Influence of Blanching on the Gene Expression Profile of Phenylpropanoid, Flavonoid and Vitamin Biosynthesis, and Their Accumulation in Oenanthe javanica
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Conditions and Blanching with the Deep Planting Method
2.2. Assays for Total Antioxidant Capacity
2.3. Determination of Polyphenol and Flavonoid Contents and DPPH Scavenging Activity
2.3.1. Extraction Protocol
2.3.2. Polyphenol Quantification
2.3.3. Flavonoid Quantification
2.3.4. DPPH Scavenging Activity Analysis
2.4. Determination of Vitamins
2.4.1. Vitamin A, B1, and B2 Quantification
2.4.2. Vitamin C Quantification
2.5. Statistical Analysis
2.6. Transcriptomic Analysis
2.6.1. RNA Extraction, Library Construction, and Illumina Sequencing
2.6.2. Quality Control and Transcriptomic Assembling
2.6.3. DEGs Identification and Their Functional Annotations
2.7. Quantitative Real-Time PCR Analysis
3. Results
3.1. Polyphenol and Flavonoid Contents and DPPH Analysis
3.2. Vitamin Analysis
3.3. Transcriptomic Sequencing and Assembly
3.4. Differentially Expressed Gene (DEG) Analysis of Blanched Water Dropwort
3.5. Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathway Enrichment Analysis
3.5.1. Phenylpropanoid Biosynthesis Pathway
3.5.2. Flavonoid Biosynthetic Pathway
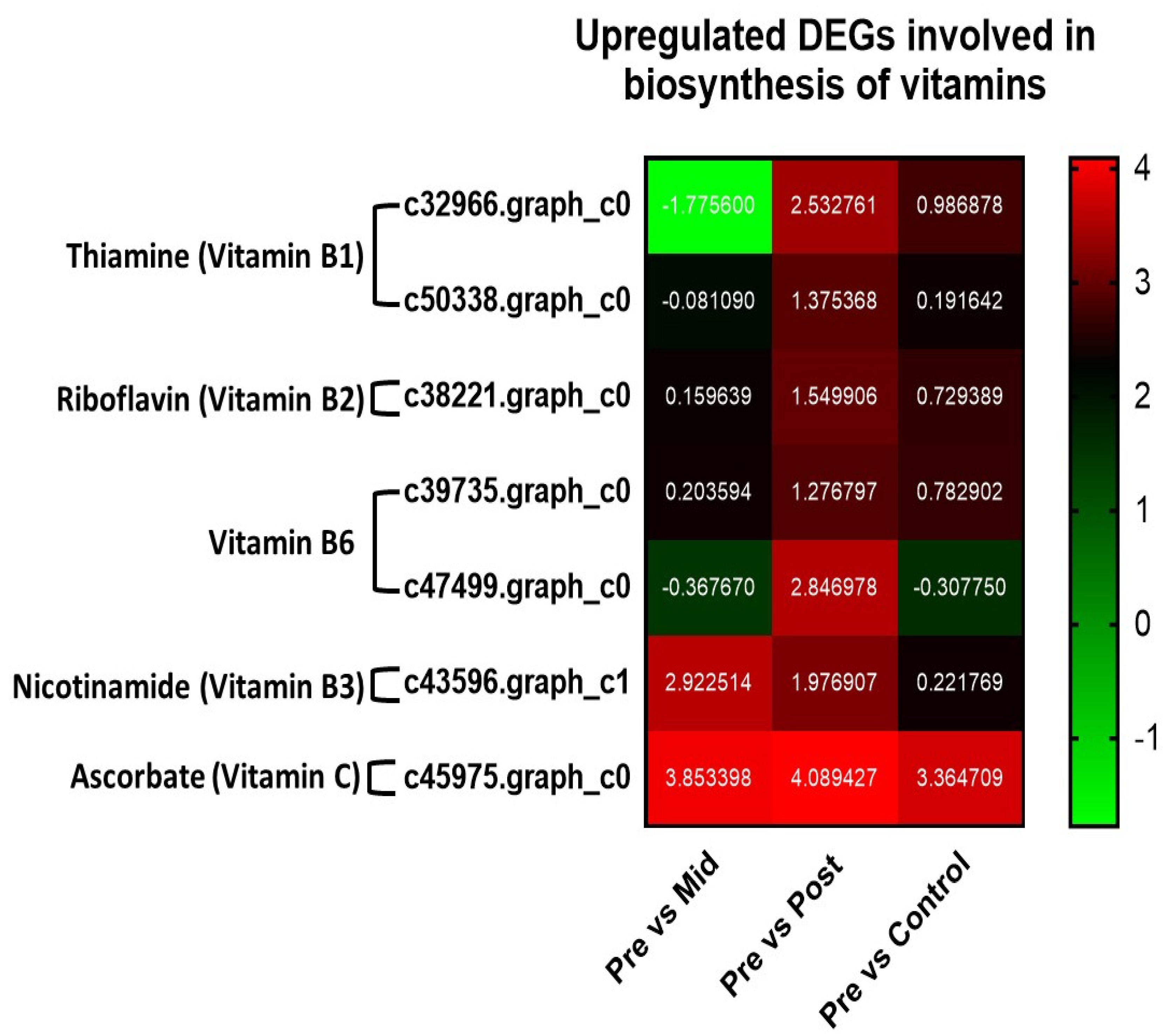
3.5.3. Upregulated DEGs in the Biosynthesis of Vitamins
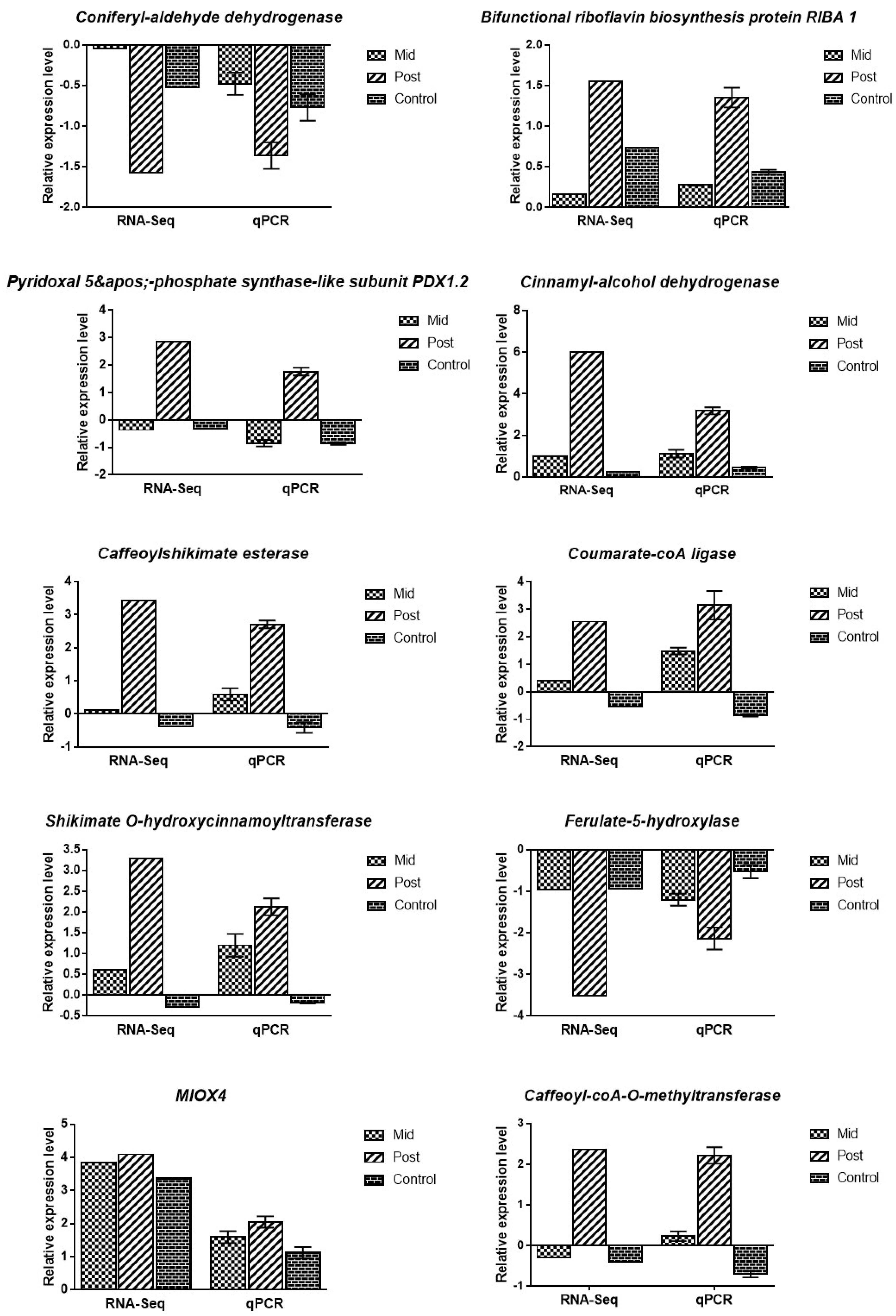
3.6. Validation of DEGs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| PBS | Phosphate-buffered saline |
| Pre-blanching | Before treatment |
| Mid-blanching | Blanched for 20 days |
| Post-blanching | Blanched for 40 days |
| Control plants | Grown under normal conditions in the field for 40 days |
| GAE | Gallic acid |
| CE | Catechin |
| TPC | Total polyphenol content |
| TFC | Total flavonoid content |
| T-AOC | Total antioxidant capacity |
| ROS | Reactive oxygen species |
| Q30 percentage | Quality score > 99.9% |
| RSEM | RNA- seq by expectation maximization |
| KOBAS | KEGG orthology-based annotation system |
| DPPH | 2,2-diphenyl-1-picryl-hydrazyl-hydrate |
| RIBA1 | Bifunctional riboflavin biosynthesis protein |
| PDX12 | Pyridoxal 5' -phosphate synthase-like subunit PDX1.2 |
| TFs | Transcription factors |
References
- Sarker, U.; Oba, S. Nutraceuticals, antioxidant pigments, and phytochemicals in the leaves of Amaranthus spinosus and Amaranthus viridis weedy species. Sci. Rep. 2019, 9, 20413. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Li, G.; Huang, X.; Ji, Q.; Zhou, K.; Hou, H.; Ke, W.; Yang, J. Phenotypic, nutritional, and antioxidant characterization of blanched Oenanthe javanica for preferable cultivar. Front. Plant Sci. 2021, 12, 231. [Google Scholar] [CrossRef]
- Sarker, U.; Hossain, M.N.; Iqbal, M.A.; Oba, S. Bioactive components and radical scavenging activity in selected advance lines of salt-tolerant vegetable Amaranth. Front. Nutr. 2020, 7, 248. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Li, G.; Yang, J.; Huang, X.; Ji, Q.; Zhou, K.; Khan, S.; Ke, W.; Hou, H. Investigation of an antioxidative system for salinity tolerance in Oenanthe javanica. Antioxidants 2020, 9, 940. [Google Scholar] [CrossRef]
- Lu, C.; Li, X. A review of Oenanthe javanica (Blume) DC. as traditional medicinal plant and its therapeutic potential. Evid.-Based Complement. Altern. Med. 2019, 2019, 6495819. [Google Scholar] [CrossRef] [Green Version]
