Can Archival Tissue Reveal Answers to Modern Research Questions?: Computer-Aided Histological Assessment of Neuroblastoma Tumours Collected over 60 Years
Abstract
:1. Introduction
2. Experimental Section
2.1. Neuroblastoma Cases and TMA Construction

2.2. Immunohistochemistry
2.3. Evaluation of TMA Immunohistochemistry
2.3.1. Digital Image Acquisition
2.3.2. Computer-Aided Image Analysis: Cell Counting
2.3.3. Computer-Aided Image Analysis: Pixel Counting for Signal Intensity
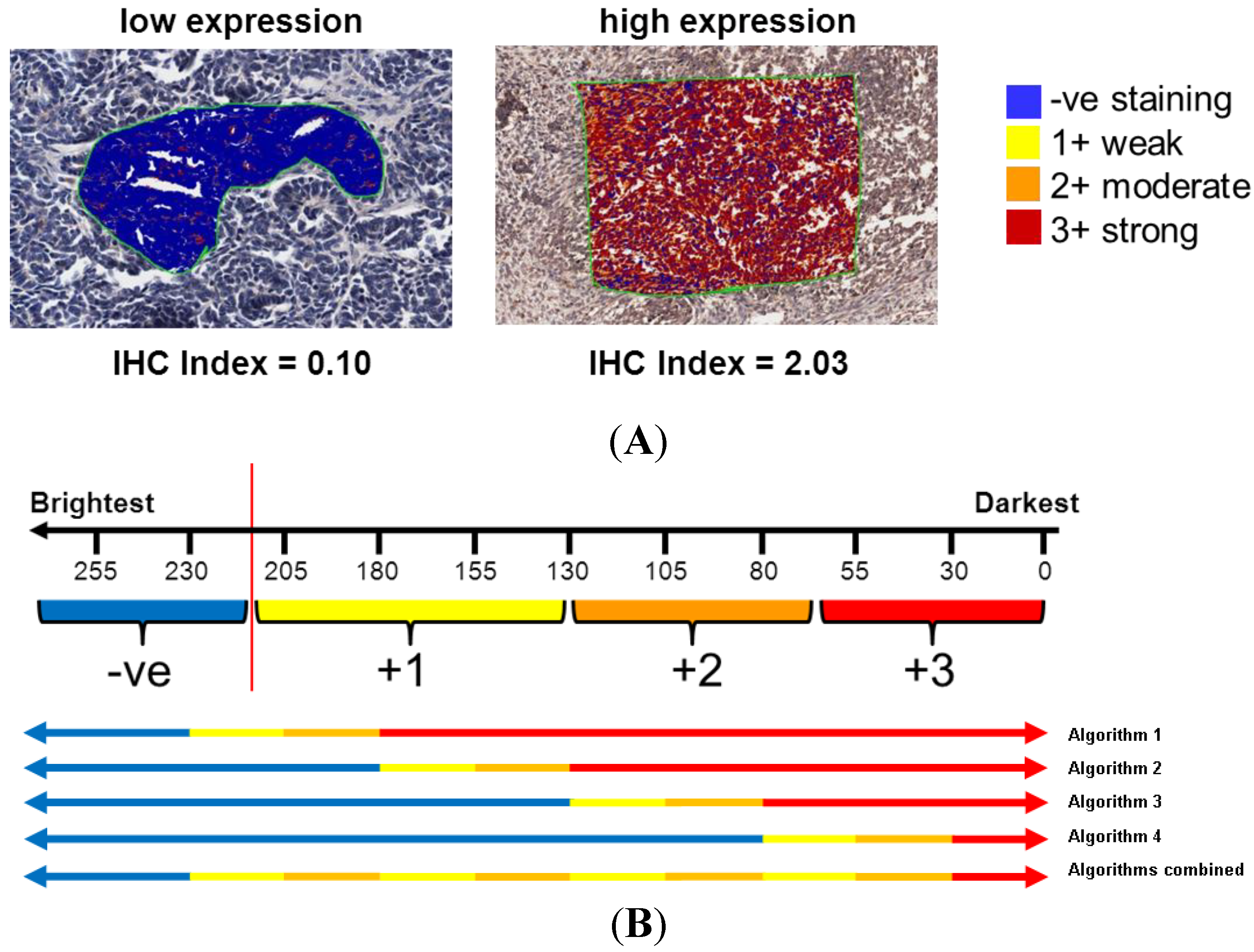
2.4. Statistical Analysis
3. Results and Discussion
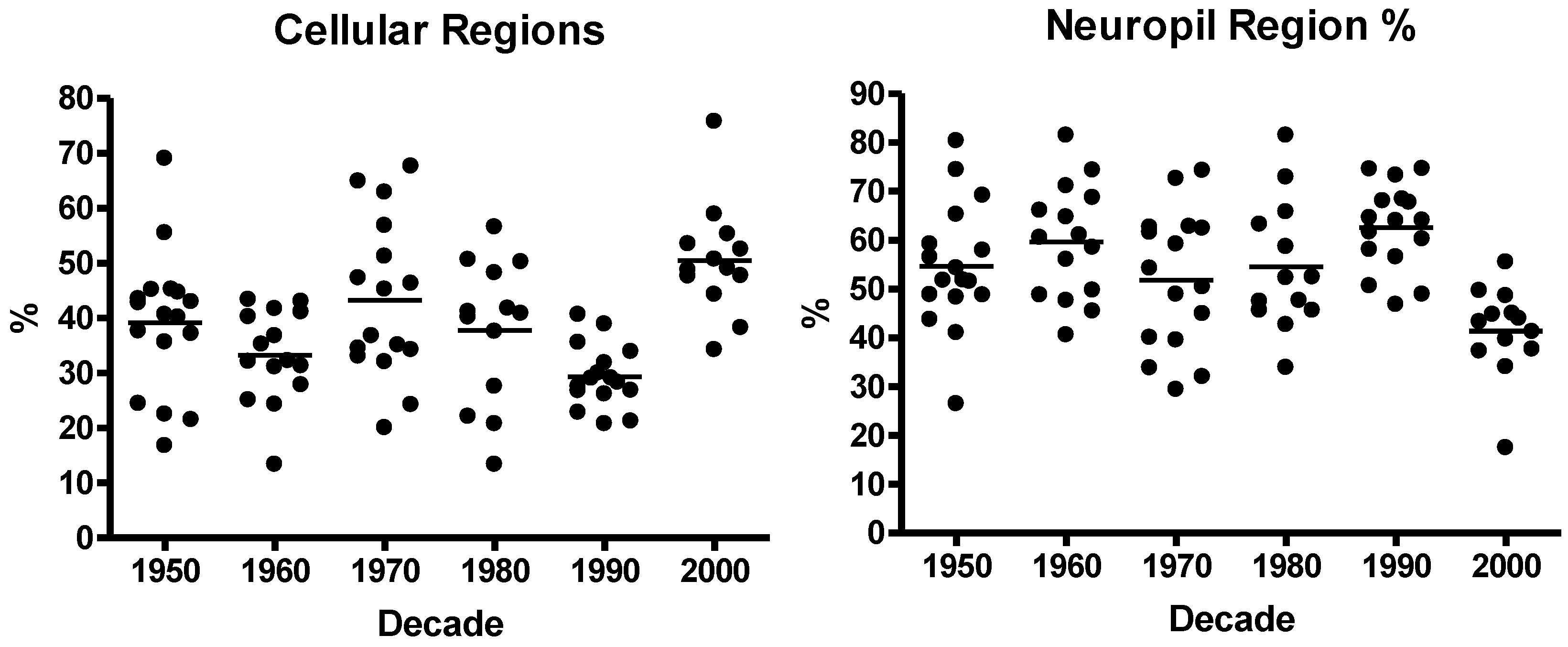
3.1. Neuroblastoma TMA Core Morphology

