Novel Antioxidant, Deethylated Ethoxyquin, Protects against Carbon Tetrachloride Induced Hepatotoxicity in Rats by Inhibiting NLRP3 Inflammasome Activation and Apoptosis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Biological Activity/Toxicity Prediction
2.3. Animals and Experimental Design
2.4. Caspase Activity Assessment
2.5. Aminotransferase Activity Assessment
2.6. Histopathological Examination
2.7. Immunofluorescent Staining
2.8. Myeloperoxidase Activity Assay
2.9. Measurements of Antioxidant Enzyme Activity
2.10. Measurement of Reduced Glutathione Concentration
2.11. Quantitative PCR
2.12. Statistical Analysis
3. Results
3.1. Toxicological Properties and Biological Activity Profile of DEQ
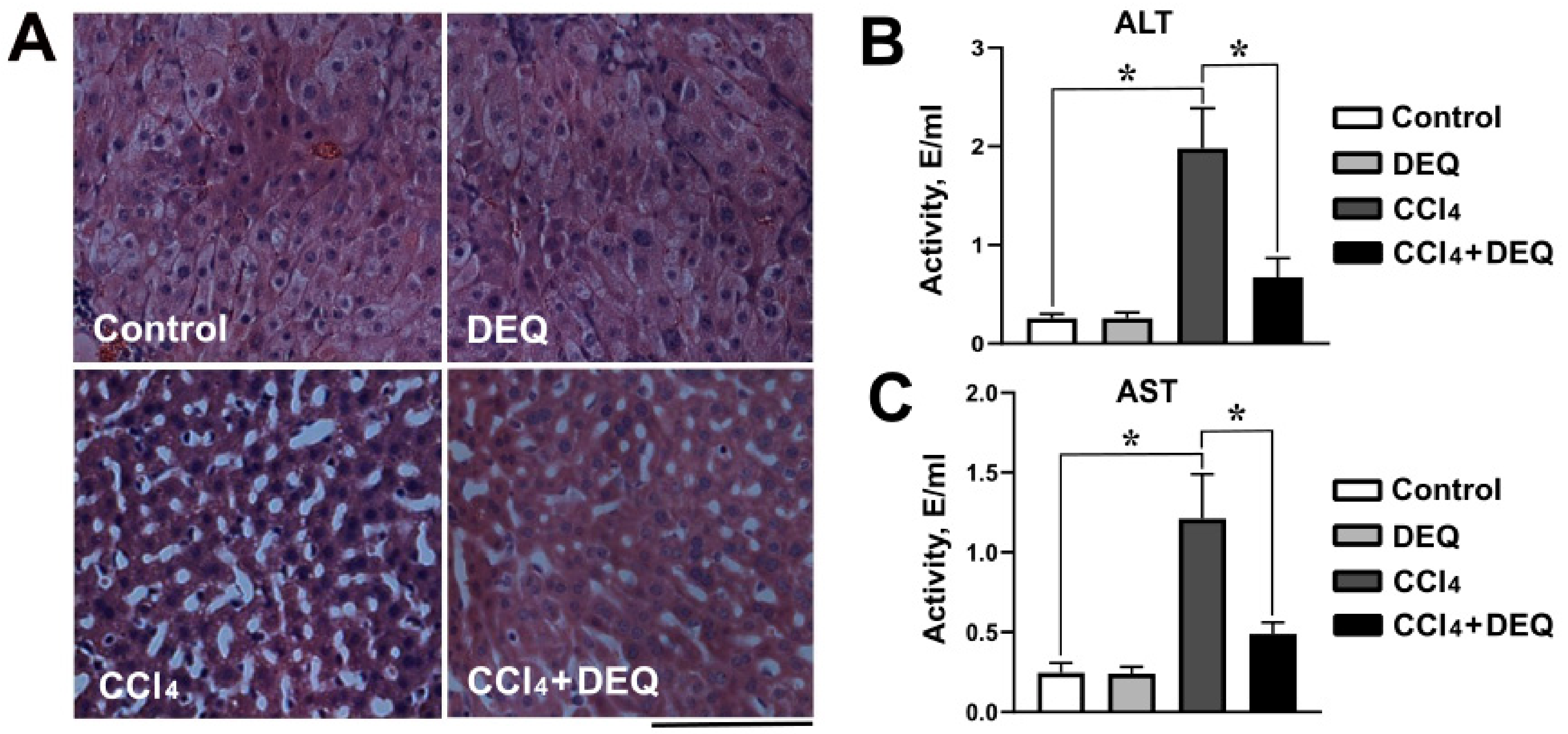
3.2. DEQ Alleviates CCl4-Induced Histological Changes in the Liver
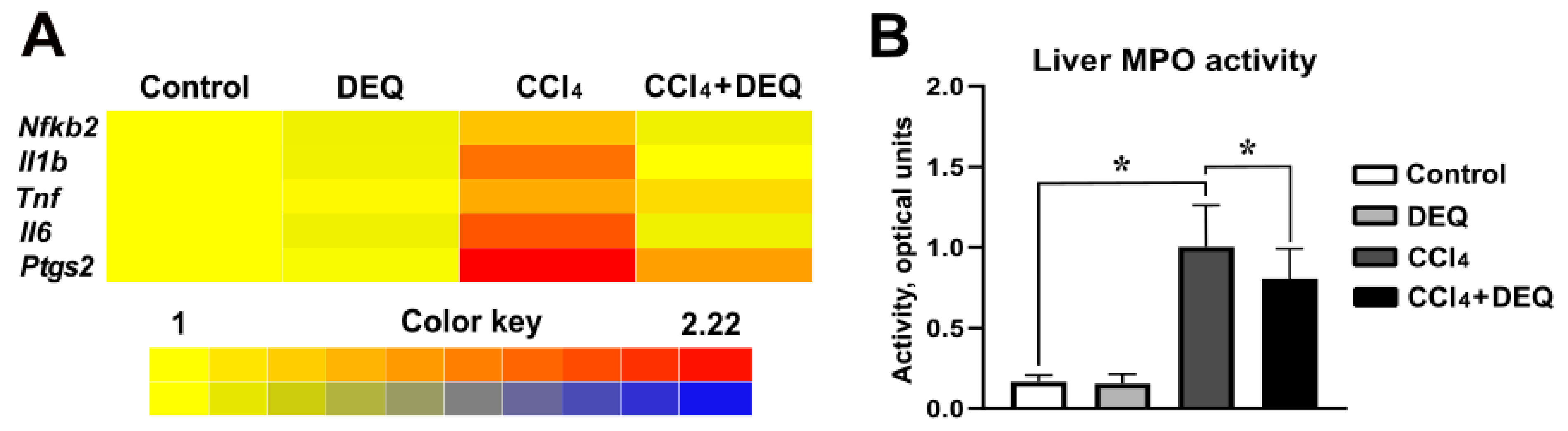
3.3. DEQ Inhibits CCl4-Induced Inflammatory Responses via NLRP3 Inflammasome Pathway in the Liver
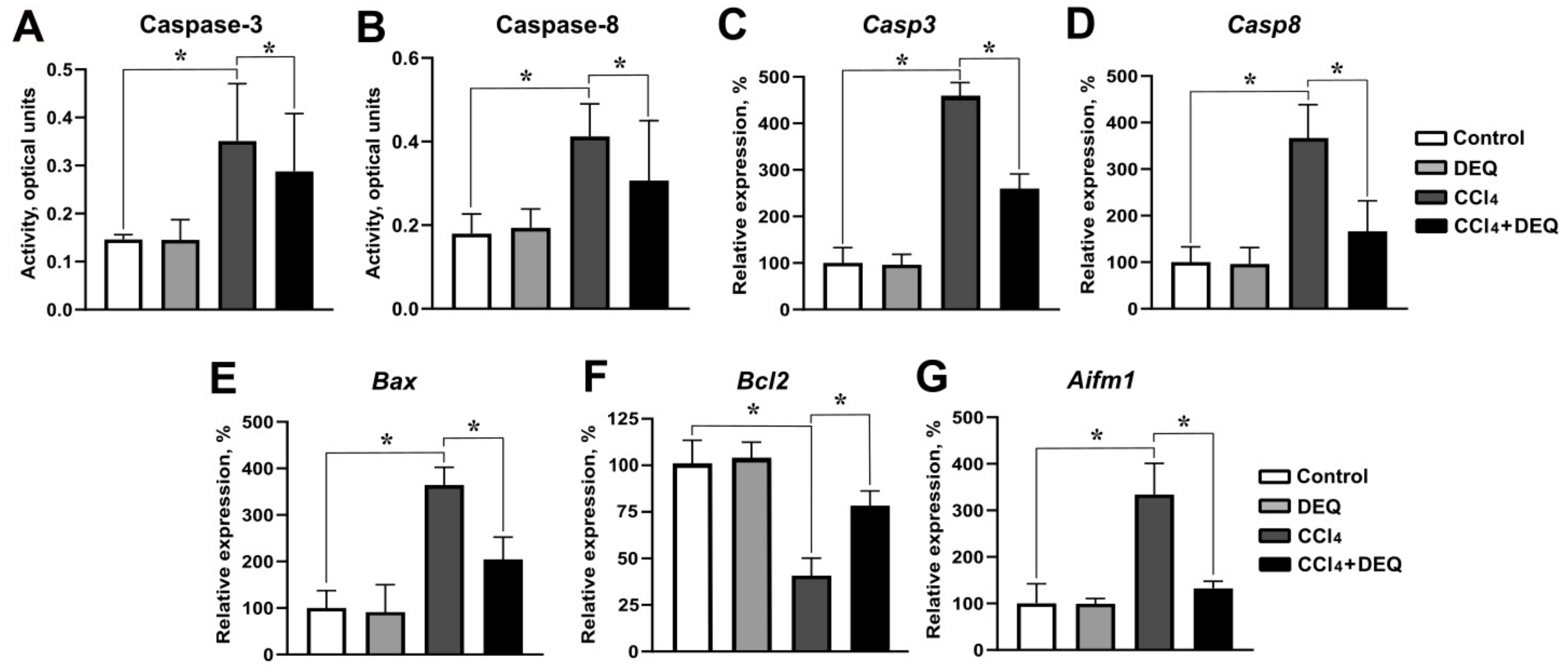
3.4. DEQ Attenuates CCl4-Induced Hepatic Apoptosis
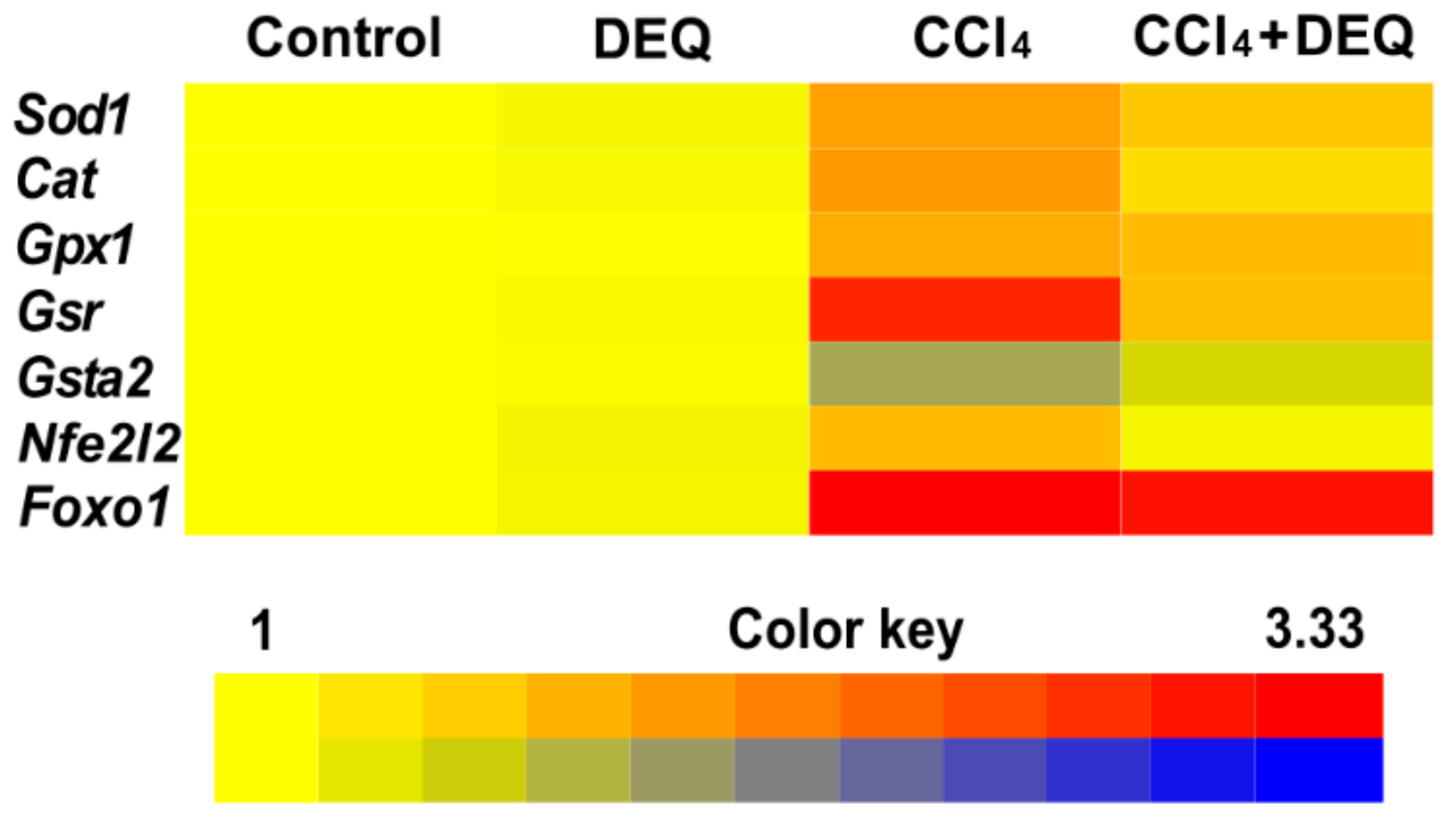
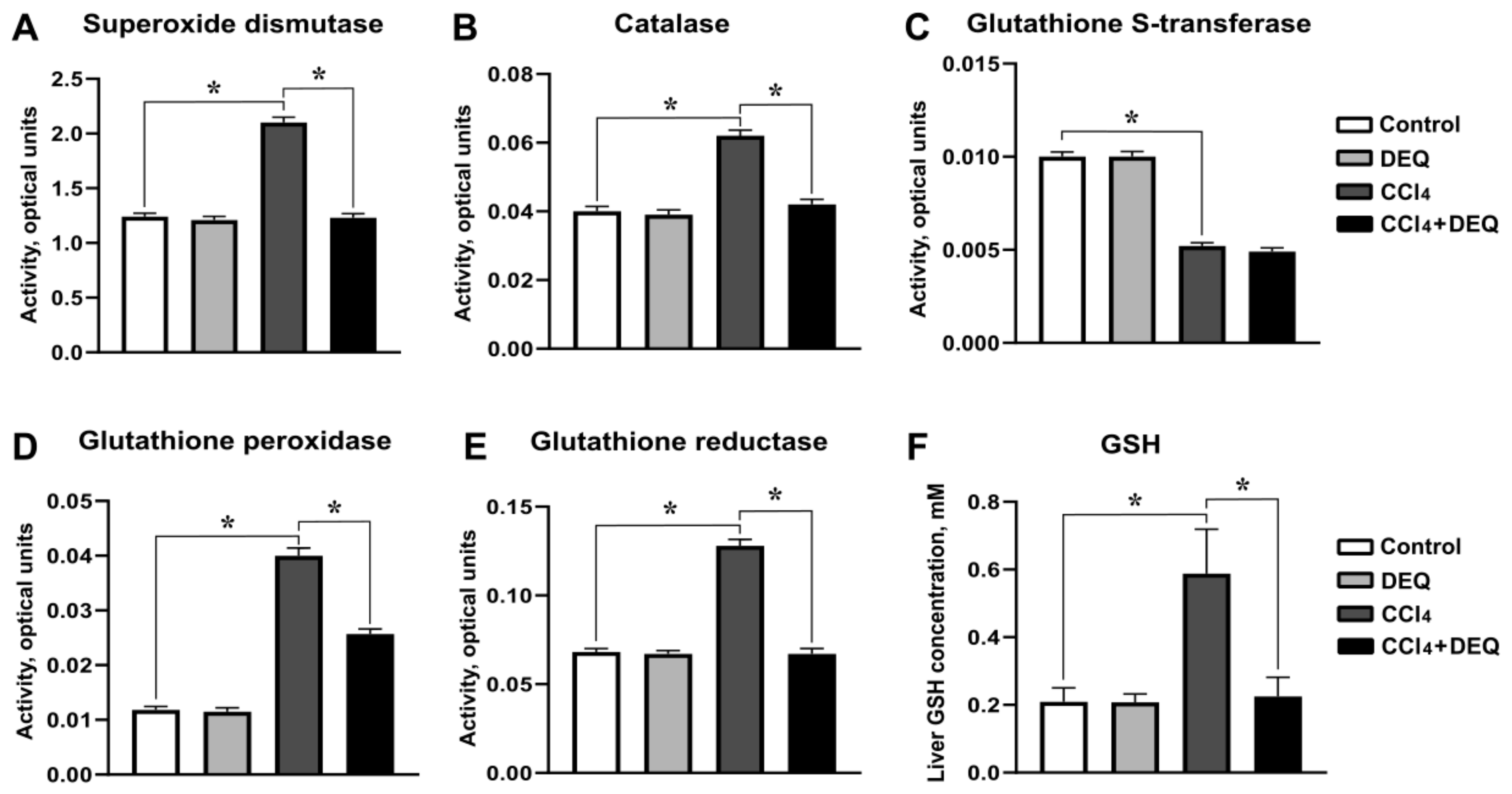
3.5. DEQ Restores the Activities of Antioxidant Enzymes and Reduced Glutathione Concentration in CCl4-Induced Liver Injury
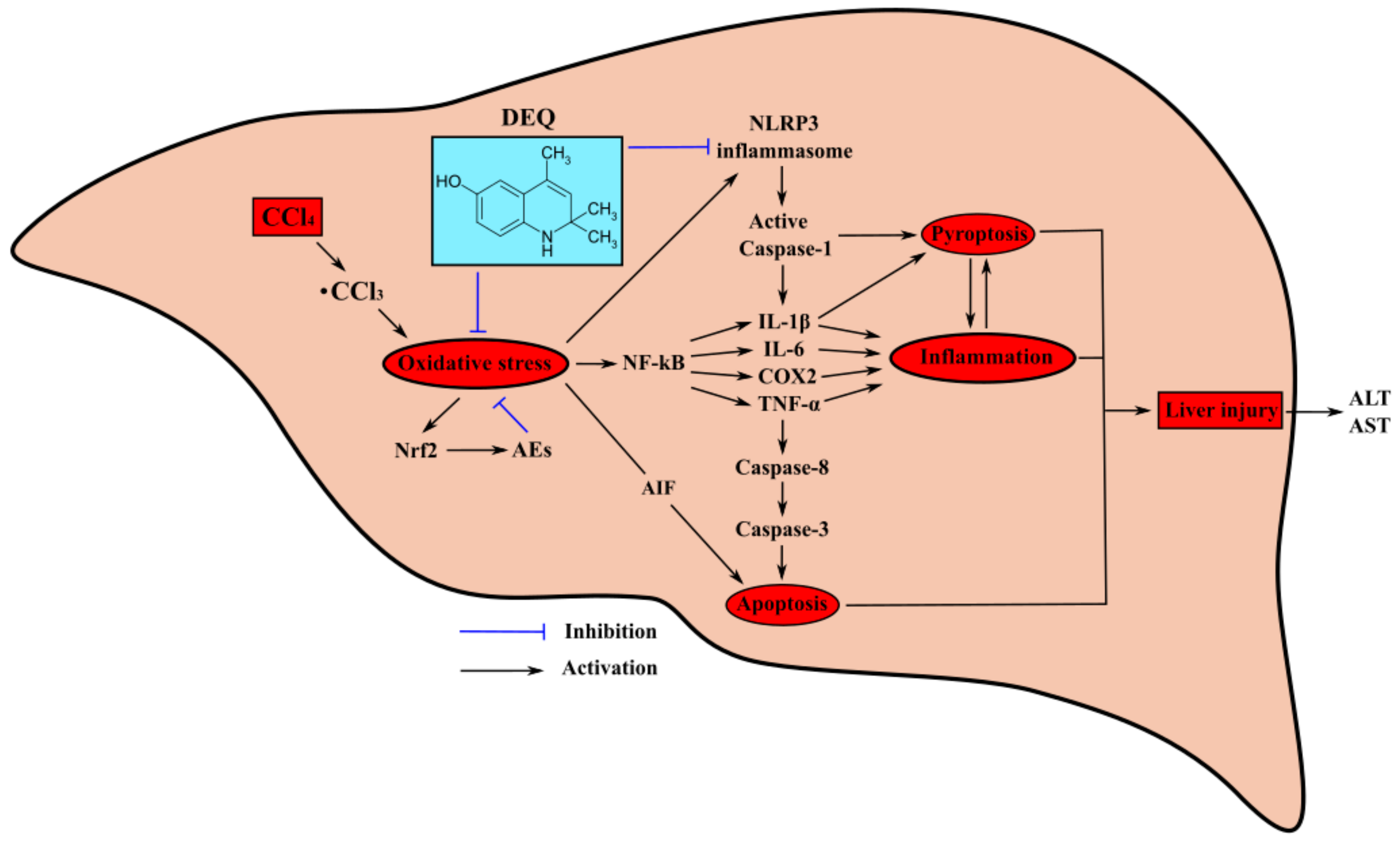
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
References
- Wu, Z.; Han, M.; Chen, T.; Yan, W.; Ning, Q. Acute liver failure: Mechanisms of immune-mediated liver injury. Liver Int. 2010, 30, 782–794. [Google Scholar] [CrossRef]
- Bernal, W.; Auzinger, G.; Dhawan, A.; Wendon, J. Acute liver failure. Lancet 2010, 376, 190–201. [Google Scholar] [CrossRef]
- Jia, R.; Cao, L.P.; Du, J.L.; Wang, J.H.; Liu, Y.J.; Jeney, G.; Xu, P.; Yin, G.J. Effects of carbon tetrachloride on oxidative stress, inflammatory response and hepatocyte apoptosis in common carp (Cyprinus carpio). Aquat. Toxicol. 2014, 152, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Wu, R.; Xiao, Z.; Zhang, X.; Liu, F.; Zhou, W.; Zhang, Y. The Cytochrome P450-mediated Metabolism Alternation of Four Effective Lignans from Schisandra chinensis in Carbon Tetrachloride-Intoxicated Rats and Patients with Advanced Hepatocellular Carcinoma. Front. Pharmacol. 2018, 9, 229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ishii, T.; Itoh, K.; Yamamoto, M. Roles of Nrf2 in activation of antioxidant enzyme genes via antioxidant responsive elements. Methods Enzymol. 2002, 348, 182–190. [Google Scholar] [CrossRef]
- Cao, M.; Wang, H.; Guo, L.; Yang, S.; Liu, C.; Khor, T.O.; Yu, S.; Kong, A.N. Dibenzoylmethane Protects Against CCl4-Induced Acute Liver Injury by Activating Nrf2 via JNK, AMPK, and Calcium Signaling. AAPS J. 2017, 19, 1703–1714. [Google Scholar] [CrossRef]