- Jiang, Q.; Wang, F.; Tan, H.W.; Li, M.Y.; Xu, Z.S.; Tan, G.F.; Xiong, A.S. De novo transcriptome assembly, gene annotation, marker development, and miRNA potential target genes validation under abiotic stresses in Oenanthe javanica. Mol. Genet. Genom. 2015, 290, 671–683. [Google Scholar] [CrossRef]
- Chan, E.W.C.; Wong, S.K.; Chan, H.T. Ulam herbs of Oenanthe javanica and Cosmos caudatus: An overview on their medicinal properties. J. Nat. Remedies 2017, 16, 137–147. [Google Scholar] [CrossRef]
- Kumar, S.; Li, G.; Yang, J.; Huang, X.; Ji, Q.; Liu, Z.; Ke, W.; Hou, H. Effect of salt stress on growth, physiological parameters, and ionic concentration of water dropwort (Oenanthe javanica) cultivars. Front. Plant Sci. 2021, 12, 660409. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.J.; Lee, K.Y.; Jeong, E.J.; Kim, S.H.; Park, J.; Choi, Y.H.; Kim, Y.C.; Sung, S.H. Persicarin from water dropwort (Oenanthe javanica) protects primary cultured rat cortical cells from glutamate-induced neurotoxicity. Phyther. Res. 2010, 24, 913–918. [Google Scholar] [CrossRef]
- Kumar, S.; Huang, X.; Li, G.; Ji, Q.; Zhou, K.; Zhu, G.; Ke, W.; Hou, H.; Zhu, H.; Yang, J. Comparative transcriptomic analysis provides novel insights into the blanched stem of Oenanthe javanica. Plants 2021, 10, 2484. [Google Scholar] [CrossRef]
- Chan, E.W.C.; Ping Tan, Y.; Jia Chin, S.; Yi Gan, L.; Xian Kang, K.; Hong Fong, C.; Qi Chang, H.; Chern How, Y. Antioxidant properties of selected fresh and processed herbs and vegetables. Free Radic. Antioxid. 2014, 4, 39–46. [Google Scholar] [CrossRef]
- Yuanying, Y.; Xinfang, H.; Weidong, K. A brief introduction of cultivation technology of water celery. Aquat. Veg. 2008, 11, 34–35. [Google Scholar] [CrossRef]
- Yuanying, Y.; Weidong, K.; Yalin, S.; Minghua, L.; Honglian, Z.; Laichun, H. Cultivar comparative trial of water dropwort (Oenanthe decumbens) under different intenerate methods. J. Chang. Veg. 2009, 16, 45–46. [Google Scholar] [CrossRef]
- Ke, W.; Li, F.; Sun, Y.; Li, M. Investigation of aquatic vegetable resources and production in the middle and south area of Anhui Province. China Veg. 2009, 20, 61–67. [Google Scholar]
- Zhu, W.; Wu, Z.; Liang, J.; Yin, X.; Li, W.; Li, M. Cultivation techniques of water celery with mulching and softening. J. Chang. Veg. 2013, 18, 183–184. [Google Scholar] [CrossRef]
- Vogt, T. Phenylpropanoid biosynthesis. Mol. Plant 2010, 3, 2–20. [Google Scholar] [CrossRef] [Green Version]
- Cuong, D.M.; Ha, T.W.; Park, C.H.; Kim, N.S.; Yeo, H.J.; Chun, S.W.; Kim, C.; Park, S.U. Effects of LED lights on expression of genes involved in phenylpropanoid biosynthesis and accumulation of phenylpropanoids in wheat sprout. Agronomy 2019, 9, 307. [Google Scholar] [CrossRef] [Green Version]
- Shahidi, F.; Ambigaipalan, P. Phenolics and polyphenolics in foods, beverages and spices: Antioxidant activity and health effects-A review. J. Funct. Foods 2015, 18, 820–897. [Google Scholar] [CrossRef]
- Jia, X.-L.; Wang, G.-L.; Xiong, F.; Yu, X.-R.; Xu, Z.-S.; Wang, F.; Xiong, A.-S. De novo assembly, transcriptome characterization, lignin accumulation and anatomic characteristics: Novel insights into lignin biosynthesis during celery leaf development. Sci. Rep. 2015, 5, 8259. [Google Scholar] [CrossRef] [Green Version]
