Aging Induces Hepatic Oxidative Stress and Nuclear Proteomic Remodeling in Liver from Wistar Rats
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals and Ethic Statements
2.2. Analytical Procedures
2.3. Total Extract from Liver and Immunoblot Analysis
2.4. Separation of Rat Liver Nuclear Enriched Fraction
2.5. RNAE and Real-Time RT-PCR
2.6. TBARS Determination
2.7. Calculations and Statistical Analysis
2.8. Protein Extraction, iTRAQ Labelling, Proteomics Data Acquisition, and Analysis
3. Results
3.1. Effect of Fasting or Fasting/Refeeding on Metabolic Characteristics of Young and Old Wistar Rats
3.2. Changes in Hepatic Lipid Peroxidation Levels and in the Expression Levels of Genes Involved in Lipid Metabolism and Oxidative Stress during Aging
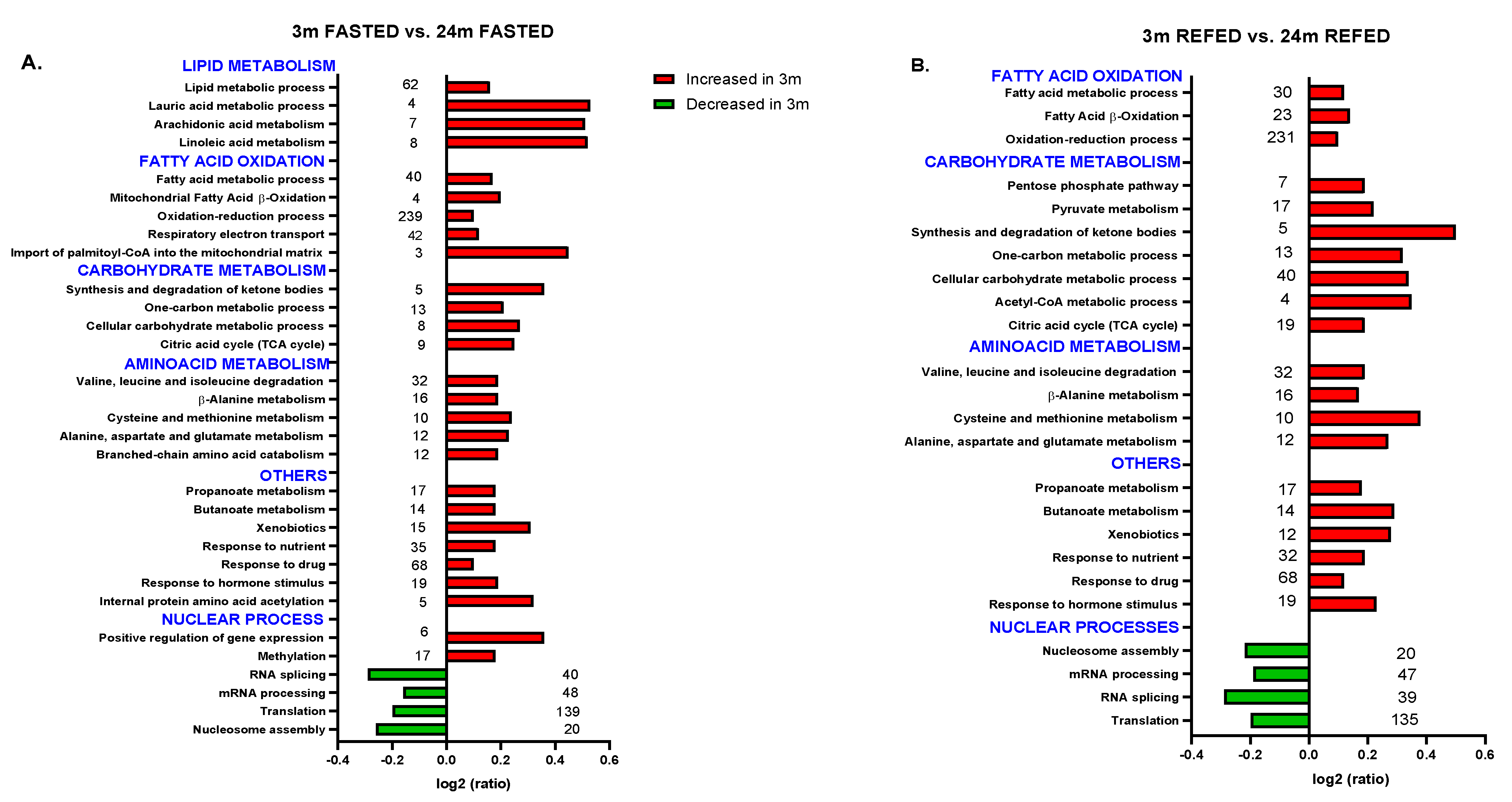
3.3. Aging Combined with Prolonged Fasting Perturbed Liver Metabolic Pathways in the Wistar Rat
3.4. Impact of Refeeding after 36 h Fasting in the Nuclear Proteome from Old Wistar Rats
3.5. Aging Combined with Prolonged Fasting Affects Nuclear Processes in Wistar Rats, Particularly RNA Alternative Splicing
3.6. Impact of Refeeding after 36 h Fasting in the Nuclear Proteome from Young Wistar Rats
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Gems, D. The Aging-Disease False Dichotomy: Understanding Senescence as Pathology. Front. Genet. 2015, 6, 212. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- López-Otín, C.; Blasco, M.A.; Partridge, L.; Serrano, M.; Kroemer, G. The Hallmarks of Aging. Cell 2013, 153, 1194–1217. [Google Scholar] [CrossRef] [Green Version]
- Furukawa, S.; Fujita, T.; Shimabukuro, M.; Iwaki, M.; Yamada, Y.; Nakajima, Y.; Nakayama, O.; Makishima, M.; Matsuda, M.; Shimomura, I. Increased Oxidative Stress in Obesity and Its Impact on Metabolic Syndrome. J. Clin. Investig. 2004, 114, 1752–1761. [Google Scholar] [CrossRef]
- Franceschi, C.; Campisi, J. Chronic Inflammation (Inflammaging) and Its Potential Contribution to Age-Associated Diseases. J. Gerontol. Ser. A Biol. Sci. Med. Sci. 2014, 69 (Suppl. S1), S4–S9. [Google Scholar] [CrossRef] [PubMed]
- Santoro, A.; Bientinesi, E.; Monti, D. Immunosenescence and Inflammaging in the Aging Process: Age-Related Diseases or Longevity? Ageing Res. Rev. 2021, 71, 101422. [Google Scholar] [CrossRef]
- Ito, F.; Sono, Y.; Ito, T. Measurement and Clinical Significance of Lipid Peroxidation as a Biomarker of Oxidative Stress: Oxidative Stress in Diabetes, Atherosclerosis, and Chronic Inflammation. Antioxidants 2019, 8, 72. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gan, L.; Chitturi, S.; Farrell, G.C. Mechanisms and Implications of Age-Related Changes in the Liver: Nonalcoholic Fatty Liver Disease in the Elderly. Curr. Gerontol. Geriatr. Res. 2011, 2011, 831536. [Google Scholar] [CrossRef] [Green Version]
- Albano, E.; Mottaran, E.; Vidali, M.; Reale, E.; Saksena, S.; Occhino, G.; Burt, A.D.; Day, C.P. Immune Response towards Lipid Peroxidation Products as a Predictor of Progression of Non-Alcoholic Fatty Liver Disease to Advanced Fibrosis. Gut 2005, 54, 987–993. [Google Scholar] [CrossRef]