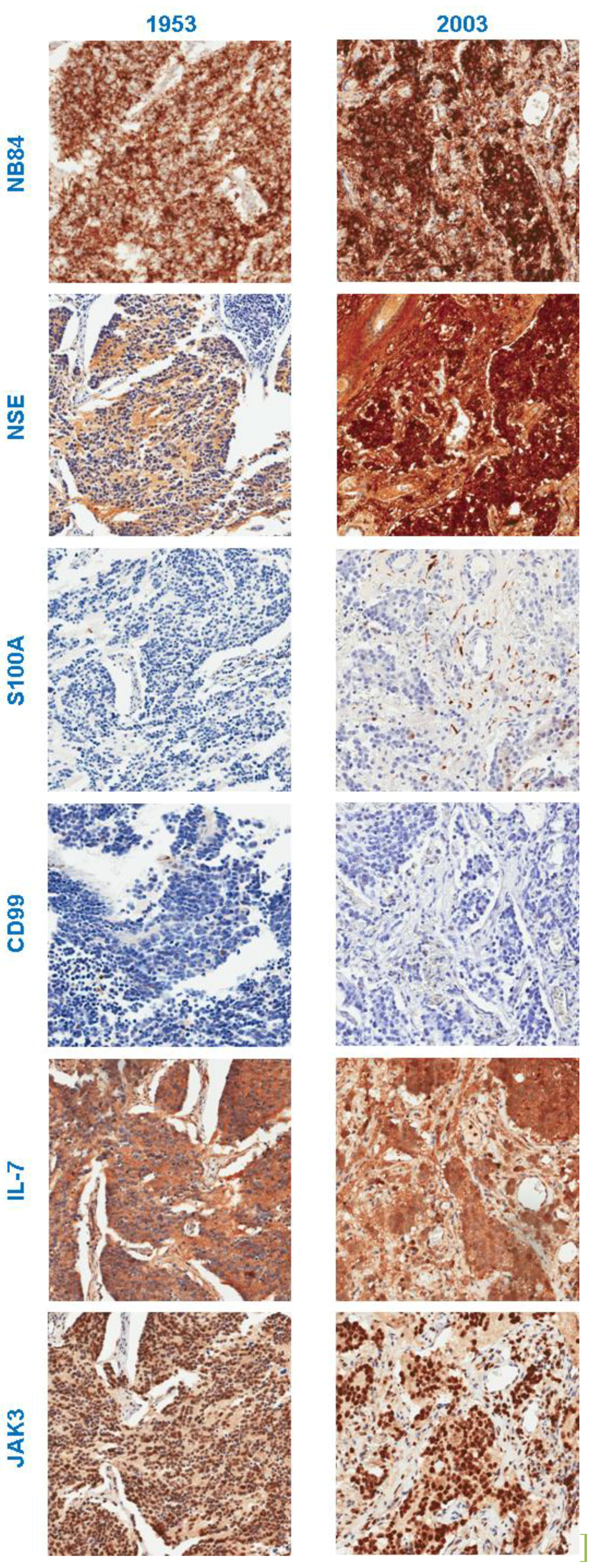
3.2. Immunohistochemistry
3.3. Computer-Aided Image Analysis
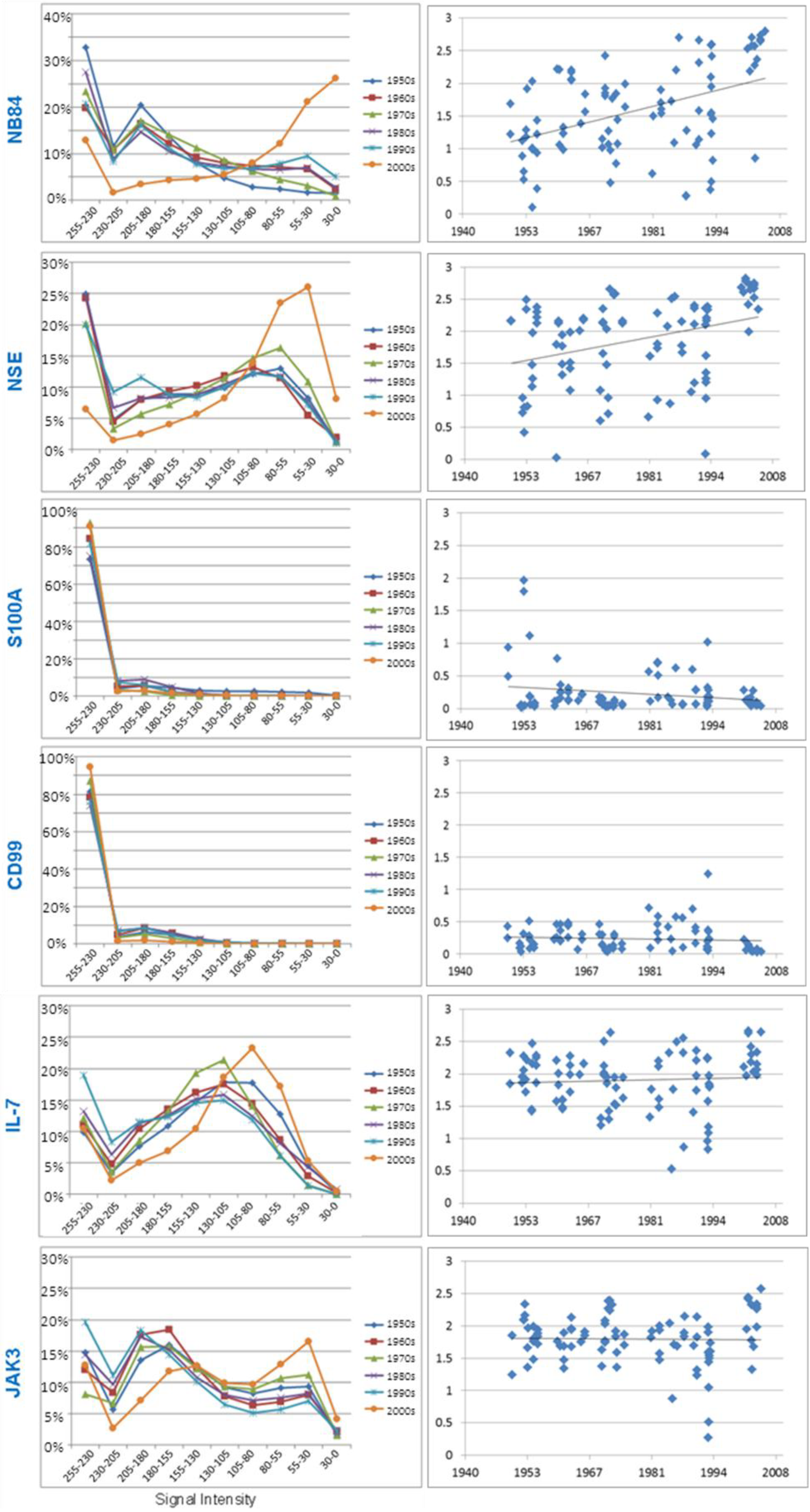
3.3.1. Neuroblastoma Marker NB84
| Marker | Linear Regression (R) | p value | |
|---|---|---|---|
| Diagnostic | NB84 | 0.432 | <0.001 |
| Chromogranin A | −0.138 | 0.144 | |
| NSE | 0.209 | 0.026 | |
| Ki-67 | 0.177 | 0.066 | |
| INI7 | 0.072 | 0.452 | |
| Neurofilament Protein | −0.050 | 0.607 | |
| S100 | −0.258 | 0.006 | |
| CD99 | −0.200 | 0.039 | |
| Synaptophysin | −0.109 | 0.265 | |
| Research | IL-7 | −0.033 | 0.724 |
| IL-7R | 0.037 | 0.698 | |
| JAK1 | 0.006 | 0.951 | |
| JAK3 | 0.065 | 0.501 | |
| STAT5 | −0.018 | 0.855 |
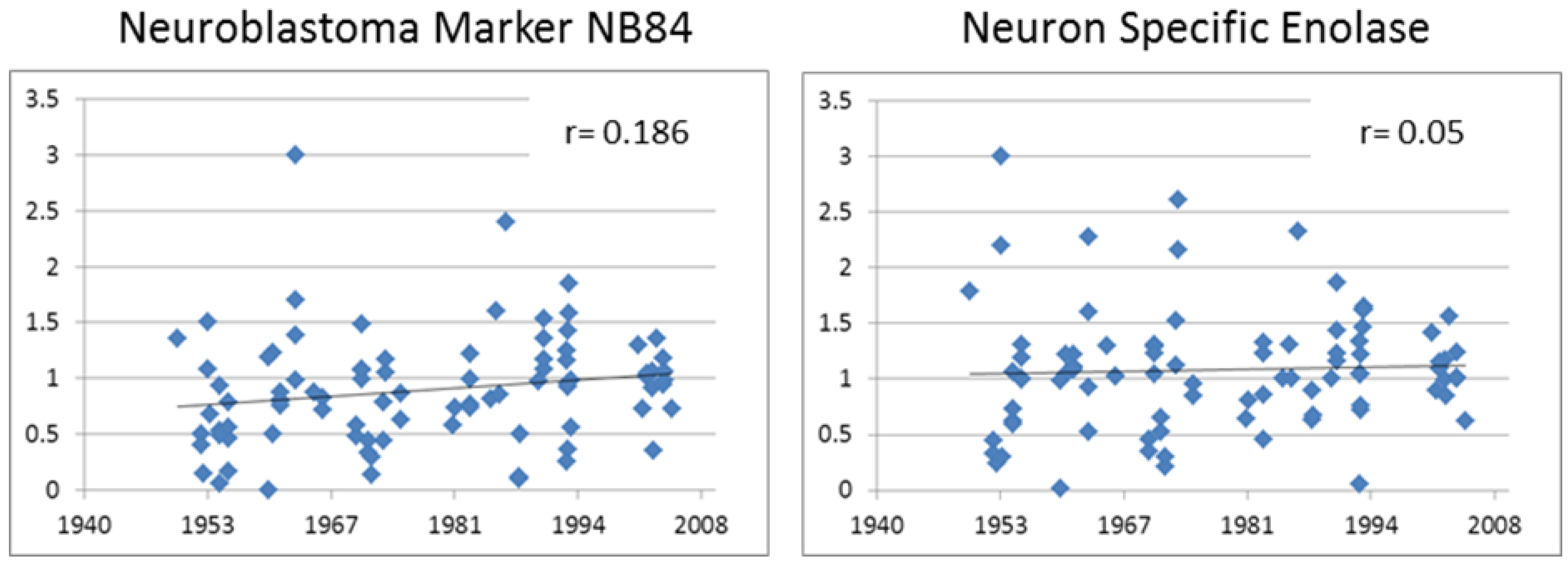
3.3.2. Neuron Specific Enolase
3.3.3. S100A
3.3.4. CD99
3.3.5. Research Antibodies: Interleukin-7, Interleukin-7 Receptor, Stat5, JAK1, and JAK3
3.3.6. Chromogranin A, Ki-67, INI1, Neurofilament Protein and Synaptophysin.
4. Conclusions
Acknowledgments
Author Contributions
Conflict of Interest
References
- Miller, R.W.; Young, J.L.; Nokavic, B. Childhood cancer. Cancer 1994, 75, 395–405. [Google Scholar] [CrossRef]
- Weinstein, J.; Katzenstein, H.; Cohn, S. Advances in the diagnosis and treatment of neuroblastoma. Oncologist 2003, 8, 278–292. [Google Scholar] [CrossRef]
- Brodeur, G.M.; Pritchard, J.; Berthold, F.; Carlsen, N.L.; Castel, V.; Castelberry, R.P.; De Bernardi, B.; Evans, A.E.; Favrot, M.; Hedborg, F. Revisions of the international criteria for neuroblastoma diagnosis, staging, and response to treatment. J. Clin. Oncol. 1993, 11, 1466–1477. [Google Scholar]