- Beckman, C.H. Phenolic-storing cells: Keys to programmed cell death and periderm formation in wilt disease resistance and in general defence responses in plants? Physiol. Mol. Plant Pathol. 2000, 57, 101–110. [Google Scholar] [CrossRef]
- Kong, Q.; Zhao, Y.; Guan, P.; Ke, W.; Lin, C. Chapter 5 Water dropwort. In The Aquatic Vegetable Cultivars and Resources of China; Kong, Q., Ed.; Hubei Science and Technology Press: Wuhan, China, 2005; pp. 175–215. ISBN 7-5352-3339-2. [Google Scholar]
- Yang, J.; Li, G.; Bishopp, A.; Heenatigala, P.P.M.; Hu, S.; Chen, Y.; Wu, Z.; Kumar, S.; Duan, P.; Yao, L.; et al. A comparison of growth on mercuric chloride for three Lemnaceae species reveals differences in growth dynamics that effect their suitability for use in either monitoring or remediating ecosystems contaminated with mercury. Front. Chem. 2018, 6, 112. [Google Scholar] [CrossRef] [PubMed]
- Dai, W.; Wang, M.; Gong, X.; Liu, J.H. The transcription factor FcWRKY40 of Fortunella crassifolia functions positively in salt tolerance through modulation of ion homeostasis and proline biosynthesis by directly regulating SOS2 and P5CS1 homologs. New Phytol. 2018, 219, 972–989. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Santos, J.; Herrero, M.; Mendiola, J.A.; Oliva-Teles, M.T.; Ibáñez, E.; Delerue-Matos, C.; Oliveira, M.B.P.P. Fresh-cut aromatic herbs: Nutritional quality stability during shelf-life. LWT-Food Sci. Technol. 2014, 59, 101–107. [Google Scholar] [CrossRef]
- Sarker, U.; Oba, S. Leaf pigmentation, its profiles and radical scavenging activity in selected Amaranthus tricolor leafy vegetables. Sci. Rep. 2020, 10, 18617. [Google Scholar] [CrossRef]
- Sarker, U.; Oba, S. Antioxidant constituents of three selected red and green color Amaranthus leafy vegetable. Sci. Rep. 2019, 9, 18233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, H.; Yu, X.; Li, K.; Klejnot, J.; Yang, H.; Lisiero, D.; Lin, C. Photoexcited CRY2 interacts with CIB1 to regulate transcription and floral initiation in Arabidopsis. Science 2008, 322, 1535–1539. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Osterlund, M.T.; Hardtke, C.S.; Wei, N.; Deng, X.W. Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 2000, 405, 462–466. [Google Scholar] [CrossRef]
- Wang, N.; Qian, Z.; Luo, M.; Fan, S.; Zhang, X.; Zhang, L. Identification of salt stress responding genes using transcriptome analysis in green alga Chlamydomonas reinhardtii. Int. J. Mol. Sci. 2018, 19, 3359. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Zhou, B.; Sun, M.; Li, Y.; Kawabata, S. UV-A light induces anthocyanin biosynthesis in a manner distinct from synergistic blue + UV-B light and UV-A/blue light responses in different parts of the hypocotyls in turnip seedlings. Plant Cell Physiol. 2012, 53, 1470–1480. [Google Scholar] [CrossRef] [Green Version]
- Raineri, J.; Ribichich, K.F.; Chan, R.L. The sunflower transcription factor HaWRKY76 confers drought and flood tolerance to Arabidopsis thaliana plants without yield penalty. Plant Cell Rep. 2015, 34, 2065–2080. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Tamang, B.G.; Fukao, T. Plant adaptation to multiple stresses during submergence and following desubmergence. Int. J. Mol. Sci. 2015, 16, 30164–30180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jackson, M.B.; Waters, I.; Setter, T.; Greenway, H. Injury to rice plants caused by complete submergence; a contribution by ethylene (ethene). J. Exp. Bot. 1987, 38, 1826–1838. [Google Scholar] [CrossRef]
