The Role of DNA Damage and Repair in Idiopathic Pulmonary Fibrosis
Abstract
:1. Introduction
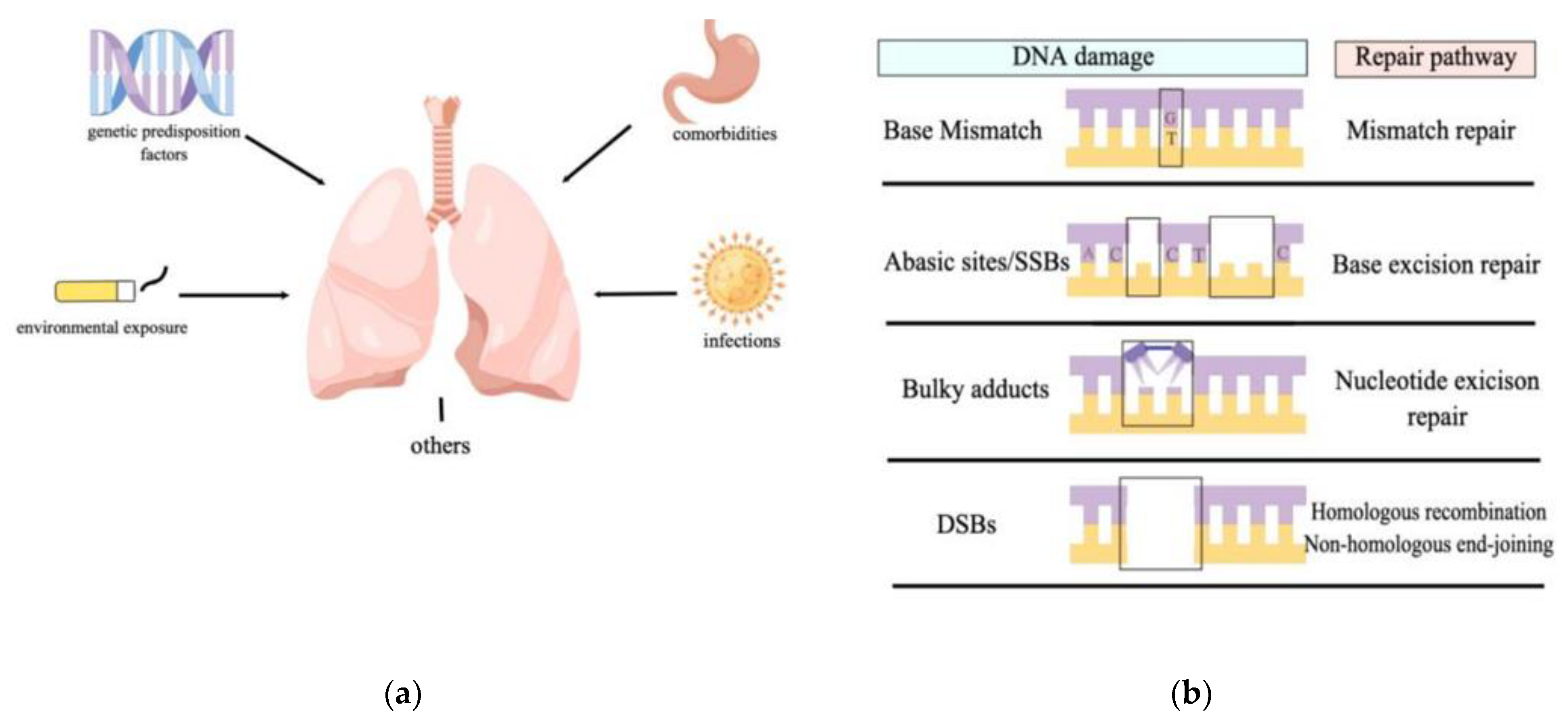
2. DNA Damage in Idiopathic Pulmonary Fibrosis
2.1. Environmental Exposures
2.2. Genetic Predisposition
2.3. Infectious Microbial Agents
2.4. Comorbidities
3. DNA Repair and Its Effects on Idiopathic Pulmonary Fibrosis
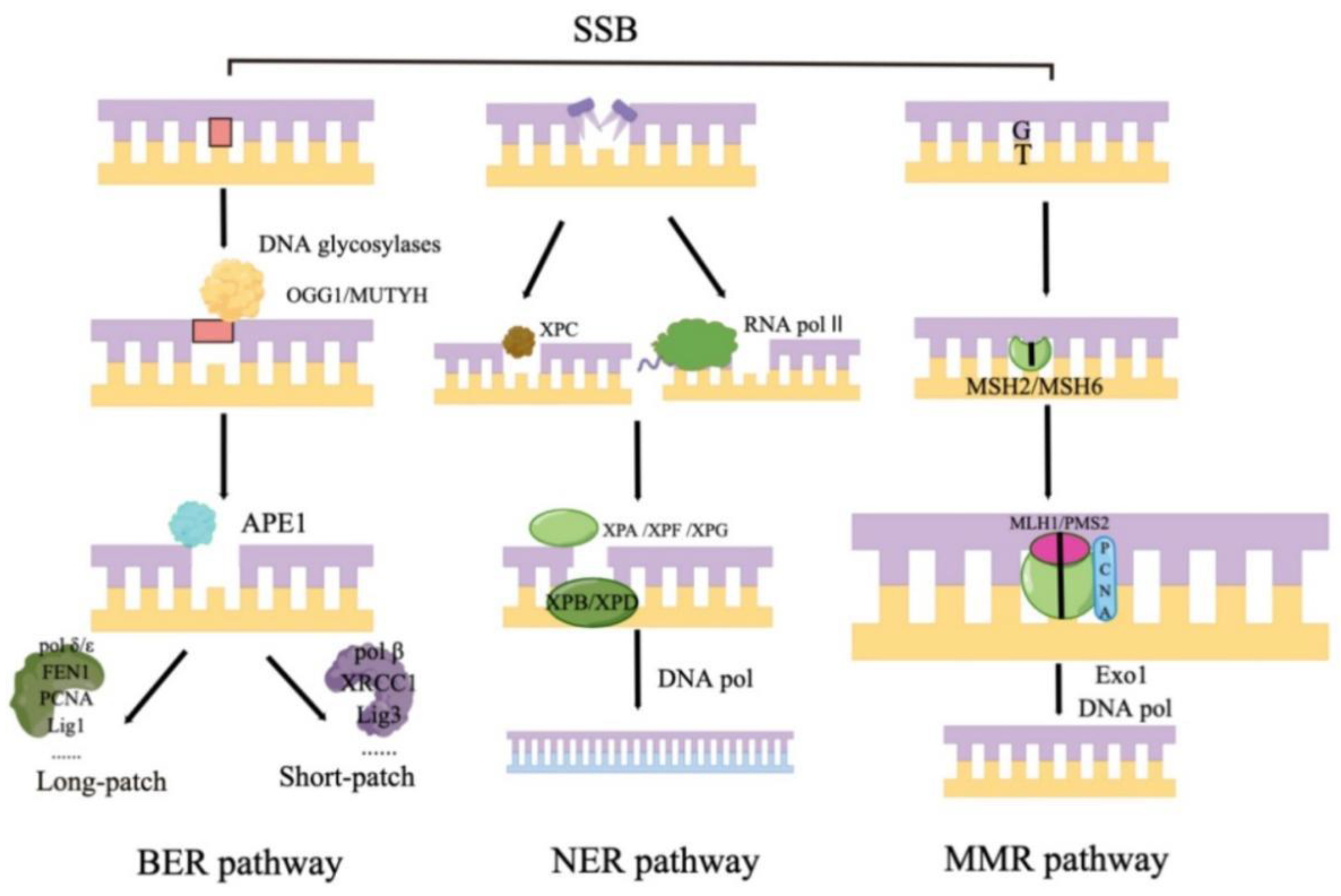
3.1. Base Excision Repair
3.1.1. DNA Glycosylases
OGG1
MUTYH
3.1.2. Apurinic/Apyrimidinic (AP) Endonucleases
3.1.3. Scaffolding Proteins
XRCC1
Poly (ADP-Ribose) Polymerases
3.2. Nucleotide Excision Repair (NER)
3.2.1. ERCC1
3.2.2. PCNA
3.3. Mismatch Repair (MMR)
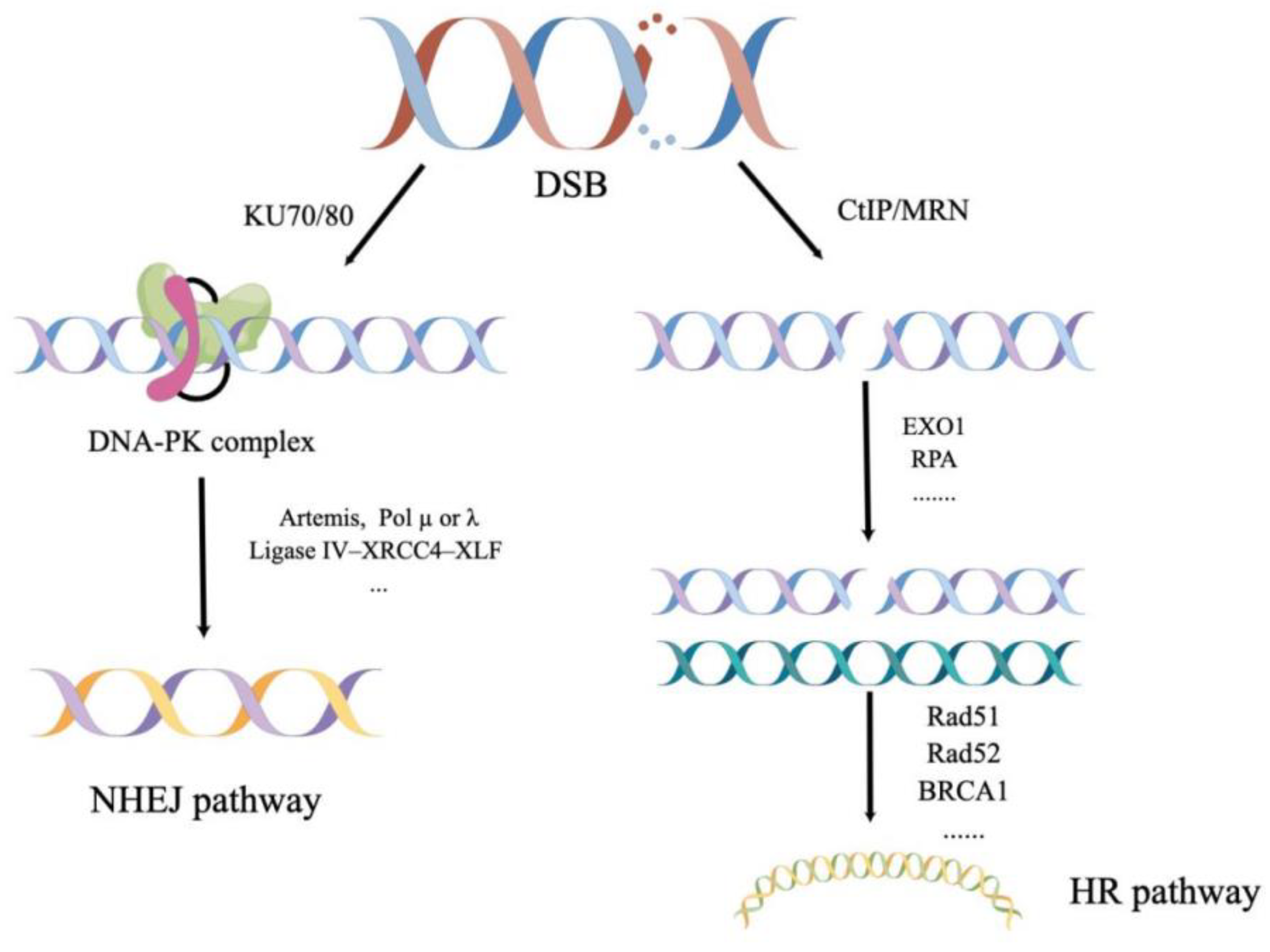
3.4. Homologous Recombination (HR)
3.5. Non-Homologous End-Joining (NHEJ)
3.5.1. Ku70/Ku80
3.5.2. DNA PKcs
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Wuyts, W.A.; Agostini, C.; Antoniou, K.M.; Bouros, D.; Chambers, R.; Cottin, V.; Egan, J.J.; Lambrecht, B.N.; Lories, R.; Parfrey, H.; et al. The pathogenesis of pulmonary fibrosis: A moving target. Eur. Respir. J. 2013, 41, 1207–1218. [Google Scholar] [CrossRef] [PubMed]
- Sgalla, G.; Iovene, B.; Calvello, M.; Ori, M.; Varone, F.; Richeldi, L. Idiopathic pulmonary fibrosis: Pathogenesis and management. Respir. Res. 2018, 19, 32. [Google Scholar] [CrossRef] [Green Version]
- van der Vliet, A.; Janssen-Heininger, Y.M.W.; Anathy, V. Oxidative stress in chronic lung disease: From mitochondrial dysfunction to dysregulated redox signaling. Mol. Asp. Med. 2018, 63, 59–69. [Google Scholar] [CrossRef] [PubMed]
- Andreadis, A.A.; Hazen, S.L.; Comhair, S.A.; Erzurum, S.C. Oxidative and nitrosative events in asthma. Free Radic. Biol. Med. 2003, 35, 213–225. [Google Scholar] [CrossRef]
- Povedano, J.M.; Martinez, P.; Flores, J.M.; Mulero, F.; Blasco, M.A. Mice with Pulmonary Fibrosis Driven by Telomere Dysfunction. Cell Rep. 2015, 12, 286–299. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wolters, P.J.; Collard, H.R.; Jones, K.D. Pathogenesis of Idiopathic Pulmonary Fibrosis. Annu. Rev. Pathol. Mech. Dis. 2014, 9, 157–179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, P.; Li, S.; Chen, H. Macrophages in Lung Injury, Repair, and Fibrosis. Cells 2021, 10, 436. [Google Scholar] [CrossRef] [PubMed]
- Carusillo, A.; Mussolino, C. DNA Damage: From Threat to Treatment. Cells 2020, 9, 1665. [Google Scholar] [CrossRef] [PubMed]
- Gillman, R.; Floro, K.L.; Wankell, M.; Hebbard, L. The role of DNA damage and repair in liver cancer. Biochim. Biophys. Acta 2020, 1875, 188493. [Google Scholar] [CrossRef] [PubMed]
- Lindahl, T. Instability and decay of the primary structure of DNA. Nature 1993, 362, 709–715. [Google Scholar] [CrossRef] [PubMed]
- De Bont, R.; van Larebeke, N. Endogenous DNA damage in humans: A review of quantitative data. Mutagenesis 2004, 19, 169–185. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ou, H.-L.; Schumacher, B. DNA damage responses and p53 in the aging process. Blood 2018, 131, 488–495. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patel, J.; Baptiste, B.A.; Kim, E.; Hussain, M.; Croteau, D.L.; Bohr, V.A. DNA damage and mitochondria in cancer and aging. Carcinogenesis 2020, 41, 1625–1634. [Google Scholar] [CrossRef] [PubMed]
- Taskar, V.S.; Coultas, D.B. Is idiopathic pulmonary fibrosis an environmental disease? Proc. Am. Thorac. Soc. 2006, 3, 293–298. [Google Scholar] [CrossRef] [PubMed]
- Seibold, M.A.; Wise, A.L.; Speer, M.C.; Steele, M.P.; Brown, K.K.; Loyd, J.E.; Fingerlin, T.E.; Zhang, W.; Gudmundsson, G.; Schwartz, D.A.; et al. A common MUC5B promoter polymorphism and pulmonary fibrosis. New Engl. J. Med. 2011, 364, 1503–1512. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tobin, R.W.; Pope, C.E.; Pellegrini, C.A.; Emond, M.J.; Sillery, J.; Raghu, G. Increased Prevalence of Gastroesophageal Reflux in Patients with Idiopathic Pulmonary Fibrosis. Am. J. Respir. Crit. Care Med. 1998, 158, 1804–1808. [Google Scholar] [CrossRef] [PubMed]
