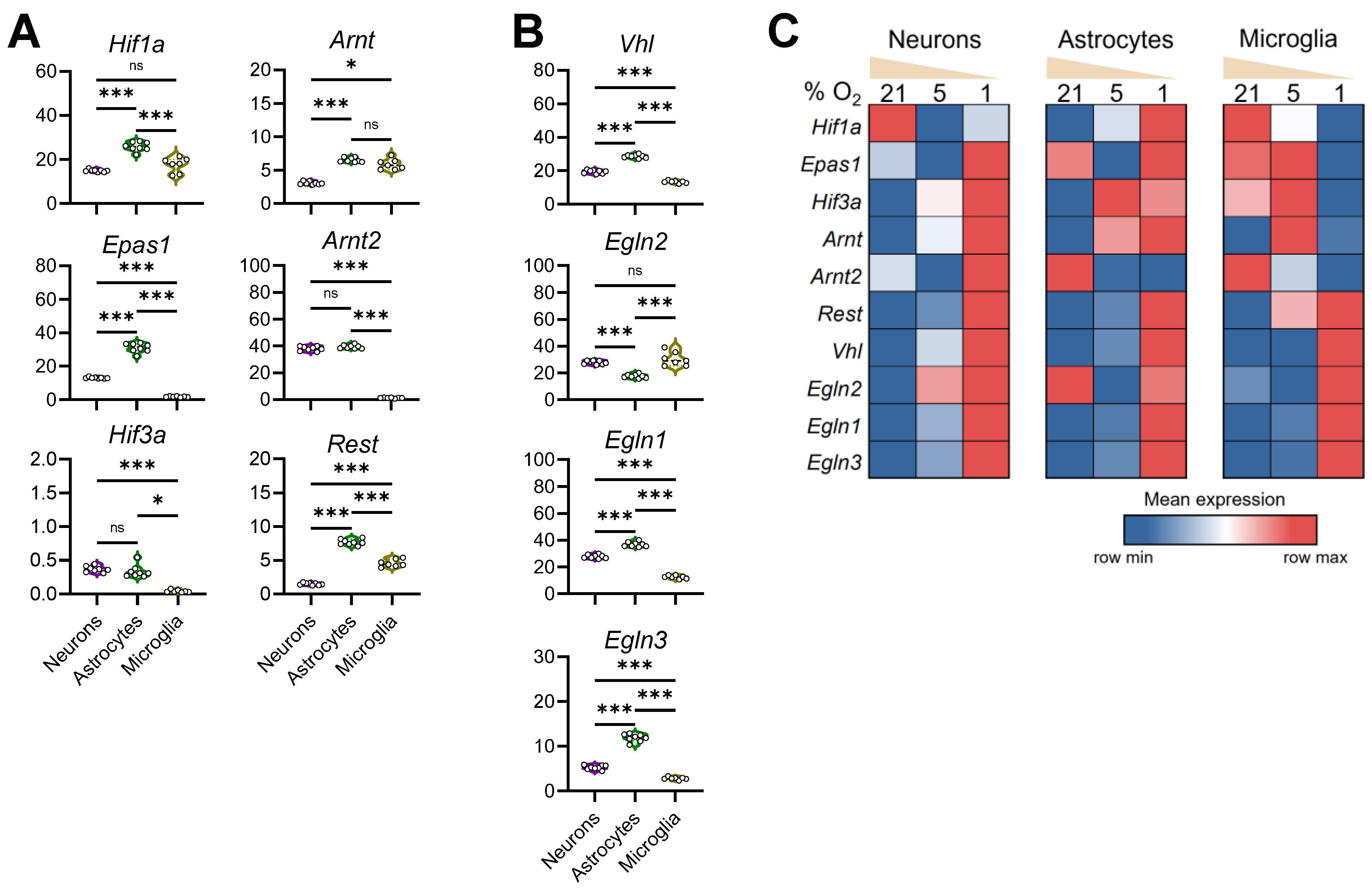
3.5. Configuration of the Hypoxia Response Machinery in Mouse Brain Cells
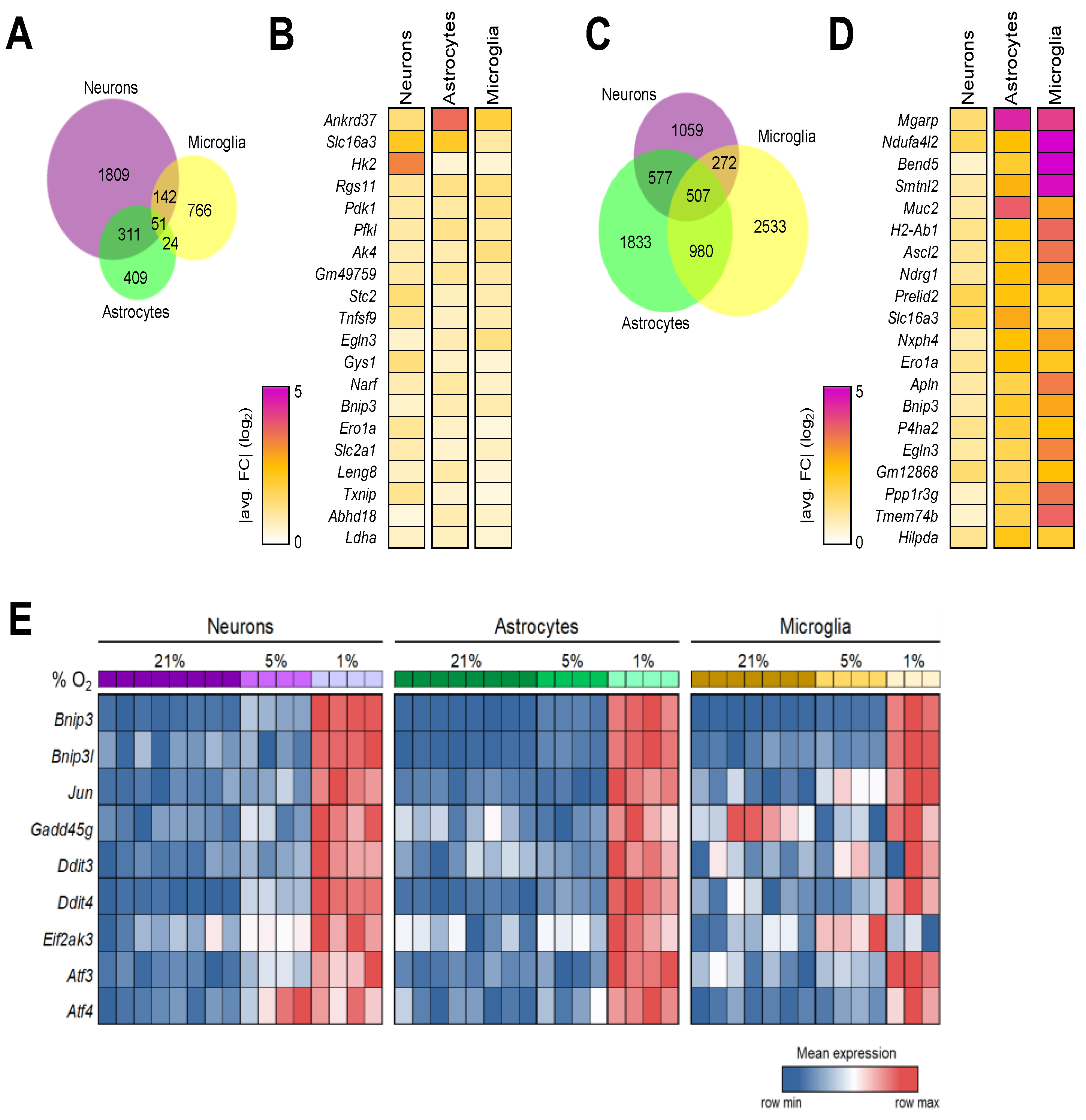
3.6. Shared Brain Cell Responses to Oxygen Decline
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bunn, H.F.; Poyton, R.O. Oxygen sensing and molecular adaptation to hypoxia. Physiol. Rev. 1996, 76, 839–885. [Google Scholar] [CrossRef]
- Kumar, H.; Choi, D.K. Hypoxia Inducible Factor Pathway and Physiological Adaptation: A Cell Survival Pathway? Mediat. Inflamm. 2015, 2015, 584758. [Google Scholar] [CrossRef] [PubMed]
- Ratcliffe, P.J. Oxygen sensing and hypoxia signalling pathways in animals: The implications of physiology for cancer. J. Physiol. 2013, 591, 2027–2042. [Google Scholar] [CrossRef] [PubMed]
- Kaelin, W.G., Jr.; Ratcliffe, P.J. Oxygen sensing by metazoans: The central role of the HIF hydroxylase pathway. Mol. Cell 2008, 30, 393–402. [Google Scholar] [CrossRef] [PubMed]
- Prabhakar, N.R.; Semenza, G.L. Adaptive and maladaptive cardiorespiratory responses to continuous and intermittent hypoxia mediated by hypoxia-inducible factors 1 and 2. Physiol. Rev. 2012, 92, 967–1003. [Google Scholar] [CrossRef] [PubMed]
- Semenza, G.L. Hypoxia-inducible factors: Mediators of cancer progression and targets for cancer therapy. Trends Pharmacol. Sci. 2012, 33, 207–214. [Google Scholar] [CrossRef] [PubMed]
- Eltzschig, H.K.; Carmeliet, P. Hypoxia and inflammation. New Engl. J. Med. 2011, 364, 656–665. [Google Scholar] [CrossRef] [PubMed]
- Colgan, S.P.; Furuta, G.T.; Taylor, C.T. Hypoxia and Innate Immunity: Keeping Up with the HIFsters. Annu. Rev. Immunol. 2020, 38, 341–363. [Google Scholar] [CrossRef] [PubMed]
- Luo, Z.; Tian, M.; Yang, G.; Tan, Q.; Chen, Y.; Li, G.; Zhang, Q.; Li, Y.; Wan, P.; Wu, J. Hypoxia signaling in human health and diseases: Implications and prospects for therapeutics. Signal Transduct. Target. Ther. 2022, 7, 218. [Google Scholar] [CrossRef] [PubMed]
- Palazon, A.; Goldrath, A.W.; Nizet, V.; Johnson, R.S. HIF transcription factors, inflammation, and immunity. Immunity 2014, 41, 518–528. [Google Scholar] [CrossRef]
- Wang, G.L.; Semenza, G.L. General involvement of hypoxia-inducible factor 1 in transcriptional response to hypoxia. Proc. Natl. Acad. Sci. USA 1993, 90, 4304–4308. [Google Scholar] [CrossRef]
- Schodel, J.; Oikonomopoulos, S.; Ragoussis, J.; Pugh, C.W.; Ratcliffe, P.J.; Mole, D.R. High-resolution genome-wide mapping of HIF-binding sites by ChIP-seq. Blood 2011, 117, e207–e217. [Google Scholar] [CrossRef]
- Downes, N.L.; Laham-Karam, N.; Kaikkonen, M.U.; Yla-Herttuala, S. Differential but Complementary HIF1alpha and HIF2alpha Transcriptional Regulation. Mol. Ther. 2018, 26, 1735–1745. [Google Scholar] [CrossRef] [PubMed]
- Strowitzki, M.J.; Cummins, E.P.; Taylor, C.T. Protein Hydroxylation by Hypoxia-Inducible Factor (HIF) Hydroxylases: Unique or Ubiquitous? Cells 2019, 8, 384. [Google Scholar] [CrossRef] [PubMed]