- Pan, C.W.; Pan, Z.Z.; Hu, J.J.; Chen, W.L.; Zhou, G.Y.; Lin, W.; Jin, L.X.; Xu, C.L. Mangiferin alleviates lipopolysaccharide and D-galactosamine-induced acute liver injury by activating the Nrf2 pathway and inhibiting NLRP3 inflammasome activation. Eur. J. Pharmacol. 2016, 770, 85–91. [Google Scholar] [CrossRef]
- Lyu, H.; Wang, H.; Li, L.; Zhu, J.; Chen, F.; Chen, Y.; Liu, C.; Fu, J.; Yang, B.; Zhang, Q.; et al. Hepatocyte-specific deficiency of Nrf2 exacerbates carbon tetrachloride-induced liver fibrosis via aggravated hepatocyte injury and subsequent inflammatory and fibrogenic responses. Free Radic. Biol. Med. 2020, 150, 136–147. [Google Scholar] [CrossRef]
- Endig, J.; Unrau, L.; Sprezyna, P.; Rading, S.; Karsak, M.; Goltz, D.; Heukamp, L.C.; Tiegs, G.; Diehl, L. Acute Liver Injury after CCl4 Administration is Independent of Smad7 Expression in Myeloid Cells. Int. J. Mol. Sci. 2019, 20, 5528. [Google Scholar] [CrossRef] [Green Version]
- Han, J.; Bae, J.; Choi, C.Y.; Choi, S.P.; Kang, H.S.; Jo, E.K.; Park, J.; Lee, Y.S.; Moon, H.S.; Park, C.G.; et al. Autophagy induced by AXL receptor tyrosine kinase alleviates acute liver injury via inhibition of NLRP3 inflammasome activation in mice. Autophagy 2016, 12, 2326–2343. [Google Scholar] [CrossRef] [Green Version]
- Sun, W.; Zeng, C.; Liu, S.; Fu, J.; Hu, L.; Shi, Z.; Yue, D.; Ren, Z.; Zhong, Z.; Zuo, Z.; et al. Ageratina adenophora induces mice hepatotoxicity via ROS-NLRP3-mediated pyroptosis. Sci. Rep. 2018, 8, 16032. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, J.; Lin, S.; Wan, B.; Velani, B.; Zhu, Y. Pyroptosis in Liver Disease: New Insights into Disease Mechanisms. Aging Dis. 2019, 10, 1094–1108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khanova, E.; Wu, R.; Wang, W.; Yan, R.; Chen, Y.; French, S.W.; Llorente, C.; Pan, S.Q.; Yang, Q.; Li, Y.; et al. Pyroptosis by caspase11/4-gasdermin-D pathway in alcoholic hepatitis in mice and patients. Hepatology 2018, 67, 1737–1753. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- DeCicco, L.A.; Rikans, L.E.; Tutor, C.G.; Hornbrook, K.R. Serum and liver concentrations of tumor necrosis factor alpha and interleukin-1beta following administration of carbon tetrachloride to male rats. Toxicol. Lett. 1998, 98, 115–121. [Google Scholar] [CrossRef]
- Kim, S.H.; Chu, H.J.; Kang, D.H.; Song, G.A.; Cho, M.; Yang, U.S.; Kim, H.J.; Chung, H.Y. NF-kappa B binding activity and cyclooxygenase-2 expression in persistent CCl(4)-treated rat liver injury. J. Korean Med. Sci. 2002, 17, 193–200. [Google Scholar] [CrossRef] [Green Version]
