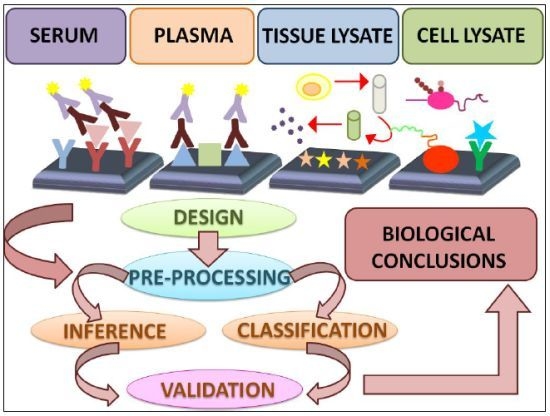
Data Analysis Strategies for Protein Microarrays
Abstract
:1. Introduction
2. Concept of Protein Microarrays and Current Applications
2.1. Types of Protein Microarrays
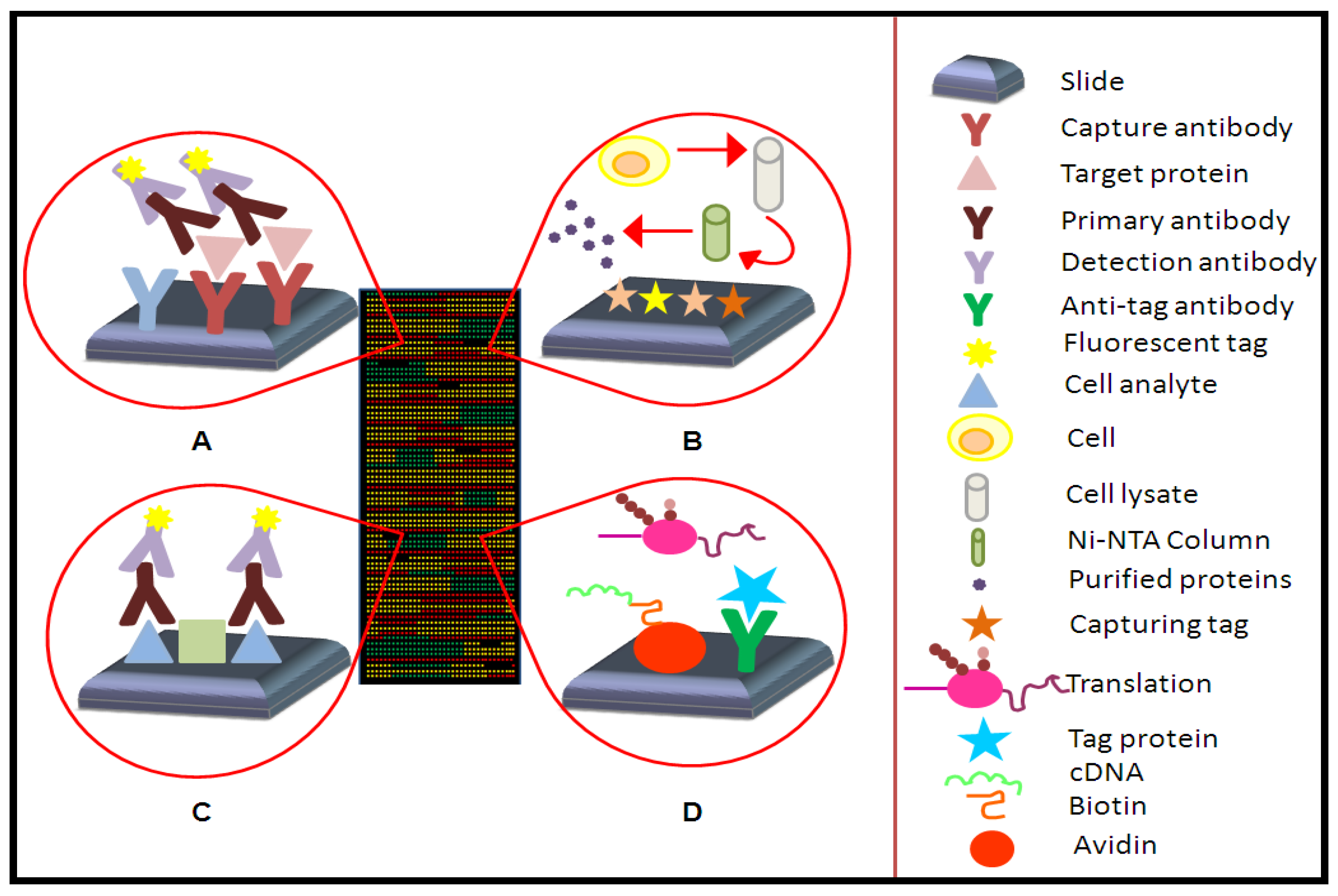
2.1.1. Analytical Microarrays
2.1.2. Reverse Phase Arrays
2.1.3. Functional Microarrays
2.2. Current Application of Protein Microarrays
| Disease | Type of microarray | Object of study | Reference |
|---|---|---|---|
| Cancer | multiplexed array | CA-125; CA19-9; EGFR; C-protein; myoglobin; APOA1; APOC3; MIP-1; IL6; IL18; tenascin-C | Amonkar et al. 2009 |
| NAPPA | p53 | Dasilva et al. 2012 | |
| Nodular thyroid disease | protein array | EGF; HGF; IL5; IL8; RANTES | Linkov et al. 2008 |
| multiplexed array | cytokines; growth factors; cell adhesion molecules | Xiaobo et al. 2010 | |
| reverse phase array | Salmonella typhimurium | Cid et al. 2011 | |
| Infectious disease | antigen microarray | Vaccinia virus; Yersinia pestis | Natesan et al. 2010 |
| antibody array | cholera; diphtheria; staphylococcal enterotoxin B; tetanus toxin; anthrax protective antigen | Rucker et al. 2005 | |
| protein array | B lymphocyte | Wadia et al. 2011; Belov et al. 2001 | |
| Systematic rheumatic disease | antibody microarray | nuclear proteins; nucleoprotein complexes | Dolores et al. 2001 |
| Diabetes (type I) | NAPPA | Sibani et al. 2011 |
2.2.1. Cancer
2.2.2. Immunology
2.2.3. Nodular Thyroid Disease
2.2.4. Infectious Disease
3. Feature Aspects of Protein Microarrays
3.1. Array Capture Agents
| Capture agent | Source of proteins | Technique |
|---|---|---|
| Mab * | mouse | Hybridoma |
| sc Fv */Fab * diabodies | antibody libraries | Phage display, in vitro evolution |