- Chioma, O.S.; Drake, W.P. Role of Microbial Agents in Pulmonary Fibrosis. Yale J. Biol. Med. 2017, 90, 219–227. [Google Scholar]
- Nakayama, T.; Kaneko, M.; Kodama, M.; Nagata, C. Cigarette smoke induces DNA single-strand breaks in human cells. Nature 1985, 314, 462–464. [Google Scholar] [CrossRef] [PubMed]
- Cheresh, P.; Kim, S.-J.; Tulasiram, S.; Kamp, D.W. Oxidative stress and pulmonary fibrosis. Biochim. Biophys. Acta (BBA)–Mol. Basis Dis. 2013, 1832, 1028–1040. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Felton, V.M.; Borok, Z.; Willis, B.C. N-acetylcysteine inhibits alveolar epithelial-mesenchymal transition. Am. J. Physiol. Cell. Mol. Physiol. 2009, 297, L805–L812. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weng, M.-W.; Lee, H.-W.; Park, S.-H.; Hu, Y.; Wang, H.-T.; Chen, L.C.; Rom, W.; Huang, W.; Lepor, H.; Wu, X.-R.; et al. Aldehydes are the predominant forces inducing DNA damage and inhibiting DNA repair in tobacco smoke carcinogenesis. Proc. Natl. Acad. Sci. USA 2018, 115, E6152–E6161. [Google Scholar] [CrossRef]
- Fingerlin, T.E.; Murphy, E.; Zhang, W.; Peljto, A.L.; Brown, K.K.; Steele, M.P.; Loyd, J.E.; Cosgrove, G.P.; Lynch, D.; Groshong, S.; et al. Steve Groshong. Genome-wide association study identifies multiple susceptibility loci for pulmonary fibrosis. Nat. Genet. 2013, 45, 613–620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Greider, C.W.; Blackburn, E.H. Identification of a specific telomere terminal transferase activity in tetrahymena extracts. Cell 1985, 43, 405–413. [Google Scholar] [CrossRef]
- Nakamura, T.M.; Morin, G.B.; Chapman, K.B.; Weinrich, S.L.; Andrews, W.H.; Lingner, J.; Harley, C.B.; Cech, T.R. Telomerase Catalytic Subunit Homologs from Fission Yeast and Human. Science 1997, 277, 955–959. [Google Scholar] [CrossRef] [PubMed]
- Armanios, M.Y.; Chen, J.J.-L.; Cogan, J.D.; Alder, J.K.; Ingersoll, R.G.; Markin, C.; Lawson, W.E.; Xie, M.; Vulto, I.; Phillips, J.A.; et al. Telomerase Mutations in Families with Idiopathic Pulmonary Fibrosis. New Engl. J. Med. 2007, 356, 1317–1326. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Egan, J.J.; Adamali, H.I.; Lok, S.S.; Stewart, J.P.; Woodcock, A. Ganciclovir Antiviral Therapy in Advanced Idiopathic Pulmonary Fibrosis: An Open Pilot Study. Pulm. Med. 2011, 2011, 240805. [Google Scholar] [CrossRef] [PubMed]
- Knippenberg, S.; Ueberberg, B.; Maus, R.; Bohling, J.; Ding, N.; Tarres, M.T.; Hoymann, H.-G.; Jonigk, D.; Izykowski, N.; Paton, J.C.; et al. Streptococcus pneumoniae triggers progression of pulmonary fibrosis through pneumolysin. Thorax 2015, 70, 636–646. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duckworth, A.; Longhurst, H.J.; Paxton, J.K.; Scotton, C.J. The Role of Herpes Viruses in Pulmonary Fibrosis. Front. Med. 2021, 8, 704222. [Google Scholar] [CrossRef] [PubMed]
- Atabati, E.; Dehghani-Samani, A.; Mortazavimoghaddam, S.G. Association of COVID-19 and other viral infections with interstitial lung diseases, pulmonary fibrosis, and pulmonary hypertension: A narrative review. Can. J. Respir. Ther. 2020, 56, 70–78. [Google Scholar] [CrossRef] [PubMed]
- Lechowicz, K.; Drożdżal, S.; Machaj, F.; Rosik, J.; Szostak, B.; Zegan-Barańska, M.; Biernawska, J.; Dabrowski, W.; Rotter, I.; Kotfis, K. COVID-19: The Potential Treatment of Pulmonary Fibrosis Associated with SARS-CoV-2 Infection. J. Clin. Med. 2020, 9, 1917. [Google Scholar] [CrossRef] [PubMed]
- Weitzman, M.D.; Fradet-Turcotte, A. Virus DNA Replication and the Host DNA Damage Response. Annu. Rev. Virol. 2018, 5, 141–164. [Google Scholar] [CrossRef] [PubMed]
- O’Meara, M.J.; Guo, J.Z.; Swaney, D.L.; Tummino, T.A.; Hüttenhain, R. A SARS-CoV-2-Human Protein-Protein Interaction Map Reveals Drug Targets and Potential Drug-Repurposing. BioRxiv 2020, 2020, 123. [Google Scholar]