- Hyogo, H.; Yamagishi, S. Advanced Glycation End Products (AGEs) and Their Involvement in Liver Disease. Curr. Pharm. Des. 2008, 14, 969–972. [Google Scholar] [CrossRef]
- Fernando, D.H.; Forbes, J.M.; Angus, P.W.; Herath, C.B. Development and Progression of Non-Alcoholic Fatty Liver Disease: The Role of Advanced Glycation End Products. Int. J. Mol. Sci. 2019, 20, 5037. [Google Scholar] [CrossRef] [Green Version]
- Bochkis, I.M.; Przybylski, D.; Chen, J.; Regev, A. Changes in Nucleosome Occupancy Associated with Metabolic Alterations in Aged Mammalian Liver. Cell Rep. 2014, 9, 996–1006. [Google Scholar] [CrossRef] [Green Version]
- Nakajima, T.; Moriguchi, M.; Katagishi, T.; Sekoguchi, S.; Nishikawa, T.; Takashima, H.; Kimura, H.; Minami, M.; Itoh, Y.; Kagawa, K.; et al. Premature Telomere Shortening and Impaired Regenerative Response in Hepatocytes of Individuals with NAFLD. Liver Int. Off. J. Int. Assoc. Study Liver 2006, 26, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Whitton, H.; Singh, L.N.; Patrick, M.A.; Price, A.J.; Osorio, F.G.; López-Otín, C.; Bochkis, I.M. Changes at the Nuclear Lamina Alter Binding of Pioneer Factor Foxa2 in Aged Liver. Aging Cell 2018, 17, e12742. [Google Scholar] [CrossRef] [PubMed]
- Mertens, J.; Paquola, A.C.M.; Ku, M.; Hatch, E.; Böhnke, L.; Ladjevardi, S.; McGrath, S.; Campbell, B.; Lee, H.; Herdy, J.R.; et al. Directly Reprogrammed Human Neurons Retain Aging-Associated Transcriptomic Signatures and Reveal Age-Related Nucleocytoplasmic Defects. Cell Stem Cell 2015, 17, 705–718. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Horrillo, D.; Gallardo, N.; Lauzurica, N.; Barrus, M.T.; San Frutos, M.G.; Andres, A.; Ros, M.; Fernandez-Agullo, T. Development of Liver Fibrosis during Aging: Effects of Caloric Restriction. J. Biol. Regul. Homeost. Agents 2013, 27, 377–388. [Google Scholar]
- Salamanca, A.; Bárcena, B.; Arribas, C.; Fernández-Agulló, T.; Martínez, C.; Carrascosa, J.M.; Ros, M.; Andrés, A.; Gallardo, N. Aging Impairs the Hepatic Subcellular Distribution of ChREBP in Response to Fasting/Feeding in Rats: Implications on Hepatic Steatosis. Exp. Gerontol. 2015, 69, 9–19. [Google Scholar] [CrossRef] [Green Version]
- Fernández, A.; Mazuecos, L.; Pintado, C.; Rubio, B.; López, V.; de Solís, A.J.; Rodríguez, M.; Andrés, A.; Gallardo, N. Effects of Moderate Chronic Food Restriction on the Development of Postprandial Dyslipidemia with Ageing. Nutrients 2019, 11, 1865. [Google Scholar] [CrossRef] [Green Version]
- Horrillo, D.; Sierra, J.; Arribas, C.; García-San Frutos, M.; Carrascosa, J.M.; Lauzurica, N.; Fernández-Agulló, T.; Ros, M. Age-Associated Development of Inflammation in Wistar Rats: Effects of Caloric Restriction. Arch. Physiol. Biochem. 2011, 117, 140–150. [Google Scholar] [CrossRef]
- Kodiha, M.; Stochaj, U. Nuclear Transport: A Switch for the Oxidative Stress-Signaling Circuit? J. Signal. Transduct. 2012, 2012, 208650. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Major, A.S.; Zienkiewicz, J.; Gabriel, C.L.; Veach, R.A.; Moore, D.J.; Collins, R.D.; Hawiger, J. Nuclear Transport Modulation Reduces Hypercholesterolemia, Atherosclerosis, and Fatty Liver. J. Am. Heart Assoc. 2013, 2, e000093. [Google Scholar] [CrossRef] [Green Version]