- Shen, H.; Sheng, L.; Chen, Z.; Jiang, L.; Su, H.; Yin, L.; Omary, M.B.; Rui, L. Mouse hepatocyte overexpression of NF-κB-inducing kinase (NIK) triggers fatal macrophage-dependent liver injury and fibrosis. Hepatology 2014, 60, 2065–2076. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, H.; Chai, Y.; Dong, D.; Zhang, N.; Liu, W.; Ma, T.; Wu, R.; Lv, Y.; Hu, L. AICAR-Induced AMPK Activation Inhibits the Noncanonical NF-κB Pathway to Attenuate Liver Injury and Fibrosis in BDL Rats. Can. J. Gastroenterol. Hepatol. 2018, 2018, 6181432. [Google Scholar] [CrossRef] [Green Version]
- Yue, S.; Xue, N.; Li, H.; Huang, B.; Chen, Z.; Wang, X. Hepatoprotective Effect of Apigenin Against Liver Injury via the Non-canonical NF-κB Pathway In Vivo and In Vitro. Inflammation 2020, 43, 1634–1648. [Google Scholar] [CrossRef]
- Lin, S.Y.; Dan, X.; Du, X.X.; Ran, C.L.; Lu, X.; Ren, S.J.; Tang, Z.T.; Yin, L.Z.; He, C.L.; Yuan, Z.X.; et al. Protective Effects of Salidroside against Carbon Tetrachloride (CCl4)-Induced Liver Injury by Initiating Mitochondria to Resist Oxidative Stress in Mice. Int. J. Mol. Sci. 2019, 20, 3187. [Google Scholar] [CrossRef] [Green Version]
- Ullah, H.; Khan, A.; Baig, M.W.; Ullah, N.; Ahmed, N.; Tipu, M.K.; Ali, H.; Khan, S. Poncirin attenuates CCL4-induced liver injury through inhibition of oxidative stress and inflammatory cytokines in mice. BMC Complement. Med. Ther. 2020, 20, 115. [Google Scholar] [CrossRef] [Green Version]
- Milani, L.; Galindo, C.M.; Turin de Oliveira, N.M.; Corso, C.R.; Adami, E.R.; Stipp, M.C.; Beltrame, O.C.; Acco, A. The GLP-1 analog liraglutide attenuates acute liver injury in mice. Ann. Hepatol. 2019, 18, 918–928. [Google Scholar] [CrossRef] [PubMed]
- Wattenberg, L.W. Inhibition of carcinogenic and toxic effects of polycyclic hydrocarbons by phenolic antioxidants and ethoxyquin. J. Natl. Cancer Inst. 1972, 48, 1425–1430. [Google Scholar] [PubMed]
- Błaszczyk, A.; Augustyniak, A.; Skolimowski, J. Ethoxyquin: An Antioxidant Used in Animal Feed. Int. J. Food Sci. 2013, 2013, 585931. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Plumb, G.W.; Lambert, N.; Chambers, S.J.; Clarke, H.L.; Manson, M.M.; Neal, G.; Williamson, G. Effect of ethoxyquin on glutathione levels and peroxidisability of rat liver membranes. Biochem. Soc. Trans. 1996, 24, 380S. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Henson, K.L.; Stauffer, G.; Gallagher, E.P. Induction of glutathione S-transferase activity and protein expression in brown bullhead (Ameiurus nebulosus) liver by ethoxyquin. Toxicol. Sci. 2001, 62, 54–60. [Google Scholar] [CrossRef] [Green Version]