- Flynn, R.A.; Belk, J.A.; Qi, Y.; Yasumoto, Y.; Wei, J.; Alfajaro, M.M.; Shi, Q.; Mumbach, M.R.; Limaye, A.; DeWeirdt, P.C.; et al. Discovery and functional interrogation of SARS-CoV-2 RNA-host protein interactions. Cell 2021, 184, 2394–2411. [Google Scholar] [CrossRef] [PubMed]
- Gordon, D.E.; Jang, G.M.; Bouhaddou, M.; Xu, J.; Obernier, K.; White, K.M.; O’Meara, M.J.; Rezelj, V.V.; Guo, J.Z.; Swaney, D.L.; et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature 2020, 583, 459–468. [Google Scholar] [CrossRef]
- Singh, N.; Bharara Singh, A. S2 subunit of SARS-nCoV-2 interacts with tumor suppressor protein p53 and BRCA: An in silico study. Tran. Oncol. 2020, 13, 100814. [Google Scholar] [CrossRef]
- Vakil, N.; Van Zanten, S.V.; Kahrilas, P.; Dent, J.; Jones, R. The Montreal definition and classification of gastroesophageal reflux disease: A global evidence-based consensus. Off. J. Am. Coll. Gastroenterol. 2006, 101, 1900–1920. [Google Scholar] [CrossRef]
- Lee, C.M.; Lee, D.H.; Ahn, B.K.; Hwang, J.J.; Yoon, H.; Shin, C.M.; Park, Y.S.; Kim, Y.S.P.A.N. Protective Effect of Proton Pump Inhibitor for Survival in Patients with Gastroesophageal Reflux Disease and Idiopathic Pulmonary Fibrosis. J. Neurogastroenterol. Motil. 2016, 22, 444–451. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nelkine, L.; Vrolijk, M.F.; Drent, M.; Bast, A. Role of antioxidants in the treatment of gastroesophageal reflux disease-associated idiopathic pulmonary fibrosis. Curr. Opin. Pulm. Med. 2020, 26, 363–371. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Beri, R.; Mueller, A.; Kamp, D.W. Molecular mechanisms of asbestos-induced lung epithelial cell apoptosis. Chem. Interactions 2010, 188, 309–318. [Google Scholar] [CrossRef]
- Achanta, G.; Huang, P. Role of p53 in Sensing Oxidative DNA Damage in Response to Reactive Oxygen Species-Generating Agents. Cancer Res. 2004, 64, 6233–6239. [Google Scholar] [CrossRef] [Green Version]
- Kliment, C.R.; Oury, T.D. Oxidative stress, extracellular matrix targets, and idiopathic pulmonary fibrosis. Free. Radic. Biol. Med. 2010, 49, 707–717. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Cheresh, P.; Kamp, D.W. Molecular Basis of Asbestos-Induced Lung Disease. Annu. Rev. Pathol. Mech. Dis. 2013, 8, 161–187. [Google Scholar] [CrossRef] [PubMed]
- Krokan, H.E.; Bjørås, M. Base excision repair. Cold Spring Harb. Perspect. Biol. 2013, 5, a012583. [Google Scholar] [CrossRef]
- Wright, G.; Gassman, N.R. Transcriptional dysregulation of base excision repair proteins in breast cancer. DNA Repair 2020, 93, 102922. [Google Scholar] [CrossRef]
- Kim, Y.J.; Wilson, D.M., III. Overview of base excision repair biochemistry. Curr. Mol. Pharmacol. 2012, 5, 3–13. [Google Scholar] [CrossRef]
- Zharkov, D.O. Base excision DNA repair. Cell. Mol. Life Sci. 2008, 65, 1544–1565. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Hosoki, K.; Bacsi, A.; Radak, Z.; Hegde, M.L.; Sur, S.; Hazra, T.K.; Brasier, A.R.; Ba, X.; Boldogh, I. 8-Oxoguanine DNA glycosylase-1-mediated DNA repair is associated with Rho GTPase activation and α-smooth muscle actin polymerization. Free Radic. Biol. Med. 2014, 73, 430–438. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Damsma, G.E.; Cramer, P. Molecular Basis of Transcriptional Mutagenesis at 8-Oxoguanine. J. Biol. Chem. 2009, 284, 31658–31663. [Google Scholar] [CrossRef] [Green Version]
- Dizdaroglu, M. Substrate specificities and excision kinetics of DNA glycosylases involved in base-excision repair of oxidative DNA damage. Mutat. Res. Mol. Mech. Mutagen. 2003, 531, 109–126. [Google Scholar] [CrossRef]