- Evans, A.E.; D’Angio, G.J.; Propert, K.; Anderson, J.; Hann, H.W. Prognostic factors in neuroblastoma. Cancer 1987, 59, 1853–1859. [Google Scholar] [CrossRef]
- Shimada, H.; Ambros, I.M.; Dehner, L.P.; Hata, J.; Joshi, V.V.; Roald, B.; Stram, D.O.; Gerbing, R.B.; Lukens, J.N.; Matthay, K.K.; et al. The international neuroblastoma pathology classification. Cancer 1999, 86, 364–372. [Google Scholar] [CrossRef]
- Maris, J.M. Recent advances in neuroblastoma. New Engl. J. Med. 2010, 362, 2202–2211. [Google Scholar] [CrossRef]
- Brodeur, G.M. Neuroblastoma: Biological insights into a clinical enigma. Nat. Rev. Canc. 2003, 3, 203–216. [Google Scholar] [CrossRef]
- Matthay, K.K.; Villablanca, J.G.; Seeger, R.C.; Stram, D.O.; Harris, R.E.; Ramsay, N.K.; Swift, P.; Shimada, H.; Black, C.T.; Brodeur, G.M.; Gerbing, R.B.; Reynolds, C.P. Treatment of high risk neuroblastoma with intensive chemotherapy, radiotherapy, autologous bone marrow transplantation, and 13-cis-retinoic acid. New Engl. J. Med. 1999, 341, 1165–1173. [Google Scholar] [CrossRef]
- Schmidt, M.L.; Lukens, J.N.; Seeger, R.C.; Brodeur, G.M.; Shimada, H.; Gerbing, R.B.; Stram, D.O.; Perez, C.; Haase, G.M.; Matthey, K.K. Biologic factors determine prognosis in infants with stage IV neuroblastoma: A prospective Children’s Cancer Group study. J. Clin. Oncol. 2000, 18, 1260–1268. [Google Scholar]
- Riley, R.D.; Heney, D.; Jones, D.R.; Sutton, A.J.; Lambert, P.C.; Abrams, K.R.; Young, B.; Wailoo, A.J.; Burchill, S.A. A systematic review of molecular and biological tumor markers in neuroblastoma. Clin. Canc. Res. 2004, 10, 4–12. [Google Scholar] [CrossRef]