- Kaelin, W.G., Jr. Von Hippel-Lindau disease: Insights into oxygen sensing, protein degradation, and cancer. J. Clin. Investig. 2022, 132, 170–181. [Google Scholar] [CrossRef] [PubMed]
- Masamoto, K.; Tanishita, K. Oxygen transport in brain tissue. J. Biomech. Eng. 2009, 131, 074002. [Google Scholar] [CrossRef] [PubMed]
- Mukamel, E.A.; Ngai, J. Perspectives on defining cell types in the brain. Curr. Opin. Neurobiol. 2019, 56, 61–68. [Google Scholar] [CrossRef] [PubMed]
- Kreiman, G.; Koch, C.; Fried, I. Imagery neurons in the human brain. Nature 2000, 408, 357–361. [Google Scholar] [CrossRef] [PubMed]
- Vasile, F.; Dossi, E.; Rouach, N. Human astrocytes: Structure and functions in the healthy brain. Brain Struct. Funct. 2017, 222, 2017–2029. [Google Scholar] [CrossRef] [PubMed]
- Tremblay, M.-È.; Stevens, B.; Sierra, A.; Wake, H.; Bessis, A.; Nimmerjahn, A. The Role of Microglia in the Healthy Brain. J. Neurosci. 2011, 31, 16064–16069. [Google Scholar] [CrossRef] [PubMed]
- Halterman, M.W.; Gill, M.; DeJesus, C.; Ogihara, M.; Schor, N.F.; Federoff, H.J. The endoplasmic reticulum stress response factor CHOP-10 protects against hypoxia-induced neuronal death. J. Biol. Chem. 2010, 285, 21329–21340. [Google Scholar] [CrossRef] [PubMed]
- Nieber, K. Hypoxia and neuronal function under in vitro conditions. Pharmacol. Ther. 1999, 82, 71–86. [Google Scholar] [CrossRef] [PubMed]
- Sher, P.K. Chronic hypoxia in neuronal cell culture metabolic consequences. Brain Dev. 1990, 12, 293–300. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Roy, E.R.; Wang, Y.; Watkins, T.; Cao, W. DLK-MAPK Signaling Coupled with DNA Damage Promotes Intrinsic Neurotoxicity Associated with Non-Mutated Tau. Mol. Neurobiol. 2023. [Google Scholar] [CrossRef] [PubMed]
- Roy, E.R.; Wang, B.; Wan, Y.-W.; Chiu, G.; Cole, A.; Yin, Z.; Propson, N.E.; Xu, Y.; Jankowsky, J.L.; Liu, Z.; et al. Type I interferon response drives neuroinflammation and synapse loss in Alzheimer disease. J. Clin. Investig. 2020, 130, 1912–1930. [Google Scholar] [CrossRef] [PubMed]
- Raudvere, U.; Kolberg, L.; Kuzmin, I.; Arak, T.; Adler, P.; Peterson, H.; Vilo, J. g:Profiler: A web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 2019, 47, W191–W198. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Yotnda, P. Induction and testing of hypoxia in cell culture. J. Vis. Exp. 2011, 54, e2899. [Google Scholar]
- Wang, G.L.; Jiang, B.H.; Rue, E.A.; Semenza, G.L. Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc. Natl. Acad. Sci. USA 1995, 92, 5510–5514. [Google Scholar] [CrossRef] [PubMed]
- Harrison, H.; Rogerson, L.; Gregson, H.J.; Brennan, K.R.; Clarke, R.B.; Landberg, G. Contrasting Hypoxic Effects on Breast Cancer Stem Cell Hierarchy Is Dependent on ER-α Status. Cancer Res. 2013, 73, 1420–1433. [Google Scholar] [CrossRef]