- Yu, D.; Chen, G.; Pan, M.; Zhang, J.; He, W.; Liu, Y.; Nian, X.; Sheng, L.; Xu, B. High Fat Diet-Induced Oxidative Stress Blocks Hepatocyte Nuclear Factor 4α and Leads to Hepatic Steatosis in Mice. J. Cell. Physiol. 2018, 233, 4770–4782. [Google Scholar] [CrossRef]
- Del Río-Moreno, M.; Alors-Pérez, E.; González-Rubio, S.; Ferrín, G.; Reyes, O.; Rodríguez-Perálvarez, M.; Sánchez-Frías, M.E.; Sánchez-Sánchez, R.; Ventura, S.; López-Miranda, J.; et al. Dysregulation of the Splicing Machinery Is Associated to the Development of Nonalcoholic Fatty Liver Disease. J. Clin. Endocrinol. Metab. 2019, 104, 3389–3402. [Google Scholar] [CrossRef]
- Van Koppen, A.; Verschuren, L.; van den Hoek, A.M.; Verheij, J.; Morrison, M.C.; Li, K.; Nagabukuro, H.; Costessi, A.; Caspers, M.P.M.; van den Broek, T.J.; et al. Uncovering a Predictive Molecular Signature for the Onset of NASH-Related Fibrosis in a Translational NASH Mouse Model. Cell. Mol. Gastroenterol. Hepatol. 2018, 5, 83–98.e10. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Lekbaby, B.; Fares, N.; Augustin, J.; Attout, T.; Schnuriger, A.; Cassard, A.-M.; Panasyuk, G.; Perlemuter, G.; Bieche, I.; et al. Alteration of Splicing Factors’ Expression during Liver Disease Progression: Impact on Hepatocellular Carcinoma Outcome. Hepatol. Int. 2019, 13, 454–467. [Google Scholar] [CrossRef]
- Bangru, S.; Arif, W.; Seimetz, J.; Bhate, A.; Chen, J.; Rashan, E.H.; Carstens, R.P.; Anakk, S.; Kalsotra, A. Alternative Splicing Rewires Hippo Signaling Pathway in Hepatocytes to Promote Liver Regeneration. Nat. Struct. Mol. Biol. 2018, 25, 928–939. [Google Scholar] [CrossRef] [PubMed]
- Akaike, Y.; Masuda, K.; Kuwano, Y.; Nishida, K.; Kajita, K.; Kurokawa, K.; Satake, Y.; Shoda, K.; Imoto, I.; Rokutan, K. HuR Regulates Alternative Splicing of the TRA2β Gene in Human Colon Cancer Cells under Oxidative Stress. Mol. Cell. Biol. 2014, 34, 2857–2873. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cote, G.J.; Zhu, W.; Thomas, A.; Martin, E.; Murad, F.; Sharina, I.G. Hydrogen Peroxide Alters Splicing of Soluble Guanylyl Cyclase and Selectively Modulates Expression of Splicing Regulators in Human Cancer Cells. PLoS ONE 2012, 7, e41099. [Google Scholar] [CrossRef] [Green Version]
- Fontana, G.A.; Rigamonti, A.; Lenzken, S.C.; Filosa, G.; Alvarez, R.; Calogero, R.; Bianchi, M.E.; Barabino, S.M.L. Oxidative Stress Controls the Choice of Alternative Last Exons via a Brahma-BRCA1-CstF Pathway. Nucleic Acids Res. 2017, 45, 902–914. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sorensen, M.; Sanz, A.; Gómez, J.; Pamplona, R.; Portero-Otín, M.; Gredilla, R.; Barja, G. Effects of Fasting on Oxidative Stress in Rat Liver Mitochondria. Free Radic. Res. 2006, 40, 339–347. [Google Scholar] [CrossRef] [PubMed]