- Miyazawa, K.; Iimori, Y.; Makino, M.; Mikami, T.; Miyasaka, K. Effects of some non-steroidal anti-inflammatory drugs and other agents on cyclooxygenase and lipoxygenase activities in some enzyme preparations. Jpn. J. Pharmacol. 1985, 38, 199–205. [Google Scholar] [CrossRef] [Green Version]
- Levy, L. The antiinflammatory action of some compounds with antioxidant properties. Inflammation 1976, 1, 333–345. [Google Scholar] [CrossRef]
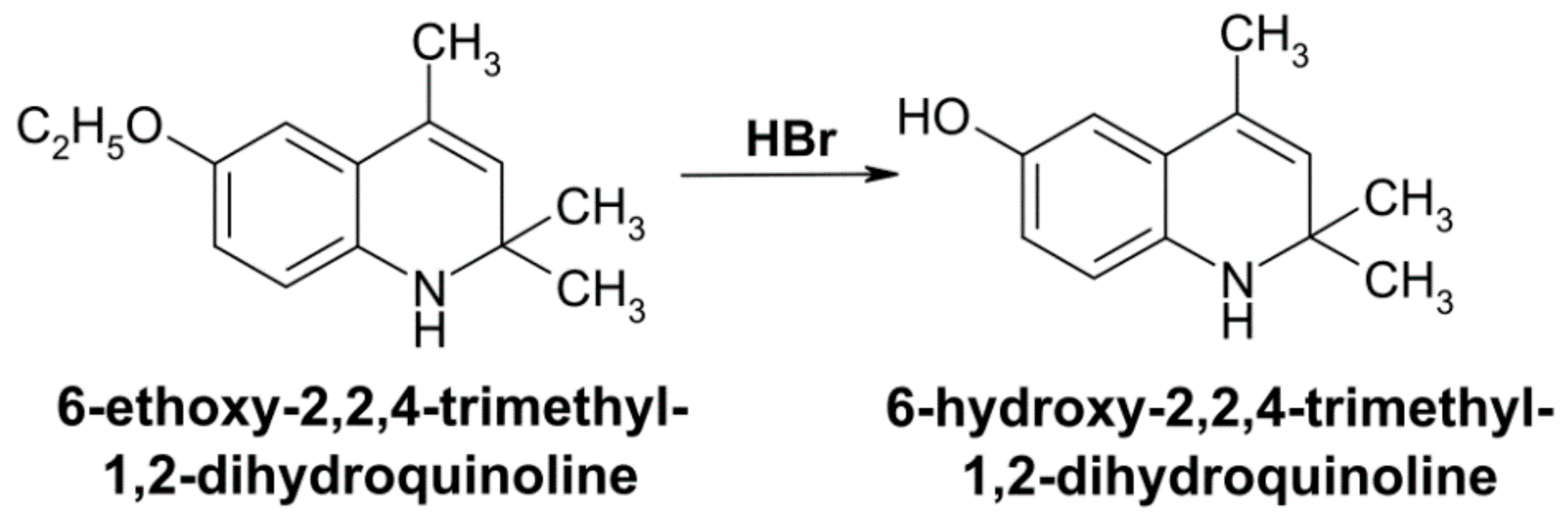
- Cawthorne, M.A.; Palmer, E.D.; Green, J. Effect of 6-ethoxy 2,2,4-trimethyl-1,2-dihydroquinoline (ethoxyquin) on carbon tetrachloride metabolism in the rat. Biochem. Pharmacol. 1973, 22, 783–788. [Google Scholar] [CrossRef]
- Brazhnikova, D.A.; Popova, T.N.; Kryl’skii, E.D.; Shulgin, K.K.; Matasova, L.V.; Shikhaliev, K.S.; Popov, S.S. Vozdeĭstvie 6-gidroksi-2,2,4-trimetil-1,2-digidro-khinolina na intensivnost’ svobodnoradikal’nykh protsessov i aktivnost’ fermentov okislitel’nogo metabolizma pri toksicheskom porazhenii pecheni u krys [Effect of 6-hydroxy-2,2,4-trimethyl-1,2-dihydroquinoline on the intensity of free radical processes and the activity of oxidative metabolism enzymes under toxic liver injury in rats]. Biomed. Khim. 2019, 65, 331–338. [Google Scholar] [CrossRef]
- Ivanov, Y.A.; Zaichenko, N.L.; Rykov, S.V.; Greenberg, O.Y.; Dubinsky, A.A.; Pirozhkov, S.D.; Rozantsev, E.G.; Pokrovskaya, I.E.; Shapiro, A.B. Synthesis of oxy-, acylo-, oxo-, N-oxides of oxo- and morpholyloxo derivatives of hydrogenated quinolines and the study of their radical analogues by EPR. Izv. USSR Acad. Sci. Ser. Chem. 1979, 8, 1800–1807. [Google Scholar]
- Lagunin, A.; Filimonov, D.; Poroikov, V. Multi-targeted natural products evaluation based on biological activity prediction with PASS. Curr. Pharm. Des. 2010, 16, 1703–1717. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Banerjee, P.; Eckert, A.O.; Schrey, A.K.; Preissner, R. ProTox-II: A webserver for the prediction of toxicity of chemicals. Nucleic Acids Res. 2018, 46, W257–W263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suzuki, K.; Ota, H.; Sasagawa, S.; Sakatani, T.; Fujikura, T. Assay method for myeloperoxidase in human polymorphonuclear leukocytes. Anal. Biochem. 1983, 132, 345–352. [Google Scholar] [CrossRef]
- Nishikimi, M.; Rao, N.A.; Yagi, K. The occurrence of superoxide anion in the reaction of reduced phenazine methosulphate and molecular oxygen. Biochem. Biophys. Res. Co. 1972, 46, 849–864. [Google Scholar] [CrossRef]
- Góth, L. A simple method for determination of serum catalase activity and revision of reference range. Clin. Chim. Acta. 1991, 196, 143–151. [Google Scholar] [CrossRef]