- Wigfield, S.M.; Winter, S.C.; Giatromanolaki, A.; Taylor, J.; Koukourakis, M.L.; Harris, A.L. PDK-1 regulates lactate production in hypoxia and is associated with poor prognosis in head and neck squamous cancer. Br. J. Cancer 2008, 98, 1975–1984. [Google Scholar] [CrossRef]
- Lee, D.C.; Sohn, H.A.; Park, Z.-Y.; Oh, S.; Kang, Y.K.; Lee, K.-M.; Kang, M.; Jang, Y.J.; Yang, S.-J.; Hong, Y.K.; et al. A Lactate-Induced Response to Hypoxia. Cell 2015, 161, 595–609. [Google Scholar] [CrossRef]
- Krock, B.L.; Skuli, N.; Simon, M.C. Hypoxia-induced angiogenesis: Good and evil. Genes Cancer 2011, 2, 1117–1133. [Google Scholar] [CrossRef] [PubMed]
- Newman, A.C.; Maddocks, O.D.K. One-carbon metabolism in cancer. Br. J. Cancer 2017, 116, 1499–1504. [Google Scholar] [CrossRef] [PubMed]
- Infantino, V.; Santarsiero, A.; Convertini, P.; Todisco, S.; Iacobazzi, V. Cancer Cell Metabolism in Hypoxia: Role of HIF-1 as Key Regulator and Therapeutic Target. Int. J. Mol. Sci. 2021, 22, 5703. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, K.A.; Murray, A.J.; Simonson, T.S. Notch Signaling and Cross-Talk in Hypoxia: A Candidate Pathway for High-Altitude Adaptation. Life 2022, 12, 437. [Google Scholar] [CrossRef] [PubMed]
- Gustafsson, M.V.; Zheng, X.; Pereira, T.; Gradin, K.; Jin, S.; Lundkvist, J.; Ruas, J.L.; Poellinger, L.; Lendahl, U.; Bondesson, M. Hypoxia Requires Notch Signaling to Maintain the Undifferentiated Cell State. Dev. Cell 2005, 9, 617–628. [Google Scholar] [CrossRef] [PubMed]
- Bonkowsky, J.L.; Son, J.-H. Hypoxia and connectivity in the developing vertebrate nervous system. Dis. Models Mech. 2018, 11, dmm037127. [Google Scholar] [CrossRef] [PubMed]
- Pan, C.-L.; Garriga, G. Fresh air is good for nerves: Hypoxia disturbs axon guidance. Nat. Neurosci. 2008, 11, 859–861. [Google Scholar] [CrossRef] [PubMed]
- Manella, G.; Aviram, R.; Bolshette, N.; Muvkadi, S.; Golik, M.; Smith, D.F.; Asher, G. Hypoxia induces a time- and tissue-specific response that elicits intertissue circadian clock misalignment. Proc. Natl. Acad. Sci. USA 2020, 117, 779–786. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Tang, D.; Liu, N.; Huang, H.; Li, Y.; Ma, Z.; Zhao, H.; Chen, P.; Qi, X.; Zhang, E.E. Reciprocal Regulation between the Circadian Clock and Hypoxia Signaling at the Genome Level in Mammals. Cell Metab. 2017, 25, 73–85. [Google Scholar] [CrossRef]
- Druker, J.; Wilson, J.W.; Child, F.; Shakir, D.; Fasanya, T.; Rocha, S. Role of Hypoxia in the Control of the Cell Cycle. Int. J. Mol. Sci. 2021, 22, 4874. [Google Scholar] [CrossRef] [PubMed]
- Hubbi, M.E.; Semenza, G.L. Regulation of cell proliferation by hypoxia-inducible factors. Am. J. Physiol.-Cell Physiol. 2015, 309, C775–C782. [Google Scholar] [CrossRef] [PubMed]