- Stankovic, M.; Mladenovic, D.; Ninkovic, M.; Vucevic, D.; Tomasevic, T.; Radosavljevic, T. Effects of Caloric Restriction on Oxidative Stress Parameters. Gen. Physiol. Biophys. 2013, 32, 277–283. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Longo, V.D.; Panda, S. Fasting, Circadian Rhythms, and Time-Restricted Feeding in Healthy Lifespan. Cell Metab. 2016, 23, 1048–1059. [Google Scholar] [CrossRef] [Green Version]
- Falcón-Pérez, J.M.; Lu, S.C.; Mato, J.M. Sub-Proteome Approach to the Knowledge of Liver. Proteom. Clin. Appl. 2010, 4, 407–415. [Google Scholar] [CrossRef]
- Wasselin, T.; Zahn, S.; Maho, Y.L.; Dorsselaer, A.V.; Raclot, T.; Bertile, F. Exacerbated Oxidative Stress in the Fasting Liver According to Fuel Partitioning. Proteomics 2014, 14, 1905–1921. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Wei, Y.; Pagliassotti, M.J. Saturated Fatty Acids Promote Endoplasmic Reticulum Stress and Liver Injury in Rats with Hepatic Steatosis. Endocrinology 2006, 147, 943–951. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Porta, E.A.; Sablan, H.M.; Joun, N.S.; Chee, G. Effects of the Type of Dietary Fat at Two Levels of Vitamin E in Wistar Male Rats during Development and Aging. IV. Biochemical and Morphometric Parameters of the Heart. Mech. Ageing Dev. 1982, 18, 159–199. [Google Scholar] [CrossRef]
- Yorke, A.; Kane, A.E.; Hancock Friesen, C.L.; Howlett, S.E.; O’Blenes, S. Development of a Rat Clinical Frailty Index. J. Gerontol. Ser. A Biol. Sci. Med. Sci. 2017, 72, 897–903. [Google Scholar] [CrossRef] [Green Version]
- Gallardo, N.; Bonzón-Kulichenko, E.; Fernández-Agulló, T.; Moltó, E.; Gómez-Alonso, S.; Blanco, P.; Carrascosa, J.M.; Ros, M.; Andrés, A. Tissue-Specific Effects of Central Leptin on the Expression of Genes Involved in Lipid Metabolism in Liver and White Adipose Tissue. Endocrinology 2007, 148, 5604–5610. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smirnova, I.V.; Bittel, D.C.; Ravindra, R.; Jiang, H.; Andrews, G.K. Zinc and Cadmium Can Promote Rapid Nuclear Translocation of Metal Response Element-Binding Transcription Factor-1. J. Biol. Chem. 2000, 275, 9377–9384. [Google Scholar] [CrossRef] [Green Version]
- Bonzon-Kulichenko, E.; Pérez-Hernández, D.; Núñez, E.; Martínez-Acedo, P.; Navarro, P.; Trevisan-Herraz, M.; Ramos, M.D.C.; Sierra, S.; Martínez-Martínez, S.; Ruiz-Meana, M.; et al. A Robust Method for Quantitative High-Throughput Analysis of Proteomes by 18O Labeling. Mol. Cell. Proteom. MCP 2011, 10, M110.003335. [Google Scholar] [CrossRef] [Green Version]
- Martínez-Bartolomé, S.; Navarro, P.; Martín-Maroto, F.; López-Ferrer, D.; Ramos-Fernández, A.; Villar, M.; García-Ruiz, J.P.; Vázquez, J. Properties of Average Score Distributions of SEQUEST: The Probability Ratio Method. Mol. Cell. Proteom. MCP 2008, 7, 1135–1145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Navarro, P.; Vázquez, J. A Refined Method to Calculate False Discovery Rates for Peptide Identification Using Decoy Databases. J. Proteome Res. 2009, 8, 1792–1796. [Google Scholar] [CrossRef]