| Affinity binding agents | recombinant fibronectin structures | In vitro evolution |
| Affibodies | ||
| Aptamers (DNA, RNA, peptide) | ||
| Receptors ligands | synthetic | Combinatorial chemistry |
| Substrates of enzymes | synthetic; pro-and eukaryotic organisms | Protein purification, recombinant protein technology(bacterial, fusion proteins, baculovirus, peptide synthesis) |
3.2. Array Surfaces
3.3. Array Detection Technologies
4. Data Analysis Methods

- • Spot intensity determination: microarray image analysis starts with the fixing of spot intensity. Generally, for this task, GenePix Pro software (Molecular Devices, Union City, CA) is used. First of all, a grid of circles must be placed over the protein spots. Their position and size have to be adjusted in order to get reliable intensity data. Finally, an output file is created by the program.
- • Z-score analysis: the Z-score equation,
, where Zs is the Z-score for the sth spot, Ss is the signal for that spot, µ is the mean signal across all spots and σ is the standard deviation across all spots, is an interesting tool to determine which signals are significantly different from the expected value and which are not.
- • Concentration-dependent analysis (CDA): due to the quantity of spotted proteins on the slide, absolute signals are affected by protein concentration. As a means to solve this issue, a different Z-score,
, can be calculated to remove outliers. This novel Z-score is calculated using an iteration process that is repeated until every spot signal measured is in accordance with the mean value. In the equation above, Zs is the Z-score for the sth spot, Ss is the signal for that spot, µw is the mean signal for the spots within the window and σw is the standard deviation for spots within the window.