- Batie, M.; Frost, J.; Frost, M.; Wilson, J.W.; Schofield, P.; Rocha, S. Hypoxia induces rapid changes to histone methylation and reprograms chromatin. Science 2019, 363, 1222–1226. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.; Ure, K.; Ding, P.; Nashaat, M.; Yuan, L.; Ma, J.; Hammer, R.E.; Hsieh, J. The Master Negative Regulator REST/NRSF Controls Adult Neurogenesis by Restraining the Neurogenic Program in Quiescent Stem Cells. J. Neurosci. 2011, 31, 9772–9786. [Google Scholar] [CrossRef] [PubMed]
- da Luz Sousa Fialho, M.; Purnama, U.; Dennis, K.M.J.H.; Aparicio, C.N.M.; Castro-Guarda, M.; Massourides, E.; Tyler, D.J.; Carr, C.A.; Heather, L.C. Activation of HIF1α Rescues the Hypoxic Response and Reverses Metabolic Dysfunction in the Diabetic Heart. Diabetes 2021, 70, 2518–2531. [Google Scholar] [CrossRef] [PubMed]
- Isaza, S.C.; Del Pozo-Maroto, E.; Domínguez-Alcón, L.; Elbouayadi, L.; González-Rodríguez, Á.; García-Monzón, C. Hypoxia and Non-alcoholic Fatty Liver Disease. Front. Med. 2020, 7, 578001. [Google Scholar] [CrossRef] [PubMed]
- Koike, T.; Kimura, N.; Miyazaki, K.; Yabuta, T.; Kumamoto, K.; Takenoshita, S.; Chen, J.; Kobayashi, M.; Hosokawa, M.; Taniguchi, A.; et al. Hypoxia induces adhesion molecules on cancer cells: A missing link between Warburg effect and induction of selectin-ligand carbohydrates. Proc. Natl. Acad. Sci. USA 2004, 101, 8132–8137. [Google Scholar] [CrossRef] [PubMed]
- Yılmaz, C.; Karali, K.; Fodelianaki, G.; Gravanis, A.; Chavakis, T.; Charalampopoulos, I.; Alexaki, V.I. Neurosteroids as regulators of neuroinflammation. Front. Neuroendocrinol. 2019, 55, 100788. [Google Scholar] [CrossRef] [PubMed]
- Bosch, M.; Sánchez-Álvarez, M.; Fajardo, A.; Kapetanovic, R.; Steiner, B.; Dutra, F.; Moreira, L.; López, J.A.; Campo, R.; Marí, M.; et al. Mammalian lipid droplets are innate immune hubs integrating cell metabolism and host defense. Science 2020, 370, eaay8085. [Google Scholar] [CrossRef]
- Metge, B.J.; Kammerud, S.C.; Pruitt, H.C.; Shevde, L.A.; Samant, R.S. Hypoxia re-programs 2′-O-Me modifications on ribosomal RNA. iScience 2021, 24, 102010. [Google Scholar] [CrossRef]
- Brumwell, A.; Fell, L.; Obress, L.; Uniacke, J. Hypoxia influences polysome distribution of human ribosomal protein S12 and alternative splicing of ribosomal protein mRNAs. RNA 2020, 26, 361–371. [Google Scholar] [CrossRef] [PubMed]
- Chandra, A.; Chakraborty, U.; Pal, J.; Karmakar, P. Silent hypoxia: A frequently overlooked clinical entity in patients with COVID-19. BMJ Case Rep. 2020, 13, e237207. [Google Scholar] [CrossRef] [PubMed]
- Jahani, M.; Dokaneheifard, S.; Mansouri, K. Hypoxia: A key feature of COVID-19 launching activation of HIF-1 and cytokine storm. J. Inflamm. 2020, 17, 33. [Google Scholar] [CrossRef] [PubMed]
- Serebrovska, Z.O.; Chong, E.Y.; Serebrovska, T.V.; Tumanovska, L.V.; Xi, L. Hypoxia, HIF-1α, and COVID-19: From pathogenic factors to potential therapeutic targets. Acta Pharmacol. Sin. 2020, 41, 1539–1546. [Google Scholar] [CrossRef] [PubMed]