- Boldogh, I.; Hajas, G.; Aguilera-Aguirre, L.; Hegde, M.L.; Radak, Z.; Bacsi, A.; Sur, S.; Hazra, T.K.; Mitra, S. Activation of Ras Signaling Pathway by 8-Oxoguanine DNA Glycosylase Bound to Its Excision Product, 8-Oxoguanine. J. Biol. Chem. 2012, 287, 20769–20773. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Chen, T.; Pan, Z.; Lin, Z.; Yang, L.; Zou, B.; Yao, W.; Feng, D.; Huangfu, C.; Lin, C.; et al. 8-Oxoguanine DNA glycosylase modulates the cell transformation process in pulmonary fibrosis by inhibiting Smad2/3 and interacting with Smad7. FASEB J. 2020, 34, 13461–13473. [Google Scholar] [CrossRef] [PubMed]
- Ling, H.; Song, C.; Fang, Y.; Yin, Y.; Wu, Z.; Wang, Y.; Xu, Z.; Gao, S.; Li, A.; Liu, G. TH5487, a small molecule inhibitor of OGG1, attenuates pulmonary fibrosis by NEDD4L-mediated OGG1 degradation. Chem. Interactions 2022, 362, 109999. [Google Scholar] [CrossRef]
- Parker, A.R.; Eshleman, J.R. Human MutY: Gene structure, protein functions and interactions, and role in carcinogenesis. Cell. Mol. Life Sci. CMLS 2003, 60, 2064–2083. [Google Scholar] [CrossRef] [PubMed]
- Banda, D.M.; Nuñez, N.N.; Burnside, M.A.; Bradshaw, K.M.; David, S.S. Repair of 8-oxoG:A mismatches by the MUTYH glycosylase: Mechanism, metals and medicine. Free. Radic. Biol. Med. 2017, 107, 202–215. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Sun, J.; Guo, W.; Zhuang, Y.; Xu, L.; Wang, Y. AluYb8 insertion polymorphism in the MUTYH gene impairs mitochondrial DNA maintenance and affects the age of onset of IPF. Aging 2019, 11, 933–949. [Google Scholar] [CrossRef] [PubMed]
- Sun, Q.; Chen, J.; Xu, L.; Kang, J.; Wu, X.; Ren, Y.; Nakabeppu, Y.; Wang, Y. MUTYH Deficiency Is Associated with Attenuated Pulmonary Fibrosis in a Bleomycin-Induced Model. Oxidative Med. Cell. Longev. 2020, 2020, 4828256. [Google Scholar] [CrossRef]
- Evans, R.A.; Limp-Foster, M.; Kelley, M.R. Going APE over ref-1. Mutat. Res. 2000, 461, 83–108. [Google Scholar] [CrossRef]
- Wilson, D.M.; Takeshita, M.; Grollman, A.P.; Demple, B. Incision activity of human apurinic endonuclease (Ape) at abasic site analogs in DNA. J. Biol. Chem. 1995, 270, 16002–16007. [Google Scholar] [CrossRef] [Green Version]
- Doetsch, P.W.; Cunningham, R.P. The enzymology of apurinic/apyrimidinic endonucleases. Mutat. Res. Repair 1990, 236, 173–201. [Google Scholar] [CrossRef]
- Wei, X. Prediction of survival prognosis of non-small cell lung cancer by APE1 through regulation of Epithelial-Mesenchymal Transition. Oncotarget 2016, 7, 28523–28539. [Google Scholar] [CrossRef]
- Yang, X.; Peng, Y.; Jiang, X.; Lu, X.; Duan, W.; Zhang, S.; Dai, N.; Shan, J.; Feng, Y.; Li, X.; et al. The regulatory role of APE1 in epithelial-to-mesenchymal transition and in determining EGFR-TKI responsiveness in non-small-cell lung cancer. Cancer Med. 2018, 7, 4406–4419. [Google Scholar] [CrossRef] [PubMed]
- Sakai, Y.; Yamamori, T.; Yasui, H.; Inanami, O. Downregulation of the DNA repair enzyme apurinic/apyrimidinic endonuclease 1 stimulates transforming growth factor-β1 production and promotes actin rearrangement. Biochem. Biophys. Res. Commun. 2015, 461, 35–41. [Google Scholar] [CrossRef]
- Thompson, L.H.; Brookman, K.W.; Jones, N.J.; Allen, S.A.; Carrano, A.V. Molecular cloning of the human XRCC1 gene, which corrects defective DNA strand break repair and sister chromatid exchange. Mol. Cell. Biol. 1990, 10, 6160–6171. [Google Scholar] [CrossRef] [PubMed]
- Thompson, L.H.; West, M.G. XRCC1 keeps DNA from getting stranded. Mutat. Res. Repair 2000, 459, 1–18. [Google Scholar] [CrossRef]
- Im, J.; Nho, R.S. Fibroblasts from patients with idiopathic pulmonary fibrosis are resistant to cisplatin-induced cell death via enhanced CK2-dependent XRCC1 activity. Apoptosis 2019, 24, 499–510. [Google Scholar] [CrossRef] [PubMed]
- Paul, O.H.; Hassa, P.O.; Hottiger, M.O. The diverse biological roles of mammalian PARPS, a small but powerful family of poly-ADP-ribose polymerases. Front. Biosci. 2008, 13, 3046–3082. [Google Scholar] [CrossRef] [Green Version]