- Bonzon-Kulichenko, E.; Garcia-Marques, F.; Trevisan-Herraz, M.; Vázquez, J. Revisiting Peptide Identification by High-Accuracy Mass Spectrometry: Problems Associated with the Use of Narrow Mass Precursor Windows. J. Proteome Res. 2015, 14, 700–710. [Google Scholar] [CrossRef]
- Navarro, P.; Trevisan-Herraz, M.; Bonzon-Kulichenko, E.; Núñez, E.; Martínez-Acedo, P.; Pérez-Hernández, D.; Jorge, I.; Mesa, R.; Calvo, E.; Carrascal, M.; et al. General Statistical Framework for Quantitative Proteomics by Stable Isotope Labeling. J. Proteome Res. 2014, 13, 1234–1247. [Google Scholar] [CrossRef] [PubMed]
- García-Marqués, F.; Trevisan-Herraz, M.; Martínez-Martínez, S.; Camafeita, E.; Jorge, I.; Lopez, J.A.; Méndez-Barbero, N.; Méndez-Ferrer, S.; Del Pozo, M.A.; Ibáñez, B.; et al. A Novel Systems-Biology Algorithm for the Analysis of Coordinated Protein Responses Using Quantitative Proteomics. Mol. Cell. Proteom. MCP 2016, 15, 1740–1760. [Google Scholar] [CrossRef] [Green Version]
- Steinhauser, M.L.; Olenchock, B.A.; O’Keefe, J.; Lun, M.; Pierce, K.A.; Lee, H.; Pantano, L.; Klibanski, A.; Shulman, G.I.; Clish, C.B.; et al. The Circulating Metabolome of Human Starvation. JCI Insight 2018, 3, 121434. [Google Scholar] [CrossRef] [PubMed]
- Sheedfar, F.; Sung, M.M.; Aparicio-Vergara, M.; Kloosterhuis, N.J.; Miquilena-Colina, M.E.; Vargas-Castrillón, J.; Febbraio, M.; Jacobs, R.L.; de Bruin, A.; Vinciguerra, M.; et al. Increased Hepatic CD36 Expression with Age Is Associated with Enhanced Susceptibility to Nonalcoholic Fatty Liver Disease. Aging 2014, 6, 281–295. [Google Scholar] [CrossRef] [Green Version]
- Ezquerro, S.; Méndez-Giménez, L.; Becerril, S.; Moncada, R.; Valentí, V.; Catalán, V.; Gómez-Ambrosi, J.; Frühbeck, G.; Rodríguez, A. Acylated and Desacyl Ghrelin Are Associated with Hepatic Lipogenesis, β-Oxidation and Autophagy: Role in NAFLD Amelioration after Sleeve Gastrectomy in Obese Rats. Sci. Rep. 2016, 6, 39942. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Höhn, A.; Weber, D.; Jung, T.; Ott, C.; Hugo, M.; Kochlik, B.; Kehm, R.; König, J.; Grune, T.; Castro, J.P. Happily (n)Ever after: Aging in the Context of Oxidative Stress, Proteostasis Loss and Cellular Senescence. Redox Biol. 2017, 11, 482–501. [Google Scholar] [CrossRef]
- Abdelmegeed, M.A.; Moon, K.-H.; Hardwick, J.P.; Gonzalez, F.J.; Song, B.-J. Role of Peroxisome Proliferator-Activated Receptor-Alpha in Fasting-Mediated Oxidative Stress. Free Radic. Biol. Med. 2009, 47, 767–778. [Google Scholar] [CrossRef] [Green Version]
- Narce, M.; Bellenger, J.; Rialland, M.; Bellenger, S. Recent Advances on Stearoyl-Coa Desaturase Regulation in Fatty Liver Diseases. Curr. Drug Metab. 2012, 13, 1454–1463. [Google Scholar] [CrossRef]
- Rudraiah, S.; Moscovitz, J.E.; Donepudi, A.C.; Campion, S.N.; Slitt, A.L.; Aleksunes, L.M.; Manautou, J.E. Differential Fmo3 Gene Expression in Various Liver Injury Models Involving Hepatic Oxidative Stress in Mice. Toxicology 2014, 325, 85–95. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Borbás, T.; Benko, B.; Dalmadi, B.; Szabó, I.; Tihanyi, K. Insulin in Flavin-Containing Monooxygenase Regulation. Flavin-Containing Monooxygenase and Cytochrome P450 Activities in Experimental Diabetes. Eur. J. Pharm. Sci. Off. J. Eur. Fed. Pharm. Sci. 2006, 28, 51–58. [Google Scholar] [CrossRef]