- Youlden, D.; Baade, P.; Ward, L.; Valery, P.; Hassall, T.; Green, A.; Aitken, J.F. Childhood Cancer Incidence in Australia, 1983–2006; Viertel Centre for Research in Cancer Control, Cancer Council Queensland and the Australian Paediatric Cancer Registry: Brisbane, Australia, 2009. [Google Scholar]
- Camp, R.L.; Charette, L.A.; Rimm, D.L. Validation of tissue microarray technology in breast carcinoma. Lab. Investig. 2000, 80, 1943–1949. [Google Scholar] [CrossRef]
- Litlekalsoy, J.; Vatne, V.; Jens, G.; Hostmark, J.G.; Laerum, O.D. Immunohistochemical markers in urinary bladder carcinomas from paraffin-embedded archival tissue after storage for 5–70 years. BJU Int. 2007, 99, 1013–1019. [Google Scholar] [CrossRef]
- Cronin, M.; Pho, M.; Dutta, D.; Stephans, J.C.; Shak, S.; Kiefer, M.C.; Esteban, J.M.; Baker, J.B. Measurement of gene expression in archival paraffin-embedded tissues. Am. J. Pathol. 2004, 164, 35–42. [Google Scholar] [CrossRef]
- Sugimura, H.; Mori, H.; Nagura, K.; Kiyose, S.; Hong, T.; Isozaki, M.; Igarashi, H.; Shinmura, K.; Hasegawa, A.; Kitayama, Y.; Tanioka, F. Fluorescence in situ hybridization analysis with a tissue microarray: ‘FISH and chips’ analysis of pathology archives. Pathol. Int. 2010, 60, 543–550. [Google Scholar]
- Kononen, J.; Bubendorf, L.; Kallioniemi, A.; Barlund, M.; Schraml, P.; Leighton, S.; Torhorst, J.; Mihatsch, M.J.; Sauter, G.; Kallioniemi, O.P. Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nat. Med. 1998, 4, 844–847. [Google Scholar] [CrossRef]
- Catchpoole, D.R.; Mackie, N.; Chetcuti, A.; McIver, S.; Henwood, T.; Kan, A.; Graf, N.; Arbuckle, S. Tape transfer sectioning of tissue microarrays may lead to false positive immunohistochemistry staining. Biotech. Histochem. 2011, 86, 421–428. [Google Scholar] [CrossRef]