- Pacher, P.; Liaudet, L.; Bai, P.; Virág, L.; Mabley, J.; Haskó, G.; Szabó, C. Activation of Poly(ADP-Ribose) Polymerase Contributes to Development of Doxorubicin-Induced Heart Failure. J. Pharmacol. Exp. Ther. 2002, 300, 862–867. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mukhopadhyay, P.; Rajesh, M.; Cao, Z.; Horváth, B.; Park, O.; Wang, H.; Erdelyi, K.; Holovac, E.; Wang, Y.; Liaudet, L.; et al. Poly (ADP-ribose) polymerase-1 is a key mediator of liver inflammation and fibrosis. Hepatology 2014, 59, 1998–2009. [Google Scholar] [CrossRef] [Green Version]
- Abdallah, Y.; Gligorievski, D.; Kasseckert, S.A.; Dieterich, L.; Kuhlmann, C.R.; Sauer, H.; Piper, H.; Schäfer, M.; Noll, T. The role of poly(ADP-ribose) polymerase (PARP) in the autonomous proliferative response of endothelial cells to hypoxia. Cardiovasc. Res. 2007, 73, 568–574. [Google Scholar] [CrossRef]
- Genovese, T.; Mazzon, E.; Di Paola, R.; Muià, C.; Threadgill, M.D.; Caputi, A.P.; Thiemermann, C.; Cuzzocrea, S. Inhibitors of Poly(ADP-Ribose) Polymerase Modulate Signal Transduction Pathways and the Development of Bleomycin-Induced Lung Injury. J. Pharmacol. Exp. Ther. 2005, 313, 529–538. [Google Scholar] [CrossRef]
- Hu, B.; Wu, Z.; Hergert, P.; Henke, C.A.; Bitterman, P.B.; Phan, S.H. Regulation of Myofibroblast Differentiation by Poly(ADP-Ribose) Polymerase 1. Am. J. Pathol. 2013, 182, 71–83. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Durante, M.; Sgambellone, S.; Lanzi, C.; Nardini, P.; Pini, A.; Moroni, F.; Masini, E.; Lucarini, L. Effects of PARP-1 Deficiency and Histamine H4 Receptor Inhibition in an Inflammatory Model of Lung Fibrosis in Mice. Front. Pharmacol. 2019, 10, 525. [Google Scholar] [CrossRef]
- Lucarini, L.; Durante, M.; Lanzi, C.; Pini, A.; Boccalini, G.; Calosi, L.; Moroni, F.; Masini, E.; Mannaioni, G. HYDAMTIQ, a selective PARP-1 inhibitor, improves bleomycin-induced lung fibrosis by dampening the TGF-β/SMAD signalling pathway. J. Cell. Mol. Med. 2017, 21, 324–335. [Google Scholar] [CrossRef] [PubMed]
- Marteijn, J.A.; Lans, H.; Vermeulen, W.; Hoeijmakers, J.H.J. Understanding nucleotide excision repair and its roles in cancer and ageing. Nat. Rev. Mol. Cell Biol. 2014, 15, 465–481. [Google Scholar] [CrossRef] [PubMed]
- Coin, F.; Oksenych, V.; Egly, J.-M. Distinct Roles for the XPB/p52 and XPD/p44 Subcomplexes of TFIIH in Damaged DNA Opening during Nucleotide Excision Repair. Mol. Cell 2007, 26, 245–256. [Google Scholar] [CrossRef] [PubMed]
- Oksenych, V.; de Jesus, B.B.; Zhovmer, A.; Egly, J.-M.; Coin, F. Molecular insights into the recruitment of TFIIH to sites of DNA damage. EMBO J. 2009, 28, 2971–2980. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Chen, Y.; Xiang, F.; Li, M.; Li, H.; Chi, J.; Ren, K. Suppression of TGF-β1 enhances chemosensitivity of cisplatin-resistant lung cancer cells through the inhibition of drug-resistant proteins. Artif. Cells Nanomed. Biotechnol. 2018, 46, 1505–1512. [Google Scholar] [CrossRef] [PubMed]
- Mocquet, V.; Lainé, J.P.; Riedl, T.; Yajin, Z.; Lee, M.Y.; Egly, J.M. Sequential recruitment of the repair factors during NER: The role of XPG in initiating the resynthesis step. EMBO J. 2008, 27, 155–167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matsumoto, Y.; Kim, K.; Hurwitz, J.; Gary, R.; Levin, D.S.; Tomkinson, A.E.; Park, M.S. Reconstitution of Proliferating Cell Nuclear Antigen-dependent Repair of Apurinic/Apyrimidinic Sites with Purified Human Proteins. J. Biol. Chem. 1999, 274, 33703–33708. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ko, R.; Bennett, S.E. Physical and functional interaction of human nuclear uracil-DNA glycosylase with proliferating cell nuclear antigen. DNA Repair 2005, 4, 1421–1431. [Google Scholar] [CrossRef] [Green Version]
- Akbari, M. Repair of U/G and U/A in DNA by UNG2-associated repair complexes takes place predominantly by short-patch repair both in proliferating and growth-arrested cells. Nucleic Acids Res. 2004, 32, 5486–5498. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, A.-L.; Bai, H.; Shi, G.; Chang, D.-Y. MutY and MutY homologs (MYH) in genome maintenance. Front. Biosci. 2006, 11, 3062–3080. [Google Scholar] [CrossRef] [PubMed]