- Cavadas, M.A.S.; Mesnieres, M.; Crifo, B.; Manresa, M.C.; Selfridge, A.C.; Keogh, C.E.; Fabian, Z.; Scholz, C.C.; Nolan, K.A.; Rocha, L.M.A.; et al. REST is a hypoxia-responsive transcriptional repressor. Sci. Rep. 2016, 6, 31355. [Google Scholar] [CrossRef] [PubMed]
- Batie, M.; Del Peso, L.; Rocha, S. Hypoxia and Chromatin: A Focus on Transcriptional Repression Mechanisms. Biomedicines 2018, 6, 47. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Xia, X.; Li, Q.; Dou, Y.; Suo, X.; Sun, Z.; Liu, N.; Han, Y.; Sun, X.; He, Y.; et al. A causal association of ANKRD37 with human hippocampal volume. Mol. Psychiatry 2022, 27, 4432–4445. [Google Scholar] [CrossRef] [PubMed]
- Benita, Y.; Kikuchi, H.; Smith, A.D.; Zhang, M.Q.; Chung, D.C.; Xavier, R.J. An integrative genomics approach identifies Hypoxia Inducible Factor-1 (HIF-1)-target genes that form the core response to hypoxia. Nucleic Acids Res. 2009, 37, 4587–4602. [Google Scholar] [CrossRef] [PubMed]
- Saraswati, S.; Guo, Y.; Atkinson, J.; Young, P.P. Prolonged hypoxia induces monocarboxylate transporter-4 expression in mesenchymal stem cells resulting in a secretome that is deleterious to cardiovascular repair. Stem Cells 2015, 33, 1333–1344. [Google Scholar] [CrossRef]
- Baek, G.; Tse, Y.F.; Hu, Z.; Cox, D.; Buboltz, N.; McCue, P.; Yeo, C.J.; White, M.A.; DeBerardinis, R.J.; Knudsen, E.S.; et al. MCT4 defines a glycolytic subtype of pancreatic cancer with poor prognosis and unique metabolic dependencies. Cell Rep. 2014, 9, 2233–2249. [Google Scholar] [CrossRef]
- Ikeda, S.; Abe, F.; Matsuda, Y.; Kitadate, A.; Takahashi, N.; Tagawa, H. Hypoxia-inducible hexokinase-2 enhances anti-apoptotic function via activating autophagy in multiple myeloma. Cancer Sci. 2020, 111, 4088–4101. [Google Scholar] [CrossRef] [PubMed]
- Minchenko, A.; Leshchinsky, I.; Opentanova, I.; Sang, N.; Srinivas, V.; Armstead, V.; Caro, J. Hypoxia-inducible factor-1-mediated expression of the 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase-3 (PFKFB3) gene. Its possible role in the Warburg effect. J. Biol. Chem. 2002, 277, 6183–6187. [Google Scholar] [CrossRef] [PubMed]
- Wujak, M.; Veith, C.; Wu, C.-Y.; Wilke, T.; Kanbagli, Z.I.; Novoyatleva, T.; Guenther, A.; Seeger, W.; Grimminger, F.; Sommer, N.; et al. Adenylate Kinase 4-A Key Regulator of Proliferation and Metabolic Shift in Human Pulmonary Arterial Smooth Muscle Cells via Akt and HIF-1α Signaling Pathways. Int. J. Mol. Sci. 2021, 22, 10371. [Google Scholar] [CrossRef] [PubMed]
- Ail, D.; Samardzija, M.; Chang, A.C.M.; Keck, J.; Reddel, R.R.; Grimm, C. Stanniocalcin2, but Not Stanniocalcin1, Responds to Hypoxia in a HIF1-Dependent Manner in the Retina. Front. Neurosci. 2022, 16, 882559. [Google Scholar] [CrossRef] [PubMed]
- Kajiwara, T.; Tanaka, T.; Kukita, K.; Kutomi, G.; Saito, K.; Okuya, K.; Takaya, A.; Kochin, V.; Kanaseki, T.; Tsukahara, T.; et al. Hypoxia augments MHC class I antigen presentation via facilitation of ERO1-α-mediated oxidative folding in murine tumor cells. Eur. J. Immunol. 2016, 46, 2842–2851. [Google Scholar] [CrossRef] [PubMed]