- Tafavogh, S.; Navarro, K.F.; Catchpoole, D.R.; Kennedy, P.J. Segmenting cellular regions of neuroblastoma tumor and splitting overlapping cells using shortest path between convex regions of cell contours. Artif. Intell. Med. 2013, 7885, 171–175. [Google Scholar]
- Tafavogh, S.; Catchpoole, D.R.; Kennedy, P.J. Determining cellularity status of tumors based on histopathology using hybrid image segmentation. In Proceedings of the 2012 International Joint Conference on Neural Networks (IJCNN), Brisbane, Australia, 10–15 June 2012; pp. 1–8.
- Tafavogh, S.; Navarro, K.F.; Catchpoole, D.R.; Kennedy, P.J. Non-parametric and integrated framework for segmenting and counting neuroblastic cells within neuroblastoma tumor images. Med. Biol. Eng. Comput. 2013, 51, 645–655. [Google Scholar] [CrossRef]
- Ryan, D.; Mulrane, L.; Rexhepaj, E.; Gallagher, W.M. Tissue microarrays and digital image analysis. In Drug Safety Evaluation: Methods and Protocols, Methods in Molecular Biology; Gautier, J.-C., Ed.; Springer Science+Business Media LLC: Philadelphia, PA, USA, 2011; Volume 691, pp. 97–112. [Google Scholar]
- Chetcuti, A.; Aktas, S.; Mackie, N.; Ulger, C.; Toruner, G.; Alkan, M.; Catchpoole, D. Expression profiling reveals MSX1 and EphB2 expression correlates with the invasion capacity of Wilms tumors. Pediatr. Blood Canc. 2011, 57, 950–957. [Google Scholar] [CrossRef]
- Krajewska, M.; Smith, L.H.; Rong, J.; Huang, X.; Hyer, M.L.; Zeps, N.; Iacopetta, B.; Linke, S.P.; Olson, A.H.; Reed, J.C.; Krajewski, S. Image analysis algorithms for immunohistochemical assessment of cell death events and fibrosis in tissue sections. J. Histochem. Cytochem. 2009, 57, 649–663. [Google Scholar] [CrossRef]