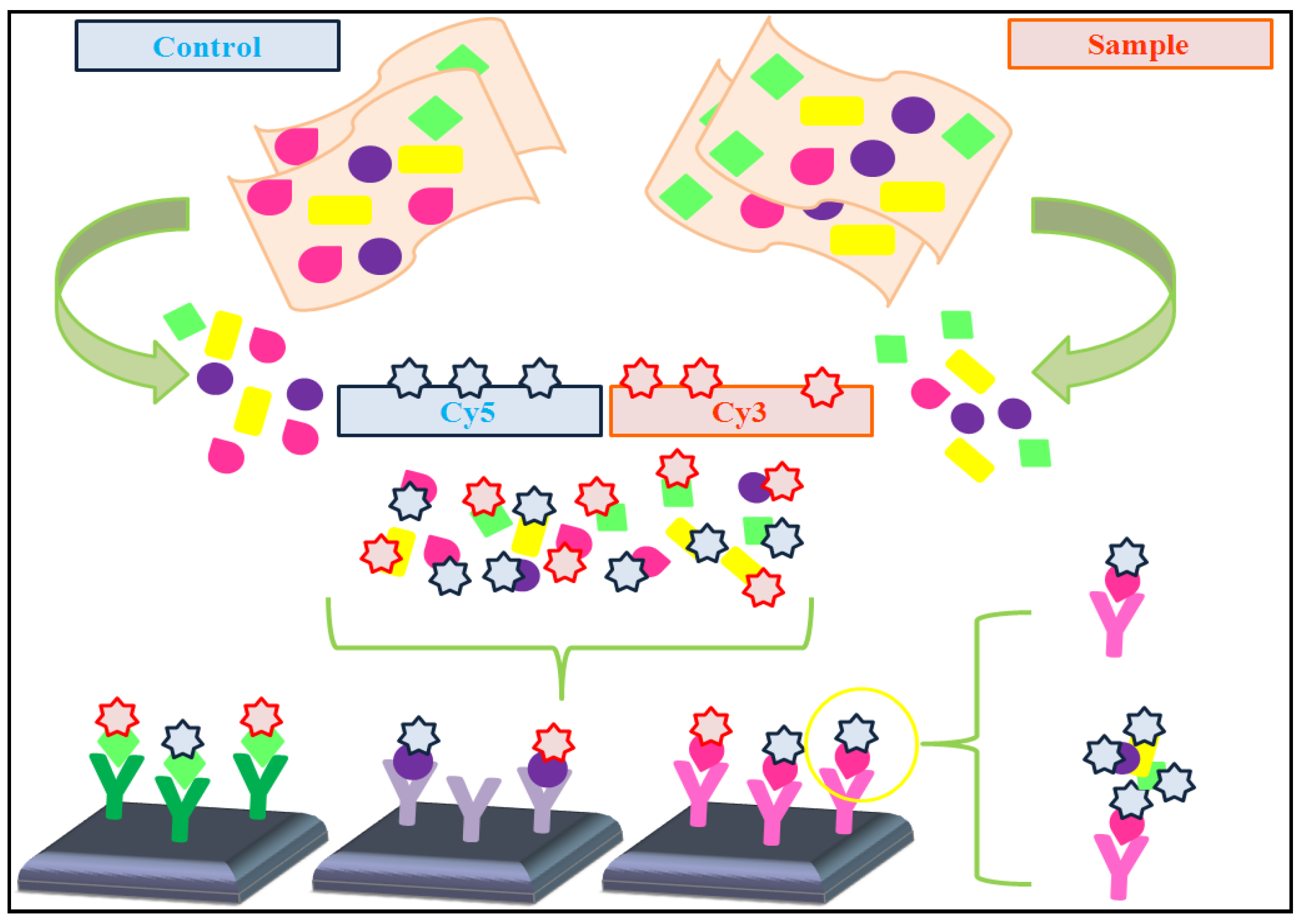
4.1. Dual-Color vs. Single-Color Assays
- * Single-color assays: Olle et al. [47] developed a single antibody-based microarray which presents standard antigen concentration. Also, it uses an internal controlled system based on two colors, one for the amount of antibody spotted and the other for the amount of the antigen used for the quantification of the level of protein expression. To validate this microarray, levels of protein expression were compared with results obtained by western blot analysis and the data were similar, although the sensitivity was higher with the microarray. In their study, they show that this microarray has not only the potential to accurately assess proteins in complex fluids, but also a large range of linearity.
- * Dual-color assays: Data pre-processing protocols are usually applied to prevent undesired technical artifacts. These protocols frequently include the following steps (adapted from [48]):
- • Filtering, in order to remove failed and low-quality spots.
- • Background correction, to avoid fluorescence signal due to non-specific binding.
- • Data normalization, aimed to reduce variations between the two samples co-hybridized on each array and also between arrays.
- • Reference design: the sample of interest is labeled with one fluorescent fluorochrome (e.g., Cy3), whereas the single reference sample is labeled with a different fluorescent fluorochrome (e.g., Cy5). In this type of design, it is necessary to calculate the log ratio of dyes intensities.