- Qunn, L.; Takemura, T.; Ikushima, S.; Ando, T.; Yanagawa, T.; Akiyama, O.; Oritsu, M.; Tanaka, N.; Kuroki, T. Hyperplastic epithelial foci in honeycomb lesions in idiopathic pulmonary fibrosis. Virchows Arch. 2002, 441, 271–278. [Google Scholar] [CrossRef] [PubMed]
- Tong, B.; Fu, L.; Hu, B.; Zhang, Z.; Tan, Z.; Li, S.; Chen, Y.; Zhang, C.; Wang, H.; Xu, D.; et al. Tauroursodeoxycholic acid alleviates pulmonary endoplasmic reticulum stress and epithelial-mesenchymal transition in bleomycin-induced lung fibrosis. BMC Pulm. Med. 2021, 21, 149. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Azad, N.; Wang, L.; Iyer, A.K.V.; Castranova, V.; Jiang, B.-H.; Rojanasakul, Y. Phosphatidylinositol-3-Kinase/Akt Regulates Bleomycin-Induced Fibroblast Proliferation and Collagen Production. Am. J. Respir. Cell Mol. Biol. 2010, 42, 432–441. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Feng, X.; Hu, B.; Xia, Q.; Ni, X.; Song, Y. P2X4R promotes airway remodeling by acting on the phenotype switching of bronchial smooth muscle cells in rats. Purinergic Signal. 2018, 14, 433–442. [Google Scholar] [CrossRef] [PubMed]
- Marudamuthu, A.S.; Bhandary, Y.P.; Fan, L.; Radhakrishnan, V.; MacKenzie, B.; Maier, E.; Shetty, S.K.; Nagaraja, M.; Gopu, V.; Tiwari, N.; et al. Caveolin-1–derived peptide limits development of pulmonary fibrosis. Sci. Transl. Med. 2019, 11, eaat2848. [Google Scholar] [CrossRef]
- Kasam, R.K.; Reddy, G.B.; Jegga, A.G.; Madala, S.K. Dysregulation of Mesenchymal Cell Survival Pathways in Severe Fibrotic Lung Disease: The Effect of Nintedanib Therapy. Front. Pharmacol. 2019, 10, 532. [Google Scholar] [CrossRef]
- Mishra, A.; Liu, J.Y.; Brody, A.R.; Morris, G.F. Inhaled asbestos fibers induce p53 expression in the rat lung. Am. J. Respir. Cell Mol. Biol. 1997, 16, 479–485. [Google Scholar] [CrossRef]
- Xu, J.; Morris, G.F. p53-Mediated Regulation of Proliferating Cell Nuclear Antigen Expression in Cells Exposed to Ionizing Radiation. Mol. Cell. Biol. 1999, 19, 12–20. [Google Scholar] [CrossRef] [Green Version]
- Waga, S.; Hannon, G.J.; Beach, D.; Stillman, B. The p21 inhibitor of cyclin-dependent kinases controls DNA replication by interaction with PCNA. Nature 1994, 369, 574–578. [Google Scholar] [CrossRef]
- Lipkin, S.M.; Wang, V.; Jacoby, R.F.; Banerjee-Basu, S.; Baxevanis, A.D.; Lynch, H.T.; Elliott, R.M.; Collins, F.S. MLH3: A DNA mismatch repair gene associated with mammalian microsatellite instability. Nat. Genet. 2000, 24, 27–35. [Google Scholar] [CrossRef]
- Vassilakis, D.A.; Sourvinos, G.; Spandidos, D.A.; Siafakas, N.M.; Bouros, D. Frequent Genetic Alterations at the Microsatellite Level in Cytologic Sputum Samples of Patients with Idiopathic Pulmonary Fibrosis. Am. J. Respir. Crit. Care Med. 2000, 162, 1115–1119. [Google Scholar] [CrossRef] [PubMed]
- Demopoulos, K.; Arvanitis, D.A.; Vassilakis, D.A.; Siafakas, N.M.; Spandidos, D.A. MYCL1, FHIT, SPARC, p16(INK4) and TP53 genes associated to lung cancer in idiopathic pulmonary fibrosis. J. Cell. Mol. Med. 2002, 6, 215–222. [Google Scholar] [CrossRef] [PubMed]
- Gao, A.; Song, S.; Wang, D.; Peng, W.; Tian, L. Effect of silicon dioxide on expression of poly (ADP-Ribose) polymerase mRNA and protein. Cell Biol. Int. 2009, 33, 749–754. [Google Scholar] [CrossRef] [PubMed]
- Ranjha, L.; Howard, S.M.; Cejka, P. Main steps in DNA double-strand break repair: An introduction to homologous recombination and related processes. Chromosoma 2018, 127, 187–214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kowalczykowski, S.C. An Overview of the Molecular Mechanisms of Recombinational DNA Repair. Cold Spring Harb. Perspect. Biol. 2015, 7, a016410. [Google Scholar] [CrossRef] [Green Version]