- Pescador, N.; Villar, D.; Cifuentes, D.; Garcia-Rocha, M.; Ortiz-Barahona, A.; Vazquez, S.; Ordoñez, A.; Cuevas, Y.; Saez-Morales, D.; Garcia-Bermejo, M.L.; et al. Hypoxia promotes glycogen accumulation through hypoxia inducible factor (HIF)-mediated induction of glycogen synthase 1. PLoS ONE 2010, 5, e9644. [Google Scholar] [CrossRef] [PubMed]
- Guo, K.; Searfoss, G.; Krolikowski, D.; Pagnoni, M.; Franks, C.; Clark, K.; Yu, K.T.; Jaye, M.; Ivashchenko, Y. Hypoxia induces the expression of the pro-apoptotic gene BNIP3. Cell Death Differ. 2001, 8, 367–376. [Google Scholar] [CrossRef]
- Krawczyk, M.A.; Kunc, M.; Styczewska, M.; Gabrych, A.; Karpinsky, G.; Izycka-Swieszewska, E.; Bien, E. High Expression of Solute Carrier Family 2 Member 1 (SLC2A1) in Cancer Cells Is an Independent Unfavorable Prognostic Factor in Pediatric Malignant Peripheral Nerve Sheath Tumor. Diagnostics 2021, 11, 598. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Lim, S.; Hoffman, D.; Aspenstrom, P.; Federoff, H.J.; Rempe, D.A. HUMMR, a hypoxia- and HIF-1alpha-inducible protein, alters mitochondrial distribution and transport. J. Cell Biol. 2009, 185, 1065–1081. [Google Scholar] [CrossRef]
- Jia, L.; Liang, T.; Yu, X.; Ma, C.; Zhang, S. MGARP regulates mouse neocortical development via mitochondrial positioning. Mol. Neurobiol. 2014, 49, 1293–1308. [Google Scholar] [CrossRef]
- Tello, D.; Balsa, E.; Acosta-Iborra, B.; Fuertes-Yebra, E.; Elorza, A.; Ordóñez, A.; Corral-Escariz, M.; Soro, I.; López-Bernardo, E.; Perales-Clemente, E.; et al. Induction of the mitochondrial NDUFA4L2 protein by HIF-1α decreases oxygen consumption by inhibiting Complex I activity. Cell Metab. 2011, 14, 768–779. [Google Scholar] [CrossRef] [PubMed]
- Dilly, A.K.; Lee, Y.J.; Zeh, H.J.; Guo, Z.S.; Bartlett, D.L.; Choudry, H.A. Targeting hypoxia-mediated mucin 2 production as a therapeutic strategy for mucinous tumors. Transl. Res. J. Lab. Clin. Med. 2016, 169, 19–30.e1. [Google Scholar] [CrossRef] [PubMed]
- Aggarwal, V.; Sahoo, S.; Donnenberg, V.S.; Chakraborty, P.; Jolly, M.K.; Sant, S. P4HA2: A link between tumor-intrinsic hypoxia, partial EMT and collective migration. Adv. Cancer Biol.-Metastasis 2022, 5, 100057. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Liu, Q.; Fang, Z.; Hu, X.; Huang, F.; Tang, L.; Zhou, S. Hypoxia induces the proliferation of endothelial progenitor cells via upregulation of Apelin/APLNR/MAPK signaling. Mol. Med. Rep. 2016, 13, 1801–1806. [Google Scholar] [CrossRef] [PubMed]
- Allan, K.C.; Hu, L.R.; Scavuzzo, M.A.; Morton, A.R.; Gevorgyan, A.S.; Cohn, E.F.; Clayton, B.L.L.; Bederman, I.R.; Hung, S.; Bartels, C.F.; et al. Non-canonical Targets of HIF1a Impair Oligodendrocyte Progenitor Cell Function. Cell Stem Cell 2021, 28, 257–272.e11. [Google Scholar] [CrossRef] [PubMed]