- Ella, E.S.; Kawano, N.; Yamauchi, Y.; Tanaka, K.; Ismail, A.M. Blocking ethylene perception enhances flooding tolerance in rice seedlings. Funct. Plant Biol. 2003, 30, 813–819. [Google Scholar] [CrossRef]
- Wang, Y.; Gao, S.; He, X.; Li, Y.; Zhang, Y.; Chen, W. Response of total phenols, flavonoids, minerals, and amino acids of four edible fern species to four shading treatments. PeerJ 2020, 8, e8354. [Google Scholar] [CrossRef]
- Kumari, R.; Singh, S.; Agrawal, S.B. Combined effects of Psoralens and ultraviolet-B on growth, pigmentation and biochemical parameters of Abelmoschus esculentus L. Ecotoxicol. Environ. Saf. 2009, 72, 1129–1136. [Google Scholar] [CrossRef]
- Deng, J.; Wu, D.; Shi, J.; Balfour, K.; Wang, H.; Zhu, G.; Liu, Y.; Wang, J.; Zhu, Z. Multiple MYB activators and repressors collaboratively regulate the juvenile red fading in leaves of sweetpotato. Front. Plant Sci. 2020, 11, 941. [Google Scholar] [CrossRef]
- Lekklar, C.; Suriya-arunroj, D.; Pongpanich, M.; Comai, L.; Kositsup, B.; Chadchawan, S.; Buaboocha, T. Comparative genomic analysis of rice with contrasting photosynthesis and grain production under salt stress. Genes 2019, 10, 562. [Google Scholar] [CrossRef] [Green Version]
- Kim, H.-J.; Chen, F.; Wang, X.; Choi, J.-H. Effect of methyl jasmonate on phenolics, isothiocyanate, and metabolic enzymes in radish sprout (Raphanus sativus L.). J. Agric. Food Chem. 2006, 54, 7263–7269. [Google Scholar] [CrossRef]
- Giorgi, A.; Mingozzi, M.; Madeo, M.; Speranza, G.; Cocucci, M. Effect of nitrogen starvation on the phenolic metabolism and antioxidant properties of yarrow (Achillea collina Becker ex Rchb.). Food Chem. 2009, 114, 204–211. [Google Scholar] [CrossRef]
- Chen, Z.; Ma, Y.; Yang, R.; Gu, Z.; Wang, P. Effects of exogenous Ca2+ on phenolic accumulation and physiological changes in germinated wheat (Triticum aestivum L.) under UV-B radiation. Food Chem. 2019, 288, 368–376. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Shan, T.; Xie, B.; Ling, C.; Shao, S.; Jin, P.; Zheng, Y. Glycine betaine reduces chilling injury in peach fruit by enhancing phenolic and sugar metabolisms. Food Chem. 2019, 272, 530–538. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Li, Q.; Liu, G.; Xu, N.; Yang, Y.; Zeng, W.; Chen, A.; Wang, S. Integrated analysis of transcriptomic and metabolomic data reveals critical metabolic pathways involved in polyphenol biosynthesis in Nicotiana tabacum under chilling stress. Funct. Plant Biol. 2019, 46, 30. [Google Scholar] [CrossRef]
- Vanholme, R.; Cesarino, I.; Rataj, K.; Xiao, Y.; Sundin, L.; Goeminne, G.; Kim, H.; Cross, J.; Morreel, K.; Araujo, P.; et al. Caffeoyl shikimate esterase (CSE) is an enzyme in the lignin biosynthetic pathway in Arabidopsis. Science 2013, 341, 1103–1106. [Google Scholar] [CrossRef] [PubMed]
- Manaa, A.; Ben Ahmed, H.; Valot, B.; Bouchet, J.-P.; Aschi-Smiti, S.; Causse, M.; Faurobert, M. Salt and genotype impact on plant physiology and root proteome variations in tomato. J. Exp. Bot. 2011, 62, 2797–2813. [Google Scholar] [CrossRef] [Green Version]