- • Balanced-block design: two samples which are hybridized, bearing two different fluorochromes (Cy3 and Cy5). Then, samples are balanced with respect to dyes. In this case, the microarray is considered as a block.
- • Incomplete-block design: more than two samples are co-hybridized on the microarray, whereas only two fluorochromes are used (Cy3 and Cy5). Despite this, samples are balanced.
- • Loop design: each sample is hybridized in a different array using a different fluorochrome. This supposes a great disadvantage because the number of arrays is duplicated.
| EXPERIMENTAL DESIGN | ARRAY #1 | ARRAY #2 | ARRAY #3 | ARRAY #4 |
|---|---|---|---|---|
| Reference | A1/R | A2/R | B1/R | B2/R |
| Balance block | A1/B1 | B2/A2 | ||
| Incomplete block | A1/B1 | B2/C1 | C2/A2 | |
| Loop | A1/B1 | B1/A2 | A2/B2 | B2/A1 |
4.1.1. Rank-Invariant Selection Algorithm (InvTseng)
 is determined by the following expression [50]:
is determined by the following expression [50]:
4.1.2. Modified Rank-Invariant Selection Algorithm (In-vMod)

4.1.3. Rank Difference Weighted Global Loess (RDWGL)

4.2. Automated Analysis of Highly Complex Flow Cytometry
- • Automated gating of color-coded microspheres: the automated tool is responsible for the specific identification and differentiation among microsphere types and the consequent allocation of the code to each one.
- • Analysis of size-MAP data: quantification of antibody-bound proteins amounts was determined with the medians of the fluorescence label signals. Next, these data will be processed through quality control (QC), normalization and analysis.
- ○ Quality control (QC): first of all, the number of microspheres of each population is checked. Next, the density function of the signal is also determined.
- ○ Normalization: it is necessary to remove background noise and to establish protein sample differences. With the purpose of correcting the noise, the signal, from empty microspheres (those without any antibody), is subtracted from the signal of the microsphere population of interest.
- ○ Analysis: each protein entity has to be established and, for this purpose, fractions constituting specific protein entities must be defined. Then, signals for each fraction are summed up, representing the final result, which is the relative amount of a particular protein entity.
4.3. Data Analysis Methods from cDNA Arrays
4.4. Reverse Phase Array Data Analysis
- • Serial dilution curve: the monotonic s-shaped response curve is described by Sips model:

- • Parameterization of the serial dilution curve: a non-linear regression model is used to find the optimal parameters.
- • Estimating protein concentrations: first of all it is necessary to check if protein concentration is saturated. This occurs if the M/r ratio is lower than signals measured. Then, the minimum and maximum of x (xmin and xmax, respectively) are estimated with these formulas:

5. Conclusions
Acknowledgements
References
- Yu, X.; Schneiderhan-Marra, N.; Joos, T.O. Protein microarrays and personalized medicine. Ann. Biol. Clin. (Paris) 2011, 69, 17–29. [Google Scholar]
- Dasgupta, A. Handbook of Drug Monitoring Methods: Therapeutics and Drugs of Abuse; Humana Press: New York, NY, USA, 2007; pp. 5400–5411. [Google Scholar]
- Merbl, Y.; Kirschner, M.W. Protein microarrays for genome-wide posttranslational modification analysis. Wiley Interdiscip. Rev. Syst. Biol. Med. 2011, 3, 347–356. [Google Scholar] [CrossRef]
- Hanash, S. Disease proteomics. Nature 2003, 422, 226–232. [Google Scholar] [CrossRef]