- Paglia, D.E.; Valentine, W.N. Studies on the quantitative and qualitative characterization of erythrocyte glutathione peroxidase. Lab. Clin. Med. 1967, 70, 158–169. [Google Scholar]
- Zanetti, G. Rabbit liver glutathione reductase. Purification and properties. Arch. Biochem. Biophys. 1979, 198, 241–246. [Google Scholar] [CrossRef]
- Warholm, M.; Guthenberg, C.; Von Bahr, C.; Mannervik, B. Glutathione transferases from human liver. Method. Enzymol. 1985, 113, 499–504. [Google Scholar] [CrossRef]
- Ellman, G.L. Tissue sulfhydryl groups. Arch. Biochem. Biophys. 1959, 82, 70–77. [Google Scholar] [CrossRef]
- Wang, M.; Niu, J.; Ou, L.; Deng, B.; Wang, Y.; Li, S. Zerumbone Protects against Carbon Tetrachloride (CCl4)-Induced Acute Liver Injury in Mice via Inhibiting Oxidative Stress and the Inflammatory Response: Involving the TLR4/NF-κB/COX-2 Pathway. Molecules 2019, 24, 1964. [Google Scholar] [CrossRef] [Green Version]
- Mridha, A.R.; Were, A.; Robertson, A.A.B.; Yeh, M.M.; Johnson, C.D.; Van Rooyen, D.M.; Haczeyni, F.; Teoh, N.C.H.; Savard, C.; Ioannou, G.N.; et al. NLRP3 inflammasome blockade reduces liver inflammation and fibrosis in experimental NASH in mice. J. Hepatol. 2017, 66, 1037–1046. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Li, J.; Zhao, Y.; Liu, D. TNFα-induced downregulation of microRNA-186 contributes to apoptosis in rat primary cardiomyocytes. Immunobiology 2017, 222, 778–784. [Google Scholar] [CrossRef] [PubMed]
- Anuja, G.I.; Shine, V.J.; Latha, P.G.; Suja, S.R. Protective effect of ethyl acetate fraction of Drynaria quercifolia against CCl4 induced rat liver fibrosis via Nrf2/ARE and NFκB signalling pathway. J. Ethnopharmacol. 2018, 216, 79–88. [Google Scholar] [CrossRef] [PubMed]
- Qi, R.; Jiang, R.; Xiao, H.; Wang, Z.; He, S.; Wang, L.; Wang, Y. Ginsenoside Rg1 protects against D-galactose induced fatty liver disease in a mouse model via FOXO1 transcriptional factor. Life Sci. 2020, 254, 117776. [Google Scholar] [CrossRef] [PubMed]
- David, S.; Hamilton, J.P. Drug-induced Liver Injury. US Gastroenterol. Hepatol. Rev. 2010, 6, 73–80. [Google Scholar] [PubMed]
- Arauz, J.; Ramos-Tovar, E.; Muriel, P. Redox state and methods to evaluate oxidative stress in liver damage: From bench to bedside. Ann. Hepatol. 2016, 15, 160–173. [Google Scholar] [CrossRef]
- Raschi, E.; De Ponti, F. Drug-induced liver injury: Towards early prediction and risk stratification. World J. Hepatol. 2017, 9, 30–37. [Google Scholar] [CrossRef]
- Zhao, P.; Hu, Z.; Ma, W.; Zang, L.; Tian, Z.; Hou, Q. Quercetin alleviates hyperthyroidism-induced liver damage via Nrf2 Signaling pathway. BioFactors 2020, 46, 608–619. [Google Scholar] [CrossRef]