- Kosová, K.; Prášil, I.; Vítámvás, P. Protein contribution to plant salinity response and tolerance acquisition. Int. J. Mol. Sci. 2013, 14, 6757–6789. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, J.; Chen, S.; Zhao, Q.; Wang, T.; Yang, C.; Diaz, C.; Sun, G.; Dai, S. Physiological and proteomic analysis of salinity tolerance in Puccinellia tenuiflora. J. Proteome Res. 2011, 10, 3852–3870. [Google Scholar] [CrossRef]
- Zheng, Z.; Sheth, U.; Nadiga, M.; Pinkham, J.L.; Shetty, K. A model for the role of the proline-linked pentose phosphate pathway in polymeric dye tolerance in oregano. Process Biochem. 2001, 36, 941–946. [Google Scholar] [CrossRef]
- Oueslati, S.; Karray-Bouraoui, N.; Attia, H.; Rabhi, M.; Ksouri, R.; Lachaal, M. Physiological and antioxidant responses of Mentha pulegium (Pennyroyal) to salt stress. Acta Physiol. Plant. 2010, 32, 289–296. [Google Scholar] [CrossRef]
- Pelletier, M.K.; Murrell, J.R.; Shirley, B.W. Characterization of flavonol synthase and leucoanthocyanidin dioxygenase genes in Arabidopsis (Further evidence for differential regulation of “Early” and “Late” genes). Plant Physiol. 1997, 113, 1437–1445. [Google Scholar] [CrossRef] [Green Version]
- Almeida, J.R.M.; D’Amico, E.; Preuss, A.; Carbone, F.; de Vos, C.H.R.; Deiml, B.; Mourgues, F.; Perrotta, G.; Fischer, T.C.; Bovy, A.G.; et al. Characterization of major enzymes and genes involved in flavonoid and proanthocyanidin biosynthesis during fruit development in strawberry (Fragaria × ananassa). Arch. Biochem. Biophys. 2007, 465, 61–71. [Google Scholar] [CrossRef] [PubMed]
- Szankowski, I.; Flachowsky, H.; Li, H.; Halbwirth, H.; Treutter, D.; Regos, I.; Hanke, M.-V.; Stich, K.; Fischer, T.C. Shift in polyphenol profile and sublethal phenotype caused by silencing of anthocyanidin synthase in apple (Malus sp.). Planta 2009, 229, 681–692. [Google Scholar] [CrossRef] [PubMed]
- Ben-Simhon, Z.; Judeinstein, S.; Trainin, T.; Harel-Beja, R.; Bar-Ya’akov, I.; Borochov-Neori, H.; Holland, D. A “White” anthocyanin-less pomegranate (Punica granatum L.) caused by an insertion in the coding region of the leucoanthocyanidin dioxygenase (LDOX; ANS) gene. PLoS ONE 2015, 10, e0142777. [Google Scholar] [CrossRef] [PubMed]
- Rafique, M.Z.; Carvalho, E.; Stracke, R.; Palmieri, L.; Herrera, L.; Feller, A.; Malnoy, M.; Martens, S. Nonsense mutation inside anthocyanidin synthase gene controls pigmentation in yellow raspberry (Rubus idaeus L.). Front. Plant Sci. 2016, 7, 1892. [Google Scholar] [CrossRef]
- Sibout, R.; Baucher, M.; Gatineau, M.; Van Doorsselaere, J.; Mila, I.; Pollet, B.; Maba, B.; Pilate, G.; Lapierre, C.; Boerjan, W.; et al. Expression of a poplar cDNA encoding a ferulate-5-hydroxylase/coniferaldehyde 5-hydroxylase increases S lignin deposition in Arabidopsis thaliana. Plant Physiol. Biochem. 2002, 40, 1087–1096. [Google Scholar] [CrossRef]
- Ferro, A.P.; Flores Júnior, R.; Finger-Teixeira, A.; Parizotto, A.V.; Bevilaqua, J.M.; de Oliveira, D.M.; Molinari, H.B.C.; Marchiosi, R.; dos Santos, W.D.; Seixas, F.A.V.; et al. Inhibition of Zea mays coniferyl aldehyde dehydrogenase by daidzin: A potential approach for the investigation of lignocellulose recalcitrance. Process Biochem. 2020, 90, 131–138. [Google Scholar] [CrossRef]