- MacBeath, G. Protein microarrays and proteomics. Nat. Genet. 2002, 32, 526–532. [Google Scholar] [CrossRef]
- Fodor, S.P.; Read, J.L.; Pirrung, M.C.; Stryer, L.; Lu, A.T.; Solas, D. Light-directed, spatially addressable parallel chemical synthesis. Science 1991, 251, 767–773. [Google Scholar]
- Chandra, H.; Reddy, P.J.; Srivastava, S. Protein microarrays and novel detection platforms. Expert Rev. Proteomics 2011, 8, 61–79. [Google Scholar] [CrossRef]
- Chandra, H.; Srivastava, S. Cell-free synthesis-based protein microarrays and their applications. Proteomics 2010, 10, 717–730. [Google Scholar] [CrossRef]
- Gonzalez-Gonzalez, M.; Jara-Acevedo, R.; Matarraz, S.; Jara-Acevedo, M.; Paradinas, S.; Sayagues, J.M.; Orfao, A.; Fuentes, M. Nanotechniques in proteomics: Protein microarrays and novel detection platforms. Eur. J. Pharm. Sci. 2012, 45, 499–506. [Google Scholar] [CrossRef]
- Hultschig, C.; Kreutzberger, J.; Seitz, H.; Konthur, Z.; Bussow, K.; Lehrach, H. Recent advances of protein microarrays. Curr. Opin. Chem. Biol. 2006, 10, 4–10. [Google Scholar] [CrossRef]
- Dasilva, N.; Diez, P.; Matarraz, S.; Gonzalez-Gonzalez, M.; Paradinas, S.; Orfao, A.; Fuentes, M. Biomarker discovery by novel sensors based on nanoproteomics approaches. Sensors 2012, 12, 2284–2308. [Google Scholar]
- Templin, M.F.; Stoll, D.; Schrenk, M.; Traub, P.C.; Vohringer, C.F.; Joos, T.O. Protein microarray technology. Drug Discov. Today 2002, 7, 815–822. [Google Scholar]
- LaBaer, J.; Ramachandran, N. Protein microarrays as tools for functional proteomics. Curr. Opin. Chem. Biol. 2005, 9, 14–19. [Google Scholar] [CrossRef]
- Poetz, O.; Schwenk, J.M.; Kramer, S.; Stoll, D.; Templin, M.F.; Joos, T.O. Protein microarrays: Catching the proteome. Mech. Ageing Dev. 2005, 126, 161–170. [Google Scholar] [CrossRef]
- Gao, L.; Uttamchandani, M.; Yao, S.Q. Comparative proteomic profiling of mammalian cell lysates using phosphopeptide microarrays. Chem. Commun. (Camb.) 2012, 48, 2240–2242. [Google Scholar]
- Uttamchandani, M.; Lu, C.H.; Yao, S.Q. Next generation chemical proteomic tools for rapid enzyme profiling. Acc. Chem. Res. 2009, 42, 1183–1192. [Google Scholar] [CrossRef]
- Matarraz, S.; Gonzalez-Gonzalez, M.; Jara, M.; Orfao, A.; Fuentes, M. New technologies in cancer. Protein microarrays for biomarker discovery. Clin. Transl. Oncol. 2011, 13, 156–161. [Google Scholar] [CrossRef]
- Natesan, M.; Ulrich, R.G. Protein microarrays and biomarkers of infectious disease. Int. J. Mol. Sci. 2010, 11, 5165–5183. [Google Scholar] [CrossRef]
- Borrebaeck, C.A.; Wingren, C. Design of high-density antibody microarrays for disease proteomics: Key technological issues. J. Proteomics 2009, 72, 928–935. [Google Scholar] [CrossRef]
- Cahill, D.J. Protein and antibody arrays and their medical applications. J. Immunol. Methods 2001, 250, 81–91. [Google Scholar] [CrossRef]
- Chen, C.S.; Zhu, H. Protein microarrays. BioTechniques 2006, 40, 423, 425, 427 passim. [Google Scholar]
- Hall, D.A.; Ptacek, J.; Snyder, M. Protein microarray technology. Mech. Ageing Dev. 2007, 128, 161–167. [Google Scholar] [CrossRef]
- Pierobon, M.; Vanmeter, A.J.; Moroni, N.; Galdi, F.; Petricoin, E.F., III. Reverse-phase protein microarrays. Methods Mol. Biol. 2012, 823, 215–235. [Google Scholar] [CrossRef]
- Ramachandran, N.; Srivastava, S.; Labaer, J. Applications of protein microarrays for biomarker discovery. Proteomics Clin. Appl. 2008, 2, 1444–1459. [Google Scholar] [CrossRef]