- Shin, E.J.; Chung, Y.H.; Le, H.L.; Jeong, J.H.; Dang, D.K.; Nam, Y.; Wie, M.B.; Nah, S.Y.; Nabeshima, Y.; Nabeshima, T.; et al. Melatonin attenuates memory impairment induced by Klotho gene deficiency via interactive signaling between MT2 receptor, ERK, and Nrf2-Related antioxidant potential. Int. J. Neuropsychopharmacol. 2014, 18, pyu105. [Google Scholar] [CrossRef] [Green Version]
- Tonelli, T.; Chio, I.; Tuveson, D. Transcriptional Regulation by Nrf2. Antioxid. Redox Signal. 2018, 29, 1727–1745. [Google Scholar] [CrossRef] [Green Version]
- Calabrese, E.J. Hormesis is central to toxicology, pharmacology and risk assessment. Hum. Exp. Toxicol. 2010, 29, 249–261. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.A.; Alsahli, M.A.; Rahmani, A.H. Myeloperoxidase as an Active Disease Biomarker: Recent Biochemical and Pathological Perspectives. Med. Sci. 2018, 6, 33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, R.; Wang, Y.; Zhao, E.; Wu, K.; Li, W.; Shi, L.; Wang, D.; Xie, G.; Yin, Y.; Deng, M.; et al. Maresin 1, a proresolving lipid mediator, mitigates carbon tetrachloride-induced liver Injury in mice. Oxid. Med. Cell Longev. 2016, 2016, 9203716. [Google Scholar] [CrossRef] [PubMed]
- Forbes, L.V.; Sjogren, T.; Kettle, A.J. Potent reversible inhibition of myeloperoxidase by aromatic hydroxamates. J. Biol. Chem. 2013, 288, 36636–36647. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, X.; Wang, T.; Liu, X.; Cai, L.; Qi, J.; Zhang, P.; Li, Y. Biochanin A protects lipopolysaccharide/D-galactosamine-induced acute liver injury in mice by activating the Nrf2 pathway and inhibiting NLRP3 inflammasome activation. Int. Immunopharmacol. 2016, 38, 324–331. [Google Scholar] [CrossRef]
- Malhi, H.; Guicciardi, M.E.; Gores, G.J. Hepatocyte death: A clear and present, danger. Physiol. Rev. 2010, 90, 1165–1194. [Google Scholar] [CrossRef] [Green Version]
- Wei, H.Y.; Ma, X. Tamoxifen reduces infiltration of inflammatory cells, apoptosis and inhibits IKK/NF-κB pathway after spinal cord injury in rats. Neurol. Sci. 2014, 35, 1763–1768. [Google Scholar] [CrossRef]
- Li, Y.; Yang, J.Y.; Xie, X.; Jie, Z.; Zhang, L.; Shi, J.; Lin, D.; Gu, M.; Zhou, X.; Li, H.S.; et al. Preventing abnormal NF-κB activation and autoimmunity by Otub1-mediated p100 stabilization. Cell Res. 2019, 29, 474–485. [Google Scholar] [CrossRef]
- Harris, T.R.; Kodani, S.; Rand, A.A.; Yang, J.; Imai, D.M.; Hwang, S.H.; Hammock, B.D. Celecoxib Does Not Protect against Fibrosis and Inflammation in a Carbon Tetrachloride–Induced Model of Liver Injury. Mol. Pharmacol. 2018, 94, 834–841. [Google Scholar] [CrossRef] [Green Version]