- Wagner, A.; Donaldson, L.; Ralph, J. Lignification and lignin manipulations in conifers. In Advances in Botanical Research; Elsevier Ltd.: Madison, WI, USA, 2012; Volume 61, pp. 37–76. ISBN 9780124160231. [Google Scholar]
- Drewnowski, A.; Gomez-Carneros, C. Bitter taste, phytonutrients, and the consumer: A review. Am. J. Clin. Nutr. 2000, 72, 1424–1435. [Google Scholar] [CrossRef]
- Sarker, U.; Oba, S. Augmentation of leaf color parameters, pigments, vitamins, phenolic acids, flavonoids and antioxidant activity in selected Amaranthus tricolor under salinity stress. Sci. Rep. 2018, 8, 12349. [Google Scholar] [CrossRef] [Green Version]
- Sarker, U.; Oba, S. Salinity stress enhances color parameters, bioactive leaf pigments, vitamins, polyphenols, flavonoids and antioxidant activity in selected Amaranthus leafy vegetables. J. Sci. Food Agric. 2019, 99, 2275–2284. [Google Scholar] [CrossRef]
- Sharma, P.; Jha, A.B.; Dubey, R.S.; Pessarakli, M. Reactive oxygen species, oxidative damage, and antioxidative defense mechanism in plants under stressful conditions. J. Bot. 2012, 2012, 217037. [Google Scholar] [CrossRef] [Green Version]
- Samuolienė, G.; Urbonavičiūtė, A.; Brazaitytė, A.; Šabajevienė, G.; Sakalauskaitė, J.; Duchovskis, P. The impact of LED illumination on antioxidant properties of sprouted seeds. Open Life Sci. 2011, 6, 68–74. [Google Scholar] [CrossRef]
- Deng, B.; Jin, X.; Yang, Y.; Lin, Z.; Zhang, Y. The regulatory role of riboflavin in the drought tolerance of tobacco plants depends on ROS production. Plant Growth Regul. 2014, 72, 269–277. [Google Scholar] [CrossRef]
- Padmanabhan, P.; Cheema, A.; Paliyath, G. Solanaceous fruits including tomato, eggplant, and peppers. In Encyclopedia of Food and Health, 1st ed.; Academic Press: Guelph, ON, Canada, 2016; pp. 24–32. ISBN 9780123849533. [Google Scholar]
- Subki, A.; Abidin, A.A.Z.; Yusof, Z.N.B. The role of thiamine in plants and current perspectives in crop improvement. In B Group Vitamins-Current Uses and Perspectives; LeBlanc, J.G., de Giori, G.S., Eds.; InTech: London, UK, 2018; pp. 33–44. [Google Scholar]
- Soares, C.; Carvalho, M.E.A.; Azevedo, R.A.; Fidalgo, F. Plants facing oxidative challenges—A little help from the antioxidant networks. Environ. Exp. Bot. 2019, 161, 4–25. [Google Scholar] [CrossRef]
- Goyer, A. Thiamine in plants: Aspects of its metabolism and functions. Phytochemistry 2010, 71, 1615–1624. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; Yang, S.; Tang, K.; Zhang, Y.; Leng, J.; Ma, J.; Wang, Q.; Feng, X. GmPGL1, a thiamine thiazole synthase, is required for the biosynthesis of thiamine in soybean. Front. Plant Sci. 2019, 10, 1546. [Google Scholar] [CrossRef]
- Black, K.A.; Dos Santos, P.C. Shared-intermediates in the biosynthesis of thio-cofactors: Mechanism and functions of cysteine desulfurases and sulfur acceptors. Biochim. Biophys. Acta-Mol. Cell Res. 2015, 1853, 1470–1480. [Google Scholar] [CrossRef] [Green Version]
- Gill, S.S.; Tuteja, N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 2010, 48, 909–930. [Google Scholar] [CrossRef]
- Taheri, P.; Tarighi, S. Riboflavin induces resistance in rice against Rhizoctonia solani via jasmonate-mediated priming of phenylpropanoid pathway. J. Plant Physiol. 2010, 167, 201–208. [Google Scholar] [CrossRef] [PubMed]