- Beare, P.A.; Chen, C.; Bouman, T.; Pablo, J.; Unal, B.; Cockrell, D.C.; Brown, W.C.; Barbian, K.D.; Porcella, S.F.; Samuel, J.E.; Felgner, P.L.; Heinzen, R.A. Candidate antigens for Q fever serodiagnosis revealed by immunoscreening of a Coxiella burnetii protein microarray. Clin. Vaccine Immunol. 2008, 15, 1771–1779. [Google Scholar] [CrossRef]
- Lopez, J.E.; Beare, P.A.; Heinzen, R.A.; Norimine, J.; Lahmers, K.K.; Palmer, G.H.; Brown, W.C. High-throughput identification of T-lymphocyte antigens from Anaplasma marginale expressed using in vitro transcription and translation. J. Immunol. Methods 2008, 332, 129–141. [Google Scholar] [CrossRef]
- Wong, J.; Sibani, S.; Lokko, N.N.; LaBaer, J.; Anderson, K.S. Rapid detection of antibodies in sera using multiplexed self-assembling bead arrays. J. Immunol. Methods 2009, 350, 171–182. [Google Scholar] [CrossRef]
- Hurst, R.; Hook, B.; Slater, M.R.; Hartnett, J.; Storts, D.R.; Nath, N. Protein-protein interaction studies on protein arrays: Effect of detection strategies on signal-to-background ratios. Anal. Biochem. 2009, 392, 45–53. [Google Scholar]
- Mei, Q.; Fredrickson, C.K.; Jin, S.; Fan, Z.H. Toxin detection by a miniaturized in vitro protein expression array. Anal. Chem. 2005, 77, 5494–5500. [Google Scholar] [CrossRef]
- Anderson, K.S.; Ramachandran, N.; Wong, J.; Raphael, J.V.; Hainsworth, E.; Demirkan, G.; Cramer, D.; Aronzon, D.; Hodi, F.S.; Harris, L.; et al. Application of protein microarrays for multiplexed detection of antibodies to tumor antigens in breast cancer. J. Proteome Res. 2008, 7, 1490–1499. [Google Scholar]
- Sreekumar, A.; Nyati, M.K.; Varambally, S.; Barrette, T.R.; Ghosh, D.; Lawrence, T.S.; Chinnaiyan, A.M. Profiling of cancer cells using protein microarrays: Discovery of novel radiation-regulated proteins. Cancer Res. 2001, 61, 7585–7593. [Google Scholar]
- Amonkar, S.D.; Bertenshaw, G.P.; Chen, T.H.; Bergstrom, K.J.; Zhao, J.; Seshaiah, P.; Yip, P.; Mansfield, B.C. Development and preliminary evaluation of a multivariate index assay for ovarian cancer. PLoS One 2009. [Google Scholar] [CrossRef]
- Roberts, P.J.; Der, C.J. Targeting the Raf-MEK-ERK mitogen-activated protein kinase cascade for the treatment of cancer. Oncogene 2007, 26, 3291–3310. [Google Scholar] [CrossRef]
- Belov, L.; Huang, P.; Barber, N.; Mulligan, S.P.; Christopherson, R.I. Identification of repertoires of surface antigens on leukemias using an antibody microarray. Proteomics 2003, 3, 2147–2154. [Google Scholar]
- Belov, L.; de la Vega, O.; dos Remedios, C.G.; Mulligan, S.P.; Christopherson, R.I. Immunophenotyping of leukemias using a cluster of differentiation antibody microarray. Cancer Res. 2001, 61, 4483–4489. [Google Scholar]
- Sibani, S.; LaBaer, J. Immunoprofiling using NAPPA protein microarrays. Methods Mol. Biol. 2011, 723, 149–161. [Google Scholar] [CrossRef]
- Linkov, F.; Ferris, R.L.; Yurkovetsky, Z.; Marrangoni, A.; Velikokhatnaya, L.; Gooding, W.; Nolan, B.; Winans, M.; Siegel, E.R.; Lokshin, A.; et al. Multiplex analysis of cytokines as biomarkers that differentiate benign and malignant thyroid diseases. Proteomics Clin. Appl. 2008, 2, 1575–1585. [Google Scholar] [CrossRef]
- Cid, V.J.; Kauffmann, E.; Molina, M. Reverse protein arrays applied to host-pathogen interaction studies. Methods Mol. Biol. 2011, 723, 37–55. [Google Scholar] [CrossRef]
- Angenendt, P. Progress in protein and antibody microarray technology. Drug Discov. Today 2005, 10, 503–511. [Google Scholar] [CrossRef]
- Bratkovic, T. Progress in phage display: Evolution of the technique and its application. Cell Mol. Life Sci. 2010, 67, 749–767. [Google Scholar] [CrossRef]