- Orthwein, A.; Noordermeer, S.M.; Wilson, M.D.; Landry, S.; Enchev, R.I.; Sherker, A.; Munro, M.; Pinder, J.; Salsman, J.; Dellaire, G.; et al. A mechanism for the suppression of homologous recombination in G1 cells. Nature 2015, 528, 422–426. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, H.H.Y.; Pannunzio, N.R.; Adachi, N.; Lieber, M.R. Non-homologous DNA end joining and alternative pathways to double-strand break repair. Nat. Rev. Mol. Cell Biol. 2017, 18, 495–506. [Google Scholar] [CrossRef]
- Mimitou, E.P.; Symington, L.S. Nucleases and helicases take center stage in homologous recombination. Trends Biochem. Sci. 2009, 34, 264–272. [Google Scholar] [CrossRef] [PubMed]
- Heyer, W.D.; Li, X.; Rolfsmeier, M.; Zhang, X.P. Rad54: The Swiss Army knife of homologous recombination? Nucleic Acids Res. 2006, 34, 4115–4125. [Google Scholar] [CrossRef] [PubMed]
- West, S.C. Molecular views of recombination proteins and their control. Nat. Rev. Mol. Cell Biol. 2003, 4, 435–445. [Google Scholar] [CrossRef]
- Krogh, B.O.; Symington, L.S. Recombination Proteins in Yeast. Annu. Rev. Genet. 2004, 38, 233–271. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takamatsu, S.; Brown, J.; Yamaguchi, K.; Hamanishi, J.; Yamanoi, K.; Takaya, H.; Kaneyasu, T.; Mori, S.; Mandai, M.; Matsumura, N. Utility of Homologous Recombination Deficiency Biomarkers Across Cancer Types. JCO Precis. Oncol. 2022, 5, 1270–1280. [Google Scholar] [CrossRef] [PubMed]
- Abaji, C.; Cousineau, I.; Belmaaza, A. BRCA2 regulates homologous recombination in response to DNA damage: Implications for genome stability and carcinogenesis. Cancer Res. 2005, 65, 4117–4125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, Y.; Raychaudhuri, P.; Costa, R.H. Chk2 Mediates Stabilization of the FoxM1 Transcription Factor To Stimulate Expression of DNA Repair Genes. Mol. Cell. Biol. 2007, 27, 1007–1016. [Google Scholar] [CrossRef] [Green Version]
- Ochi, T.; Blackford, A.N.; Coates, J.; Jhujh, S.; Mehmood, S.; Tamura, N.; Travers, J.; Wu, Q.; Draviam, V.M.; Robinson, C.V.; et al. PAXX, a paralog of XRCC4 and XLF, interacts with Ku to promote DNA double-strand break repair. Science 2015, 347, 185–188. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghosh, D.; Raghavan, S.C. Nonhomologous end joining: New accessory factors fine tune the machinery. Trends Genet. 2021, 37, 582–599. [Google Scholar] [CrossRef]
- Ghosh, D.; Raghavan, S.C. 20 years of DNA Polymerase μ, the polymerase that still surprises. FEBS J. 2021, 288, 7230–7242. [Google Scholar] [CrossRef]
- Deriano, L.; Roth, D.B. Modernizing the Nonhomologous End-Joining Repertoire: Alternative and Classical NHEJ Share the Stage. Annu. Rev. Genet. 2013, 47, 433–455. [Google Scholar] [CrossRef] [PubMed]
- Pucci, S.; Mazzarelli, P.; Rabitti, C.; Giai, M.; Gallucci, M.; Flammia, G.; Alcini, A.; Altomare, V.; Fazio, V.M. Tumor specific modulation of KU70/80 DNA binding activity in breast and bladder human tumor biopsies. Oncogene 2001, 20, 739–747. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zahid, S.; Seif El Dahan, M.; Iehl, F.; Fernandez-Varela, P.; Le Du, M.H.; Ropars, V.; Charbonnier, J.B. The Multifaceted Roles of Ku70/80. Int. J. Mol. Sci. 2021, 22, 4134. [Google Scholar] [CrossRef]
- Bulvik, R.; Breuer, R.; Dvir-Ginzberg, M.; Reich, E.; Berkman, N.; Wallach-Dayan, S.B. SIRT1 Deficiency, Specifically in Fibroblasts, Decreases Apoptosis Resistance and Is Associated with Resolution of Lung-Fibrosis. Biomolecules 2020, 10, 996. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Lee, J.-H.; Jiang, W.; Crowe, J.L.; Zha, S.; Paull, T.T. Regulation of the DNA Damage Response by DNA-PKcs Inhibitory Phosphorylation of ATM. Mol. Cell 2016, 65, 91–104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davis, A.J.; Chen, B.P.; Chen, D.J. DNA-PK: A dynamic enzyme in a versatile DSB repair pathway. DNA Repair 2014, 17, 21–29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peters, N.; Ferguson, B.; Mazzon, M.; Fahy, A.S.; Krysztofinska, E.; Arribas-Bosacoma, R.; Pearl, L.; Ren, H.; Smith, G.L. A Mechanism for the Inhibition of DNA-PK-Mediated DNA Sensing by a Virus. PLOS Pathog. 2013, 9, e1003649. [Google Scholar] [CrossRef] [PubMed]