- Hiltunen, H.-M.; Illarionov, B.; Hedtke, B.; Fischer, M.; Grimm, B. Arabidopsis RIBA proteins: Two out of three isoforms have lost their bifunctional activity in riboflavin biosynthesis. Int. J. Mol. Sci. 2012, 13, 14086–14105. [Google Scholar] [CrossRef] [PubMed]
- Herrero, S.; González, E.; Gillikin, J.W.; Vélëz, H.; Daub, M.E. Identification and characterization of a pyridoxal reductase involved in the vitamin B6 salvage pathway in Arabidopsis. Plant Mol. Biol. 2011, 76, 157–169. [Google Scholar] [CrossRef] [PubMed]
- May, A.; Berger, S.; Hertel, T.; Köck, M. The Arabidopsis thaliana phosphate starvation responsive gene AtPPsPase1 encodes a novel type of inorganic pyrophosphatase. Biochim. Biophys. Acta-Gen. Subj. 2011, 1810, 178–185. [Google Scholar] [CrossRef] [PubMed]
- Berglund, T.; Wallström, A.; Nguyen, T.-V.; Laurell, C.; Ohlsson, A.B. Nicotinamide; antioxidative and DNA hypomethylation effects in plant cells. Plant Physiol. Biochem. 2017, 118, 551–560. [Google Scholar] [CrossRef] [PubMed]
- Mura, C.; Katz, J.E.; Clarke, S.G.; Eisenberg, D. Structure and function of an archaeal homolog of survival protein E (SurEα): An acid phosphatase with purine nucleotide specificity. J. Mol. Biol. 2003, 326, 1559–1575. [Google Scholar] [CrossRef]
- Zhang, R.-G.; Skarina, T.; Katz, J.E.; Beasley, S.; Khachatryan, A.; Vyas, S.; Arrowsmith, C.H.; Clarke, S.; Edwards, A.; Joachimiak, A.; et al. Structure of Thermotoga maritima stationary phase survival protein SurE. Structure 2001, 9, 1095–1106. [Google Scholar] [CrossRef] [Green Version]
- Hameed, A.; Iqbal, N.; Malik, S.A.; Syed, H.; Ahsanul-Haq, M. Age and organ specific accumulation of ascorbate in wheat (Triticum aestivum L.) seedlings grown under etiolation alone and in combination with oxidative stress. Cad. Pesqui. Ser. Biol. 2005, 17, 51–63. [Google Scholar]
- Lee, S.K.; Kader, A.A. Preharvest and postharvest factors influencing vitamin C content of horticultural crops. Postharvest Biol. Technol. 2000, 20, 207–220. [Google Scholar] [CrossRef] [Green Version]
- Jain, A.K.; Nessler, C.L. Metabolic engineering of an alternative pathway for ascorbic acid biosynthesis in plants. Mol. Breed. 2000, 6, 73–78. [Google Scholar] [CrossRef]
- Lorence, A.; Chevone, B.I.; Mendes, P.; Nessler, C.L. Myo-inositol oxygenase offers a possible entry point into plant ascorbate biosynthesis. Plant Physiol. 2004, 134, 1200–1205. [Google Scholar] [CrossRef] [Green Version]
- Bin Saari, N.; Fujita, S.; Miyazoe, R.; Okugawa, M. Distribution of ascorbate oxidase activities in the fruits of family Cucurbitaceae and some of their properties. J. Food Biochem. 1995, 19, 321–327. [Google Scholar] [CrossRef]
- Loewus, F.A.; Seib, P.A. Biosynthesis and metabolism of ascorbic acid in plants. CRC Crit. Rev. Plant Sci. 1987, 5, 101–119. [Google Scholar] [CrossRef]
- Wu, M.-C.; Hou, C.-Y.; Jiang, C.-M.; Wang, Y.-T.; Wang, C.-Y.; Chen, H.-H.; Chang, H.-M. A novel approach of LED light radiation improves the antioxidant activity of pea seedlings. Food Chem. 2007, 101, 1753–1758. [Google Scholar] [CrossRef]
- FAO. Human Vitamin and Mineral Requirements: Report of a Joint FAO/WHO Expert Consultation, Bangkok, Thailand; FAO: Bangkok, Thailand, 2001; pp. 1–263. ISBN 1014-9228. [Google Scholar]