- Ramachandran, N.; Larson, D.N.; Stark, P.R.; Hainsworth, E.; LaBaer, J. Emerging tools for real-time label-free detection of interactions on functional protein microarrays. FEBS J. 2005, 272, 5412–5425. [Google Scholar]
- Stuchly, J.; Kanderova, V.; Fiser, K.; Cerna, D.; Holm, A.; Wu, W.; Hrusak, O.; Lund-Johansen, F.; Kalina, T. An automated analysis of highly complex flow cytometry-based proteomic data. Cytometry A 2012, 81, 120–129. [Google Scholar]
- Schweitzer, B.; Wiltshire, S.; Lambert, J.; O'Malley, S.; Kukanskis, K.; Zhu, Z.; Kingsmore, S.F.; Lizardi, P.M.; Ward, D.C. Immunoassays with rolling circle DNA amplification: A versatile platform for ultrasensitive antigen detection. Proc. Natl. Acad. Sci. USA 2000, 97, 10113–10119. [Google Scholar]
- Varnum, S.M.; Woodbury, R.L.; Zangar, R.C. A protein microarray ELISA for screening biological fluids. Methods Mol. Biol. 2004, 264, 161–172. [Google Scholar]
- Huang, R.P. Detection of multiple proteins in an antibody-based protein microarray system. J. Immunol. Methods 2001, 255, 1–13. [Google Scholar] [CrossRef]
- DeLuca, D.S.; Marina, O.; Ray, S.; Zhang, G.L.; Wu, C.J.; Brusic, V. Data processing and analysis for protein microarrays. Methods Mol. Biol. 2011, 723, 337–347. [Google Scholar] [CrossRef]
- Olle, E.W.; Sreekumar, A.; Warner, R.L.; McClintock, S.D.; Chinnaiyan, A.M.; Bleavins, M.R.; Anderson, T.D.; Johnson, K.J. Development of an internally controlled antibody microarray. Mol. Cell. Proteomics 2005, 4, 1664–1672. [Google Scholar] [CrossRef]
- Eckel-Passow, J.E.; Hoering, A.; Therneau, T.M.; Ghobrial, I. Experimental design and analysis of antibody microarrays: Applying methods from cDNA arrays. Cancer Res. 2005, 65, 2985–2989. [Google Scholar]
- Dobbin, K.; Shih, J.H.; Simon, R. Questions and answers on design of dual-label microarrays for identifying differentially expressed genes. J. Natl. Cancer Inst. 2003, 95, 1362–1369. [Google Scholar] [CrossRef]
- Sill, M.; Schroder, C.; Hoheisel, J.D.; Benner, A.; Zucknick, M. Assessment and optimisation of normalisation methods for dual-color antibody microarrays. BMC Bioinforma. 2010. [Google Scholar] [CrossRef]
- Zhang, L.; Wei, Q.; Mao, L.; Liu, W.; Mills, G.B.; Coombes, K. Serial dilution curve: A new method for analysis of reverse phase protein array data. Bioinformatics 2009, 25, 650–654. [Google Scholar] [CrossRef]
- Allison, D.B.; Cui, X.; Page, G.P.; Sabripour, M. Microarray data analysis: From disarray to consolidation and consensus. Nat. Rev. Genet. 2006, 7, 55–65. [Google Scholar]
© 2012 by the authors; licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Díez, P.; Dasilva, N.; González-González, M.; Matarraz, S.; Casado-Vela, J.; Orfao, A.; Fuentes, M. Data Analysis Strategies for Protein Microarrays. Microarrays 2012, 1, 64-83. https://doi.org/10.3390/microarrays1020064
Díez P, Dasilva N, González-González M, Matarraz S, Casado-Vela J, Orfao A, Fuentes M. Data Analysis Strategies for Protein Microarrays. Microarrays. 2012; 1(2):64-83. https://doi.org/10.3390/microarrays1020064
Chicago/Turabian StyleDíez, Paula, Noelia Dasilva, María González-González, Sergio Matarraz, Juan Casado-Vela, Alberto Orfao, and Manuel Fuentes. 2012. "Data Analysis Strategies for Protein Microarrays" Microarrays 1, no. 2: 64-83. https://doi.org/10.3390/microarrays1020064
APA StyleDíez, P., Dasilva, N., González-González, M., Matarraz, S., Casado-Vela, J., Orfao, A., & Fuentes, M. (2012). Data Analysis Strategies for Protein Microarrays. Microarrays, 1(2), 64-83. https://doi.org/10.3390/microarrays1020064