- Genter, M.B.; Clay, C.D.; Dalton, T.P.; Dong, H.; Nebert, D.W.; Shertzer, H.G. Comparison of Mouse Hepatic Mitochondrial versus Microsomal Cytochromes P450 Following TCDD Treatment. Biochem. Biophys. Res. Commun. 2006, 342, 1375–1381. [Google Scholar] [CrossRef]
- De Luca, C.; Olefsky, J.M. Inflammation and Insulin Resistance. FEBS Lett. 2008, 582, 97–105. [Google Scholar] [CrossRef] [Green Version]
- Höhn, A.; Grune, T. Lipofuscin: Formation, Effects and Role of Macroautophagy. Redox Biol. 2013, 1, 140–144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oarada, M.; Miki, T.; Kohno, S.; Sakai, K.; Nikawa, T.; Yoneyama, M.; Gonoi, T. Refeeding with a Standard Diet after a 48-h Fast Elicits an Inflammatory Response in the Mouse Liver. J. Nutr. Biochem. 2013, 24, 1314–1323. [Google Scholar] [CrossRef] [PubMed]
- Korovila, I.; Hugo, M.; Castro, J.P.; Weber, D.; Höhn, A.; Grune, T.; Jung, T. Proteostasis, Oxidative Stress and Aging. Redox Biol. 2017, 13, 550–567. [Google Scholar] [CrossRef] [PubMed]
- Lebeaupin, C.; Vallée, D.; Hazari, Y.; Hetz, C.; Chevet, E.; Bailly-Maitre, B. Endoplasmic Reticulum Stress Signalling and the Pathogenesis of Non-Alcoholic Fatty Liver Disease. J. Hepatol. 2018, 69, 927–947. [Google Scholar] [CrossRef] [PubMed]
- Park, E.C.; Kim, G.-H.; Yun, S.-H.; Lim, H.L.; Hong, Y.; Kwon, S.-O.; Kwon, J.; Chung, Y.-H.; Kim, S.I. Analysis of the Endoplasmic Reticulum Subproteome in the Livers of Type 2 Diabetic Mice. Int. J. Mol. Sci. 2012, 13, 17230–17243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, S.; Kim, S.; Hwang, S.; Cherrington, N.J.; Ryu, D.-Y. Dysregulated Expression of Proteins Associated with ER Stress, Autophagy and Apoptosis in Tissues from Nonalcoholic Fatty Liver Disease. Oncotarget 2017, 8, 63370–63381. [Google Scholar] [CrossRef] [Green Version]
- Chang, E.T.; Parekh, P.R.; Yang, Q.; Nguyen, D.M.; Carrier, F. Heterogenous Ribonucleoprotein A18 (HnRNP A18) Promotes Tumor Growth by Increasing Protein Translation of Selected Transcripts in Cancer Cells. Oncotarget 2016, 7, 10578–10593. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cammas, A.; Pileur, F.; Bonnal, S.; Lewis, S.M.; Lévêque, N.; Holcik, M.; Vagner, S. Cytoplasmic Relocalization of Heterogeneous Nuclear Ribonucleoprotein A1 Controls Translation Initiation of Specific MRNAs. Mol. Biol. Cell 2007, 18, 5048–5059. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, X.; Peng, Q.; Wang, L.; Zhang, X.; Huang, L.; Wang, J.; Qin, Z. Serine/Arginine-Rich Splicing Factors: The Bridge Linking Alternative Splicing and Cancer. Int. J. Biol. Sci. 2020, 16, 2442–2453. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Fang, C.; Bao, H.; Fan, H.; Shen, H.; Yang, P. Nuclear Proteome Profile of C57BL/6J Mouse Liver. Sci. China Life Sci. 2013, 56, 513–523. [Google Scholar] [CrossRef] [Green Version]