- Jette, N.; Lees-Miller, S.P. The DNA-dependent protein kinase: A multifunctional protein kinase with roles in DNA double strand break repair and mitosis. Prog. Biophys. Mol. Biol. 2015, 117, 194–205. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Habiel, D.M.; Camelo, A.; Espindola, M.; Burwell, T.; Hanna, R.; Miranda, E.; Carruthers, A.; Bell, M.; Coelho, A.L.; Liu, H.; et al. Divergent roles for Clusterin in Lung Injury and Repair. Sci. Rep. 2017, 7, 15444. [Google Scholar] [CrossRef] [Green Version]
- Habiel, D.M.; Hohmann, M.S.; Espindola, M.S.; Coelho, A.L.; Jones, I.; Jones, H.; Carnibella, R.; Pinar, I.; Werdiger, F.; Hogaboam, C.M. DNA-PKcs modulates progenitor cell proliferation and fibroblast senescence in idiopathic pulmonary fibrosis. BMC Pulm. Med. 2019, 19, 165. [Google Scholar] [CrossRef] [PubMed]
| DNA Repair Pathways | Research Evidence | Research Gaps |
|---|---|---|
| BER * | OGG1 promotes TGF-β1-induced cell transformation/OGG1 led to a reduction in pulmonary fibrosis development | The mechanisms of OGG1 in different injury environments need to be further investigated, with a view to targeted clinical application. |
| MUTYH deficiency was associated with attenuated pulmonary fibrosis | ||
| APE1 promotes TGF-β transition/ APE1 knockdown upregulated the expression of TGF-β1 | The clinical feasibility of APE1 in pulmonary fibrosis remains to be investigated. | |
| XRCC1 deficiency decreased IPF fibroblast viability | Given that little is known about the mechanism of XRCC1 in pulmonary fibrosis, its clinical therapeutic potential needs to be further verified. | |
| PARP-1 induced lung fibroblasts activation | PARP-1 inhibitor could have a therapeutic potential. | |
| NER | ERCC1 expression showed a significant positive correlation with vimentin | What is the relationship between ERCC1, PCNA and pulmonary fibrosis is still an open question. |
| MMR | Ineffective MMR markers were used to investigate genetic alterations in IPF | Whether MMR is successful or not may be involved in the pathogenesis of pulmonary fibrosis, but these have not been confirmed. |
| HR | Selective targets of HR deficiency may contribute to mitigating fibrosis | Selective targets of HR deficiency may contribute to mitigating fibrosis, but specific targets remain blank. |
| NHEJ | Decreased deacetylation activity on Ku70 was associated with the attenuation of fibroblast accumulation | The evidence of the association between Ku70 and pulmonary fibrosis is limited and needs to be supplemented. |
| Loss of DNA-PKcs promotes the senescence of fibroblasts in IPF | Loss of DNA-PKcs promotes the senescence of fibroblasts in IP, additional studies are warranted to further explore the DNA-PKcs-dependent mechanisms |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, J.; Liu, L.; Ma, X.; Cao, X.; Chen, Y.; Qu, X.; Ji, M.; Liu, H.; Liu, C.; Qin, X.; et al. The Role of DNA Damage and Repair in Idiopathic Pulmonary Fibrosis. Antioxidants 2022, 11, 2292. https://doi.org/10.3390/antiox11112292
Zhu J, Liu L, Ma X, Cao X, Chen Y, Qu X, Ji M, Liu H, Liu C, Qin X, et al. The Role of DNA Damage and Repair in Idiopathic Pulmonary Fibrosis. Antioxidants. 2022; 11(11):2292. https://doi.org/10.3390/antiox11112292
Chicago/Turabian StyleZhu, Jiahui, Lexin Liu, Xiaodi Ma, Xinyu Cao, Yu Chen, Xiangping Qu, Ming Ji, Huijun Liu, Chi Liu, Xiaoqun Qin, and et al. 2022. "The Role of DNA Damage and Repair in Idiopathic Pulmonary Fibrosis" Antioxidants 11, no. 11: 2292. https://doi.org/10.3390/antiox11112292