- Copple, B.L.; Bai, S.; Burgoon, L.D.; Moon, J.-O. Hypoxia-inducible factor-1α regulates the expression of genes in hypoxic hepatic stellate cells important for collagen deposition and angiogenesis. Liver Int. 2011, 31, 230–244. [Google Scholar] [CrossRef] [PubMed]
- Marallano, V.; Sattiraju, A.; Zou, H.; Friedel, R. TAMI-61. Examining the Role of Hypoxia Induced Genes CXCR4 and NXPH4 in Invasion of Hypoxic Glioblastoma Cells. Neuro-Oncology 2021, 23 (Suppl. S6), vi211-vi. [Google Scholar] [CrossRef]
- Ono, Y.; Bono, H. Multi-Omic Meta-Analysis of Transcriptomes and the Bibliome Uncovers Novel Hypoxia-Inducible Genes. Biomedicines 2021, 9, 582. [Google Scholar] [CrossRef] [PubMed]
- Park, J.S.; Gabel, A.M.; Kassir, P.; Kang, L.; Chowdhary, P.K.; Osei-Ntansah, A.; Tran, N.D.; Viswanathan, S.; Canales, B.; Ding, P.; et al. N-myc downstream regulated gene 1 (ndrg1) functions as a molecular switch for cellular adaptation to hypoxia. eLife 2022, 11, e74031. [Google Scholar] [CrossRef]
- Burton, T.R.; Gibson, S.B. The role of Bcl-2 family member BNIP3 in cell death and disease: NIPping at the heels of cell death. Cell Death Differ. 2009, 16, 515–523. [Google Scholar] [CrossRef]
- Zhang, J.; Ney, P.A. Role of BNIP3 and NIX in cell death, autophagy, and mitophagy. Cell Death Differ. 2009, 16, 939–946. [Google Scholar] [CrossRef] [PubMed]
- Mazure, N.M.; Pouysségur, J. Hypoxia-induced autophagy: Cell death or cell survival? Curr. Opin. Cell Biol. 2010, 22, 177–180. [Google Scholar] [CrossRef] [PubMed]
- Tracy, K.; Dibling, B.C.; Spike, B.T.; Knabb, J.R.; Schumacker, P.; Macleod, K.F. BNIP3 is an RB/E2F target gene required for hypoxia-induced autophagy. Mol. Cell Biol. 2007, 27, 6229–6242. [Google Scholar] [CrossRef] [PubMed]
- Bellot, G.; Garcia-Medina, R.; Gounon, P.; Chiche, J.; Roux, D.; Pouysségur, J.; Mazure, N.M. Hypoxia-induced autophagy is mediated through hypoxia-inducible factor induction of BNIP3 and BNIP3L via their BH3 domains. Mol. Cell Biol. 2009, 29, 2570–2581. [Google Scholar] [CrossRef] [PubMed]
- Leppä, S.; Bohmann, D. Diverse functions of JNK signaling and c-Jun in stress response and apoptosis. Oncogene 1999, 18, 6158–6162. [Google Scholar] [CrossRef] [PubMed]
- Vargas, R.; Abbott, L.; Bower, D.; Frahm, N.; Shaffer, M.; Yu, W.-H. Gene signature discovery and systematic validation across diverse clinical cohorts for TB prognosis and response to treatment. PLoS Comp. Biol. 2023, 19, e1010770. [Google Scholar] [CrossRef] [PubMed]
- Walker, C.J.; Mrózek, K.; Ozer, H.G.; Nicolet, D.; Kohlschmidt, J.; Papaioannou, D.; Genutis, L.K.; Bill, M.; Powell, B.L.; Uy, G.L.; et al. Gene expression signature predicts relapse in adult patients with cytogenetically normal acute myeloid leukemia. Blood Adv. 2021, 5, 1474–1482. [Google Scholar] [CrossRef] [PubMed]
- Haynes, W.A.; Haddon, D.J.; Khatri, A.; Bongen, E.; Yiu, G.; Balboni, I.; Bolen, C.R.; Mao, R.; Utz, P.J.; Khatri, P. Integrated, multicohort analysis reveals unified signature of systemic lupus erythematosus. JCI Insight 2020, 5, e122312. [Google Scholar] [CrossRef] [PubMed]