- Insel, P.; Roth, W. Nutrition basics. In Connect Core Concepts in Health 13th Brief Edition; Insel, P., Roth, W., Eds.; The McGraw-Hill: New York, NY, USA; Stanford University: Stanford, CA, USA, 2013; pp. 194–225. ISBN 9780077613310. [Google Scholar]



| Gene ID | Gene Name | Tm |
|---|---|---|
| c41123.graph_c0 | Actin | 54.4 |
| TTGAGCGGAGGGAGTACTAT | ||
| GCGATGCCTTCTTCCAAATA | ||
| c38480.graph_c0 | Coniferyl-aldehyde dehydrogenase | 55.0 |
| AGTAGCAGAAGGTGACAAAG | ||
| CAGCATAGTAACGCAAAGTG | ||
| c38221.graph_c0 | Bifunctional riboflavin biosynthesis protein RIBA 1 | 55.3 |
| GGTTCTATGGCTCGATTACC | ||
| TCATGCAACTGGAAAGGATT | ||
| c47499.graph_c0 | Pyridoxal 5'-phosphate synthase-like subunit PDX1.2 | 54.9 |
| TTGAAGCCCAGATTCTTGAA | ||
| CATCACTTTCCTCACATTGC | ||
| c28110.graph_c0 | Cinnamyl-alcohol dehydrogenase | 54.7 |
| GTTGGAATAGGGTGCTTAGT | ||
| CAGCACATAATAGAGGAGCA | ||
| c45323.graph_c0 | Caffeoylshikimate esterase | 55.0 |
| CACGTGGACTGGACTTATAG | ||
| CTAGCTTTGCCCGAATATCT | ||
| c48210.graph_c0 | Coumarate-coA ligase | 54.8 |
| CCTACAATTTCTGATGCTGC | ||
| GGATGCTATTCTTGCTCTCA | ||
| c48821.graph_c0 | Shikimate O-hydroxycinnamoyltransferase | 53.4 |
| TTATATTCACTGCCACGCCT | ||
| CAAGGTTTGGGCACTGAAAA | ||
| c29968.graph_c0 | Ferulate-5-hydroxylase | 54.4 |
| ATGCTGATGATGGACCAGTT | ||
| GCCCGGTCATATGAAAGGTA | ||
| c45975.graph_c0 | Inositol oxygenase 4 (MIOX4) | 55.4 |
| GCCAAAGAAAACGGGACTAC | ||
| CAACTCGGACTTTGCTCTTG | ||
| c37977.graph_c0 | Caffeoyl-coA-O-methyltransferase | 54.9 |
| TTTGCCCGTTCTTGATCATA | ||
| CTCGATCACAAAGTCTCTGT | ||
| Treatment | TPC mg GAE/ 100 mg FW | TFC mg CAE/ 100 mg FW | %DPPH Scavenging Activity | T-AOC (U/mg Protein) |
|---|---|---|---|---|
| Pre | 447.1 ± 27.1 b | 220.2 ± 19.3 b | 51.1 ± 4.4 ab | 1.04 ± 0.11 a |
| Mid | 475.7 ± 8.9 c | 236.4 ± 20.7 b | 44.9 ± 2.4 a | 3.88 ± 0.24 b |
| Post | 307.9 ± 26.3 a | 126.3 ± 10.3 a | 56.3 ± 3.5 ab | 4.29 ± 0.39 b |
| Control | 386.0 ± 30.2 bc | 168.1 ± 13.1 b | 63.1 ± 4.7 b | 0.93 ± 0.06 a |
| Treatment | Vitamin A µg/g FW | Vitamin B1 µg/g FW | Vitamin B2 µg/g FW | Vitamin C mg/g FW |
|---|---|---|---|---|
| Pre | 0.680 ± 0.05 a | 1.065 ± 0.06 a | 0.870 ± 0.05 a | 0.068 ± 0.004 b |
| Mid | 0.755 ± 0.03 a | 1.390 ± 0.03 b | 1.054 ± 0.04 b | 0.106 ± 0.006 c |
| Post | 0.984 ± 0.04 b | 1.593 ± 0.06 c | 1.164 ± 0.09 b | 0.120 ± 0.005 d |
| Control | 0.657 ± 0.03 a | 1.111 ± 0.07 a | 0.885 ± 0.06 a | 0.024 ± 0.001 a |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kumar, S.; Huang, X.; Ji, Q.; Qayyum, A.; Zhou, K.; Ke, W.; Zhu, H.; Zhu, G. Influence of Blanching on the Gene Expression Profile of Phenylpropanoid, Flavonoid and Vitamin Biosynthesis, and Their Accumulation in Oenanthe javanica. Antioxidants 2022, 11, 470. https://doi.org/10.3390/antiox11030470
Kumar S, Huang X, Ji Q, Qayyum A, Zhou K, Ke W, Zhu H, Zhu G. Influence of Blanching on the Gene Expression Profile of Phenylpropanoid, Flavonoid and Vitamin Biosynthesis, and Their Accumulation in Oenanthe javanica. Antioxidants. 2022; 11(3):470. https://doi.org/10.3390/antiox11030470
Chicago/Turabian StyleKumar, Sunjeet, Xinfang Huang, Qun Ji, Abdul Qayyum, Kai Zhou, Weidong Ke, Honglian Zhu, and Guopeng Zhu. 2022. "Influence of Blanching on the Gene Expression Profile of Phenylpropanoid, Flavonoid and Vitamin Biosynthesis, and Their Accumulation in Oenanthe javanica" Antioxidants 11, no. 3: 470. https://doi.org/10.3390/antiox11030470