- Zheng, Q.; Ren, Y.; Reinach, P.S.; She, Y.; Xiao, B.; Hua, S.; Qu, J.; Chen, W. Reactive oxygen species activated NLRP3 inflammasomes prime environment-induced murine dry eye. Exp. Eye Res. 2014, 125, 1–8. [Google Scholar] [CrossRef]
- Rouzer, C.A.; Marnett, L.J. Cyclooxygenases: Structural and functional insights. J. Lipid Res. 2009, 50, S29–S34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, Y.C.; Yang, S.F.; Huang, F.M.; Liu, C.M.; Tai, K.W.; Hsieh, Y.S. Proinflammatory cytokines induce cyclooxygenase-2 mRNA and protein expression in human pulp cell cultures. J. Endod. 2003, 29, 201–204. [Google Scholar] [CrossRef]
- Zahid, A.; Li, B.; Kombe, A.J.K.; Jin, T.; Tao, J. Pharmacological Inhibitors of the NLRP3 Inflammasome. Front. Immunol. 2019, 10, 2538. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, K. Molecular mechanisms of hepatic apoptosis. Cell Death Dis. 2014, 5, e996. [Google Scholar] [CrossRef] [PubMed]
- Iorga, A.; Dara, L.; Kaplowitz, N. Drug-Induced Liver Injury: Cascade of Events Leading to Cell Death, Apoptosis or Necrosis. Int. J. Mol. Sci. 2017, 18, 1018. [Google Scholar] [CrossRef]
- Lee, Y.S.; Cho, I.J.; Kim, J.W.; Lee, M.K.; Ku, S.K.; Choi, J.S.; Lee, H.J. Hepatoprotective effects of blue honeysuckle on CCl4-induced acute liver damaged mice. Food Sci. Nutr. 2018, 7, 322–338. [Google Scholar] [CrossRef] [Green Version]
- Beaudouin, J.; Liesche, C.; Aschenbrenner, S.; Hörner, M.; Eils, R. Caspase-8 cleaves its substrates from the plasma membrane upon CD95-induced apoptosis. Cell Death Differ. 2013, 20, 599–610. [Google Scholar] [CrossRef] [Green Version]
- Ulicná, O.; Greksák, M.; Vancová, O.; Zlatos, L.; Galbavý, S.; Bozek, P.; Nakano, M. Hepatoprotective effect of rooibos tea (Aspalathus linearis) on CCl4-induced liver damage in rats. Physiol. Res. 2003, 52, 461–466. [Google Scholar]
- Yang, C.; Li, L.; Ma, Z.; Zhong, Y.; Pang, W.; Xiong, M.; Fang, S.; Li, Y. Hepatoprotective effect of methyl ferulic acid against carbon tetrachloride-induced acute liver injury in rats. Exp. Ther. Med. 2018, 15, 2228–2238. [Google Scholar] [CrossRef]
- Sevrioukova, I.F. Apoptosis-inducing factor: Structure, function, and redox regulation. Antioxid. Redox. Signal. 2011, 14, 2545–2579. [Google Scholar] [CrossRef] [Green Version]
- Sanchez, A.; Espinosa, P.; García, T.; Mancilla, R. The 19 kDa Mycobacterium tuberculosis lipoprotein (LpqH) induces macrophage apoptosis through extrinsic and intrinsic pathways: A role for the mitochondrial apoptosis-inducing factor. Clin. Dev. Immunol. 2012, 2012, 950503. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, Y.L.; Zhang, Z.H.; Liu, X.J.; Liu, X.Q.; Tao, L.; Zhang, Y.F.; Wang, H.; Zhang, C.; Chen, X.; Xu, D.X. Melatonin protects against apoptosis-inducing factor (AIF)-dependent cell death during acetaminophen-induced acute liver failure. PLoS ONE 2012, 7, e51911. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Sl.no | Biological Activities | Deethylated Ethoxyquin |
