Molecular Properties of β-Carotene Oxygenases and Their Potential in Industrial Production of Vitamin A and Its Derivatives
Abstract
:1. Introduction
2. Biochemical Properties of BCO1 and BCO2
3. Substrate Specificities of BCO1 and BCO62
4. Retinal Production by Biological Methods
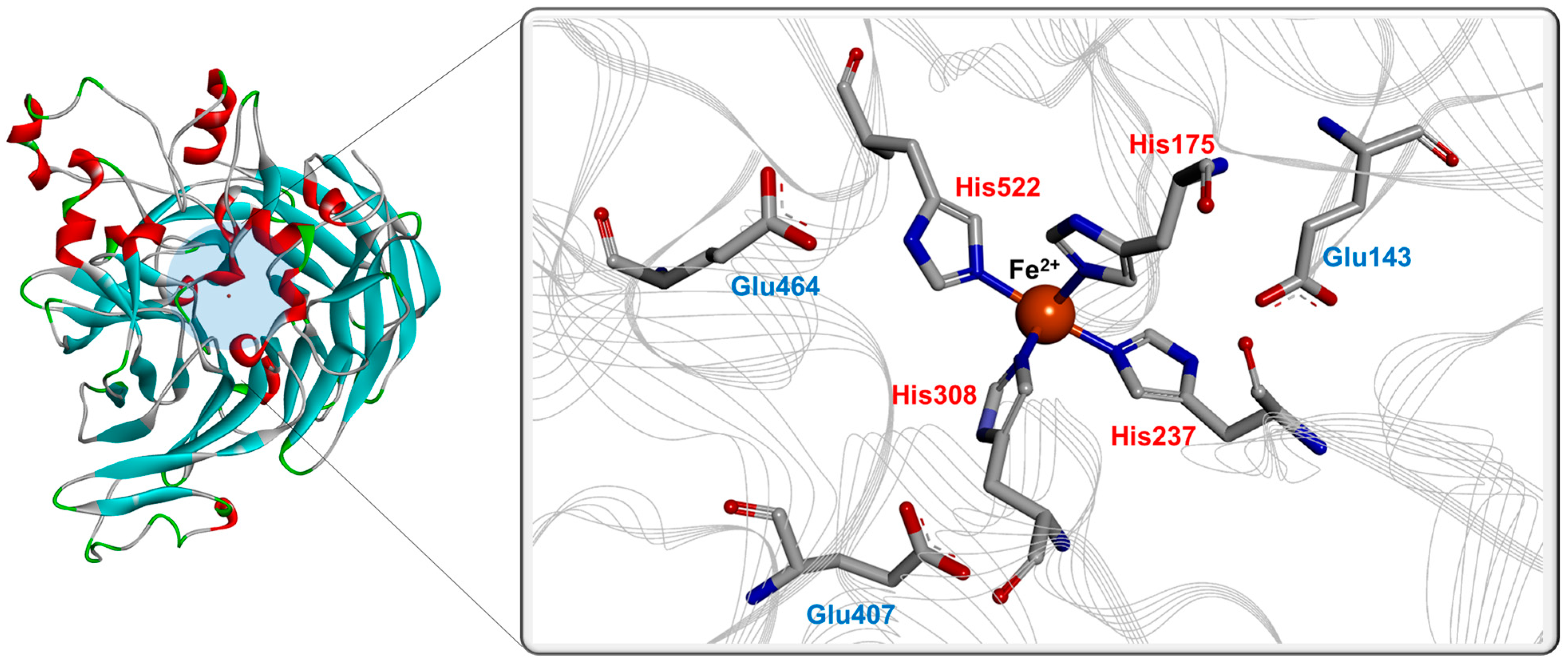
5. Proposed Roles of Amino Acids in BCOs
6. Future Perspectives of BCO in Synthetic Bacteria
7. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Blaner, W.S. Chapter 5—Vitamin A and Provitamin A Carotenoids. Present Knowledge in Nutrition, 11th ed.; Wiley-Blackwell: Cambridge, MA, USA, 2020; pp. 73–91. [Google Scholar]
- Corbo, J.C. Vitamin A1/A2 chromophore exchange: Its role in spectral tuning and visual plasticity. Dev. Biol. 2021, 475, 145–155. [Google Scholar] [CrossRef] [PubMed]
- Carazo, A.; Macakova, K.; Matousova, K.; Krcmova, L.K.; Protti, M.; Mladenka, P. Vitamin A Update: Forms, Sources, Kinetics, Detection, Function, Deficiency, Therapeutic Use and Toxicity. Nutrients 2021, 13, 1703. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.S.; Oh, D.K. Biotransformation of carotenoids to retinal by carotenoid 15,15′-oxygenase. Appl. Microbiol. Biotechnol. 2010, 88, 807–816. [Google Scholar] [CrossRef] [PubMed]
- Fisher, G.J.; Voorhees, J.J. Molecular mechanisms of retinoid actions in skin. FASEB J. 1996, 10, 1002–1013. [Google Scholar] [CrossRef] [Green Version]
- De Luca, L.M. Retinoids and their receptors in differentiation, embryogenesis, and neoplasia. FASEB J. 1991, 5, 2924–2933. [Google Scholar] [CrossRef]
- Semba, R.D. Vitamin A as “anti-infective” therapy, 1920–1940. J. Nutr. 1999, 129, 783–791. [Google Scholar] [CrossRef] [Green Version]
- Dela Sena, C.; Narayanasamy, S.; Riedl, K.M.; Curley, R.W., Jr.; Schwartz, S.J.; Harrison, E.H. Substrate specificity of purified recombinant human β-carotene 15,15′-oxygenase (BCO1). J. Biol. Chem. 2013, 288, 37094–37103. [Google Scholar] [CrossRef] [Green Version]
- Sui, X.; Kiser, P.D.; Lintig, J.; Palczewski, K. Structural basis of carotenoid cleavage: From bacteria to mammals. Arch. Biochem. Biophys. 2013, 539, 203–213. [Google Scholar] [CrossRef] [Green Version]
- Babino, D.; Golczak, M.; Kiser, P.D.; Wyss, A.; Palczewski, K.; von Lintig, J. The Biochemical Basis of Vitamin A3 Production in Arthropod Vision. ACS Chem. Biol. 2016, 11, 1049–1057. [Google Scholar] [CrossRef] [Green Version]