- Gold, S.J.; Ni, Y.G.; Dohlman, H.G.; Nestler, E.J. Regulators of G-protein signaling (RGS) proteins: Region-specific expression of nine subtypes in rat brain. J. Neurosci. 1997, 17, 8024–8037. [Google Scholar] [CrossRef]
- Li, F.; Qiao, Z.; Duan, Q.; Nevo, E. Adaptation of mammals to hypoxia. Anim. Models Exp. Med. 2021, 4, 311–318. [Google Scholar] [CrossRef]
- Gan, E.S.; Ooi, E.E. Oxygen: Viral friend or foe? Virol. J. 2020, 17, 115. [Google Scholar] [CrossRef]
- Rybnikova, E.A.; Nalivaeva, N.N.; Zenko, M.Y.; Baranova, K.A. Intermittent Hypoxic Training as an Effective Tool for Increasing the Adaptive Potential, Endurance and Working Capacity of the Brain. Front. Neurosci. 2022, 16, 941740. [Google Scholar] [CrossRef] [PubMed]
- Yue, X.; Zhou, Y.; Qiao, M.; Zhao, X.; Huang, X.; Zhao, T.; Cheng, X.; Fan, M.; Zhao, Y.; Chen, R.; et al. Intermittent hypoxia treatment alleviates memory impairment in the 6-month-old APPswe/PS1dE9 mice and reduces amyloid beta accumulation and inflammation in the brain. Alzheimer’s Res. Ther. 2021, 13, 194. [Google Scholar] [CrossRef] [PubMed]
- Yuan, H.; Liu, J.; Gu, Y.; Ji, X.; Nan, G. Intermittent hypoxia conditioning as a potential prevention and treatment strategy for ischemic stroke: Current evidence and future directions. Front. Neurosci. 2022, 16, 1067411. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Shi, X.; Schenck, H.; Hall, J.R.; Ross, S.E.; Kline, G.P.; Chen, S.; Mallet, R.T.; Chen, P. Intermittent Hypoxia Training for Treating Mild Cognitive Impairment: A Pilot Study. Am. J. Alzheimers Dis. Other Demen 2020, 35, 1533317519896725. [Google Scholar] [CrossRef]
- Gella, A.; Durany, N. Oxidative stress in Alzheimer disease. Cell Adh Migr. 2009, 3, 88–93. [Google Scholar] [CrossRef] [PubMed]
- Marola, O.J.; Syc-Mazurek, S.B.; Libby, R.T. DDIT3 (CHOP) contributes to retinal ganglion cell somal loss but not axonal degeneration in DBA/2J mice. Cell Death Discov. 2019, 5, 140. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ravel-Godreuil, C.; Roy, E.R.; Puttapaka, S.N.; Li, S.; Wang, Y.; Yuan, X.; Eltzschig, H.K.; Cao, W. Transcriptional Responses of Different Brain Cell Types to Oxygen Decline. Brain Sci. 2024, 14, 341. https://doi.org/10.3390/brainsci14040341
Ravel-Godreuil C, Roy ER, Puttapaka SN, Li S, Wang Y, Yuan X, Eltzschig HK, Cao W. Transcriptional Responses of Different Brain Cell Types to Oxygen Decline. Brain Sciences. 2024; 14(4):341. https://doi.org/10.3390/brainsci14040341
Chicago/Turabian StyleRavel-Godreuil, Camille, Ethan R. Roy, Srinivas N. Puttapaka, Sanming Li, Yanyu Wang, Xiaoyi Yuan, Holger K. Eltzschig, and Wei Cao. 2024. "Transcriptional Responses of Different Brain Cell Types to Oxygen Decline" Brain Sciences 14, no. 4: 341. https://doi.org/10.3390/brainsci14040341