- Sen, S.; Langiewicz, M.; Jumaa, H.; Webster, N.J.G. Deletion of Serine/Arginine-Rich Splicing Factor 3 in Hepatocytes Predisposes to Hepatocellular Carcinoma in Mice. Hepatology 2015, 61, 171–183. [Google Scholar] [CrossRef]
- Wang, J.; Mauvoisin, D.; Martin, E.; Atger, F.; Galindo, A.N.; Dayon, L.; Sizzano, F.; Palini, A.; Kussmann, M.; Waridel, P.; et al. Nuclear Proteomics Uncovers Diurnal Regulatory Landscapes in Mouse Liver. Cell Metab. 2017, 25, 102–117. [Google Scholar] [CrossRef] [Green Version]
- Longo, V.D.; Mattson, M.P. Fasting: Molecular Mechanisms and Clinical Applications. Cell Metab. 2014, 19, 181–192. [Google Scholar] [CrossRef] [Green Version]
- Ipsen, D.H.; Lykkesfeldt, J.; Tveden-Nyborg, P. Molecular Mechanisms of Hepatic Lipid Accumulation in Non-Alcoholic Fatty Liver Disease. Cell. Mol. Life Sci. CMLS 2018, 75, 3313–3327. [Google Scholar] [CrossRef] [Green Version]
- Hunt, N.J.; Kang, S.W.S.; Lockwood, G.P.; Le Couteur, D.G.; Cogger, V.C. Hallmarks of Aging in the Liver. Comput. Struct. Biotechnol. J. 2019, 17, 1151–1161. [Google Scholar] [CrossRef]
- Romero-Bueno, R.; de la Cruz Ruiz, P.; Artal-Sanz, M.; Askjaer, P.; Dobrzynska, A. Nuclear Organization in Stress and Aging. Cells 2019, 8, 664. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Zhou, D.; Strakovsky, R.; Zhang, Y.; Pan, Y.-X. Hepatic Cellular Senescence Pathway Genes Are Induced through Histone Modifications in a Diet-Induced Obese Rat Model. Am. J. Physiol. Gastrointest. Liver Physiol. 2012, 302, G558–G564. [Google Scholar] [CrossRef]
- Jin, J.; Valanejad, L.; Nguyen, T.P.; Lewis, K.; Wright, M.; Cast, A.; Stock, L.; Timchenko, L.; Timchenko, N.A. Activation of CDK4 Triggers Development of Non-Alcoholic Fatty Liver Disease. Cell Rep. 2016, 16, 744–756. [Google Scholar] [CrossRef] [Green Version]
- Webster, N.J.G. Alternative RNA Splicing in the Pathogenesis of Liver Disease. Front. Endocrinol. 2017, 8, 133. [Google Scholar] [CrossRef] [Green Version]
- Maya-Mendoza, A.; Hernández-Muñoz, R.; Gariglio, P.; Aranda-Anzaldo, A. Natural Ageing in the Rat Liver Correlates with Progressive Stabilisation of DNA-Nuclear Matrix Interactions and Withdrawal of Genes from the Nuclear Substructure. Mech. Ageing Dev. 2005, 126, 767–782. [Google Scholar] [CrossRef]
- Arrigo, T.; Leonardi, S.; Cuppari, C.; Manti, S.; Lanzafame, A.; D’Angelo, G.; Gitto, E.; Marseglia, L.; Salpietro, C. Role of the Diet as a Link between Oxidative Stress and Liver Diseases. World J. Gastroenterol. 2015, 21, 384–395. [Google Scholar] [CrossRef]
- Baumeier, C.; Kaiser, D.; Heeren, J.; Scheja, L.; John, C.; Weise, C.; Eravci, M.; Lagerpusch, M.; Schulze, G.; Joost, H.-G.; et al. Caloric Restriction and Intermittent Fasting Alter Hepatic Lipid Droplet Proteome and Diacylglycerol Species and Prevent Diabetes in NZO Mice. Biochim. Biophys. Acta 2015, 1851, 566–576. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Drinda, S.; Grundler, F.; Neumann, T.; Lehmann, T.; Steckhan, N.; Michalsen, A.; de Toledo, F.W. Effects of Periodic Fasting on Fatty Liver Index-A Prospective Observational Study. Nutrients 2019, 11, 2601. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, P.; Zhang, M.; Webster, N.J.G. Alternative RNA Splicing in Fatty Liver Disease. Front. Endocrinol. 2021, 12, 613213. [Google Scholar] [CrossRef] [PubMed]