- Miettinen, M.; Chatten, J.; Paetau, A.; Stevenson, A. Monoclonal antibody NB84 in the differential diagnosis of neuroblastoma and other small round cell tumors. Am. J. Surg. Pathol. 1998, 22, 327–332. [Google Scholar] [CrossRef]
- Tsokos, M.; Linnoila, R.I.; Chandra, R.S.; Triche, T.J. Neuron-specific enolase in the diagnosis of neuroblastoma and other small, round-cell tumors in children. Human Pathol. 1984, 15, 575–584. [Google Scholar] [CrossRef]
- Misugi, K.; Aoki, I.; Kikyo, S.; Sasaki, Y.; Tsunoda, A.; Nakajima, T. Immunohistochemical study of neuroblastoma and related tumors with anti-S-100 protein antibody. Pediatr. Pathol. 1985, 3, 217–226. [Google Scholar] [CrossRef]
- Sugita, H.; Kusano, K.; Tokunaga, O.; Mineta, T.; Abe, M.; Harada, H.; Shigemori, M. Olfactory neuroepithelioma: An immunohistochemical and ultrastructural study. Neuropathology 2006, 26, 400–408. [Google Scholar] [CrossRef]
- Parham, D.M. Neuroectodermal and neuroendocrine tumors principally seen in children. Am. J. Clin. Pathol. 2001, 115, S113–S128. [Google Scholar]
- Rozental, R.; Morales, M.; Mehler, M.F.; Urban, M.; Kremer, M.; Dermietzel, R.; Kessler, J.A.; Spray, D.C. Changes in the properties of gap junctions during neuronal differentiation of hippocampal progenitor cells. J. Neurosci. 1998, 18, 1753–1762. [Google Scholar]
- Prasad, L.; Gayagay, A.; Charlton, A.; Henwood, A.; Graf, N.; Arbuckle, S.; Catchpoole, D. Expression of interleukin-7 and its signalling intermediates in human neuroblastoma tumours. In Proceedings of the 103rd Annual Meeting of the American Association for Cancer Research, Chicago, IL, USA, 31 March–4 April USA 2012.