|---|---|---|
| Predicted LD50: 300 < LD50 ≤ 2000 mg/kg | ||
| Toxicity Class: 4 | ||
| Pa | ||
| 1 | Reductant | 0.888 |
| 2 | Cytochrome P450 stimulant | 0.870 |
| 3 | CYP2C12 substrate | 0.823 |
| 4 | Progesterone antagonist | 0.791 |
| 5 | Chlordecone reductase inhibitor | 0.790 |
| 6 | Autoimmune disorders treatment | 0.737 |
| 9 | Membrane integrity agonist | 0.736 |
| 10 | CYP2J substrate | 0.725 |
| 7 | Kidney function stimulant | 0.707 |
| 8 | Lipid peroxidase inhibitor | 0.697 |
| 13 | CYP2J2 substrate | 0.645 |
| 11 | Radiosensitizer | 0.642 |
| 12 | Alkane 1-monooxygenase inhibitor | 0.628 |
| 15 | Lysase inhibitor | 0.616 |
| 14 | Glucocorticoid antagonist | 0.593 |
| 23 | Anti-inflammatory | 0.583 |
| 16 | Chemosensitizer | 0.578 |
| 17 | Menopausal disorders treatment | 0.575 |
| 21 | CYP2F1 substrate | 0.572 |
| 22 | MAP kinase stimulant | 0.571 |
| 18 | Free radical scavenger | 0.567 |
| 19 | Ophthalmic drug | 0.565 |
| 20 | Estrogen receptor beta antagonist | 0.561 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Iskusnykh, I.Y.; Kryl’skii, E.D.; Brazhnikova, D.A.; Popova, T.N.; Shikhaliev, K.S.; Shulgin, K.K.; Matasova, L.V.; Popov, S.S.; Zhaglin, D.A.; Zakharova, A.A.; et al. Novel Antioxidant, Deethylated Ethoxyquin, Protects against Carbon Tetrachloride Induced Hepatotoxicity in Rats by Inhibiting NLRP3 Inflammasome Activation and Apoptosis. Antioxidants 2021, 10, 122. https://doi.org/10.3390/antiox10010122
Iskusnykh IY, Kryl’skii ED, Brazhnikova DA, Popova TN, Shikhaliev KS, Shulgin KK, Matasova LV, Popov SS, Zhaglin DA, Zakharova AA, et al. Novel Antioxidant, Deethylated Ethoxyquin, Protects against Carbon Tetrachloride Induced Hepatotoxicity in Rats by Inhibiting NLRP3 Inflammasome Activation and Apoptosis. Antioxidants. 2021; 10(1):122. https://doi.org/10.3390/antiox10010122
Chicago/Turabian StyleIskusnykh, Igor Y., Evgenii D. Kryl’skii, Darya A. Brazhnikova, Tatyana N. Popova, Khidmet S. Shikhaliev, Konstantin K. Shulgin, Larisa V. Matasova, Sergey S. Popov, Dmitry A. Zhaglin, Anastasia A. Zakharova, and et al. 2021. "Novel Antioxidant, Deethylated Ethoxyquin, Protects against Carbon Tetrachloride Induced Hepatotoxicity in Rats by Inhibiting NLRP3 Inflammasome Activation and Apoptosis" Antioxidants 10, no. 1: 122. https://doi.org/10.3390/antiox10010122