| 3m | 24m | 2-way-ANOVA | |||||
|---|---|---|---|---|---|---|---|
| 36 h Fasting | 36 h Fast + 30 min Refeed | 36 h Fasting | 36 h Fast + 30 min Refeed | Young vs. Old | Fast vs. Refeed | Interaction | |
| Liver TAG (mg/g) | 4.7 ± 0.8 | 3.4 ± 0.4 | 12.7 ± 2 * | 12.4 ± 1 | p < 0.0001 | p = 0.5361 | p = 0.6998 |
| Liver Glycogen (mg/g) | 2.0 ± 0.008 | 4.0 ± 0.3 ++++ | 4.9 ± 0.1 **** | 5.7 ± 0.2 | p < 0.0001 | p < 0.0001 | p = 0.0376 |
| Serum glucose (mM) | 4.9 ± 0.8 | 6.1 ± 0.5 ++ | 5.12 ± 0.4 | 5.6 ± 0.4 | p = 0.6141 | p = 0.0043 | p = 0.0762 |
| Serum TAG (mg/dL) | 29 ± 2 | 33 ± 4 | 52 ± 5 * | 57 ± 4 | p = 0.0003 | p = 0.3750 | p = 0.9387 |
| Serum NEFA (mm/L) | 0.58 ± 0.04 | 0.52 ± 0.06 | 0.55 ± 0.03 | 0.97 ± 0.1 ++ | p = 0.0215 | p = 0.0465 | p = 0.0106 |
| Serum TKB (mm/L) | 2.3 ± 0.1 | 0.18 ± 0.06 ++++ | 1.48 ± 0.1 * | 0.34 ± 0.06 ++ | p = 0.0174 | p < 0.0001 | p = 0.0016 |
| Serum insulin (ng/mL) | 0.71 ± 0.2 | 2.73 ± 0.1 ++ | 2.5 ± 0.1 ** | 2.39 ± 0.2 | p = 0.0069 | p = 0.0021 | p = 0.0008 |
| Serum glucagon (pg/mL) | 318 ± 9 | 355 ± 6 | 538 ± 14 ** | 251 ± 19 ++++ | p = 0.0039 | p < 0.0001 | p < 0.0001 |
| Serum leptin (ng/mL) | 1.5 ± 0.06 | 1.4 ± 0.2 | 4.9 ± 0.5 ** | 4.6 ± 0.84 | p < 0.0001 | p = 0.5402 | p = 0.9577 |
| Acetylated ghrelin (ng/mL) | 0.13 ± 1.9 | 0.13 ± 1.3 | 0.23 ± 1.8 ** | 0.18 ± 2.4 | p = 0.0005 | p = 0.1968 | p = 0.1337 |
| Nonacetylated ghrelin (ng/mL) | 1.26 ± 0.2 | 1.24 ± 0.1 | 0.8 ± 0.03 | 0.7 ± 0.03 | p = 0.0045 | p = 0.6772 | p = 0.7902 |
| Acetylated/nonacetylated ghrelin ratio | 0.12 ± 0.06 | 0.11 ± 0.01 | 0.29 ± 0.02 ** | 0.26 ± 0.04 | ------ | ------ | ------ |
| Serum ALT (IU/L) | 5.01 ± 0.8 | 6.6 ± 0.4 | 12.0 ± 1 * | 15.1 ± 1 | p < 0.0001 | p = 0.1240 | p = 0.6111 |
| Serum CRP (µg/mL) | 209 ± 1 | 212 ± 35 | 463 ± 12 **** | 382 ± 9 | p < 0.0001 | p = 0.0412 | p = 0.2369 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bárcena, B.; Salamanca, A.; Pintado, C.; Mazuecos, L.; Villar, M.; Moltó, E.; Bonzón-Kulichenko, E.; Vázquez, J.; Andrés, A.; Gallardo, N. Aging Induces Hepatic Oxidative Stress and Nuclear Proteomic Remodeling in Liver from Wistar Rats. Antioxidants 2021, 10, 1535. https://doi.org/10.3390/antiox10101535
Bárcena B, Salamanca A, Pintado C, Mazuecos L, Villar M, Moltó E, Bonzón-Kulichenko E, Vázquez J, Andrés A, Gallardo N. Aging Induces Hepatic Oxidative Stress and Nuclear Proteomic Remodeling in Liver from Wistar Rats. Antioxidants. 2021; 10(10):1535. https://doi.org/10.3390/antiox10101535
Chicago/Turabian StyleBárcena, Brenda, Aurora Salamanca, Cristina Pintado, Lorena Mazuecos, Margarita Villar, Eduardo Moltó, Elena Bonzón-Kulichenko, Jesús Vázquez, Antonio Andrés, and Nilda Gallardo. 2021. "Aging Induces Hepatic Oxidative Stress and Nuclear Proteomic Remodeling in Liver from Wistar Rats" Antioxidants 10, no. 10: 1535. https://doi.org/10.3390/antiox10101535