- Hirose, T.; Scheithauer, B.W.; Lopes, M.B.S.; Gerber, S.H.A.; Altermatt, H.I.; Harner, S.G.; VandenBerg, S.R. Olfactory neuroblastoma: An immunohistochemical, ultrastructural, and flow cytometric study. Cancer 2006, 76, 4–19. [Google Scholar]
- Wilson, B.S.; Lloyd, R.V. Detection of chromogranin in neuroendocrine cells with a monoclonal antibody. Am. J. Pathol. 1984, 115, 458–468. [Google Scholar]
- Takeuchi, T.; Nicole, S.; Misaki, A.; Furihata, M.; Iwata, J.; Sonobe, H.; Ohtsuki, Y. Expression of SMARCF1, a truncated form of SWI1, in neuroblastoma. Am. J. Pathol. 2001, 158, 663–672. [Google Scholar] [CrossRef]
- Chiriboga, L.; Osman, I.; Mikhail, M.; Lau, C. Tissue microarrays, tread carefully. Lab. Investig. 2004, 84, 1677. [Google Scholar] [CrossRef]
- Gomes, L.; Mackie, N.; Catchpoole, D.R.; Henwood, T. Test and teach—In a fix about immunohistochemistry on 60 year old tissue blocks? J. Histotechnol. 2008, 31, 183–184. [Google Scholar] [CrossRef]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Chetcuti, A.; Mackie, N.; Tafavogh, S.; Graf, N.; Henwood, T.; Charlton, A.; Catchpoole, D. Can Archival Tissue Reveal Answers to Modern Research Questions?: Computer-Aided Histological Assessment of Neuroblastoma Tumours Collected over 60 Years. Microarrays 2014, 3, 72-88. https://doi.org/10.3390/microarrays3010072
Chetcuti A, Mackie N, Tafavogh S, Graf N, Henwood T, Charlton A, Catchpoole D. Can Archival Tissue Reveal Answers to Modern Research Questions?: Computer-Aided Histological Assessment of Neuroblastoma Tumours Collected over 60 Years. Microarrays. 2014; 3(1):72-88. https://doi.org/10.3390/microarrays3010072
Chicago/Turabian StyleChetcuti, Albert, Nicole Mackie, Siamak Tafavogh, Nicole Graf, Tony Henwood, Amanda Charlton, and Daniel Catchpoole. 2014. "Can Archival Tissue Reveal Answers to Modern Research Questions?: Computer-Aided Histological Assessment of Neuroblastoma Tumours Collected over 60 Years" Microarrays 3, no. 1: 72-88. https://doi.org/10.3390/microarrays3010072
APA StyleChetcuti, A., Mackie, N., Tafavogh, S., Graf, N., Henwood, T., Charlton, A., & Catchpoole, D. (2014). Can Archival Tissue Reveal Answers to Modern Research Questions?: Computer-Aided Histological Assessment of Neuroblastoma Tumours Collected over 60 Years. Microarrays, 3(1), 72-88. https://doi.org/10.3390/microarrays3010072