- Kiefer, C.; Hessel, S.; Lampert, J.M.; Vogt, K.; Lederer, M.O.; Breithaupt, D.E.; von Lintig, J. Identification and characterization of a mammalian enzyme catalyzing the asymmetric oxidative cleavage of provitamin A. J. Biol. Chem. 2001, 276, 14110–14116. [Google Scholar] [CrossRef] [Green Version]
- Amengual, J.; Widjaja-Adhi, M.A.K.; Rodriguez-Santiago, S.; Hessel, S.; Golczak, M.; Palczewski, K.; von Lintig, J. Two carotenoid oxygenases contribute to mammalian provitamin A metabolism. J. Biol. Chem. 2013, 288, 34081–34096. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jin, Y.; Yu, Y.; Zhang, C.; Li, S.; Zhang, X.; Li, F. Characterization and function analysis of the β-carotene oxygenase-like genes in carotenoids metabolism of the ridgetail white prawn Exopalaemon carinicauda. Front. Physiol. 2020, 11, 745. [Google Scholar] [CrossRef]
- Kim, Y.S.; Park, C.S.; Oh, D.K. Retinal production from β-carotene by β-carotene 15,15′-dioxygenase from an unculturable marine bacterium. Biotechnol. Lett. 2010, 32, 957–961. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.S.; Yeom, S.J.; Oh, D.K. Production of β-apo-10′-carotenal from β-carotene by human β-carotene-9′,10′-oxygenase expressed in E. coli. Biotechnol. Lett. 2011, 33, 1195–1200. [Google Scholar] [CrossRef]
- Olson, J.A.; Hayaishi, O. The enzymatic cleavage of β-carotene into vitamin A by soluble enzymes of rat liver and intestine. Proc. Natl. Acad. Sci. USA 1965, 54, 1364–1370. [Google Scholar] [CrossRef] [Green Version]
- Goodman, D.S.; Huang, H.S. Biosynthesis of Vitamin a with Rat Intestinal Enzymes. Science 1965, 149, 879–880. [Google Scholar] [CrossRef]
- Lakshmanan, M.R.; Chansang, H.; Olson, J.A. Purification and properties of carotene 15,15′-dioxygenase of rabbit intestine. J. Lipid Res. 1972, 13, 477–482. [Google Scholar] [CrossRef]
- Devery, J.; Milborrow, B.V. β-Carotene-15,15′-dioxygenase (EC 1.13.11.21) isolation reaction mechanism and an improved assay procedure. Br. J. Nutr. 1994, 72, 397–414. [Google Scholar] [CrossRef] [Green Version]
- During, A.; Nagao, A.; Hoshino, C.; Terao, J. Assay of β-carotene 15,15′-dioxygenase activity by reverse-phase high-pressure liquid chromatography. Anal. Biochem. 1996, 241, 199–205. [Google Scholar] [CrossRef]
- Pan, W.M.; Zhou, Y.L.; Wang, J.; Dai, H.E.; Wang, X.; Liu, L. Structural and functional analysis of nonheme iron enzymes BCMO-1 and BCMO-2 from Caenorhabditis elegans. Front. Mol. Biosci. 2022, 9, 844453. [Google Scholar] [CrossRef]
- Wyss, A.; Wirtz, G.; Woggon, W.; Brugger, R.; Wyss, M.; Friedlein, A.; Bachmann, H.; Hunziker, W. Cloning and expression of β,β-carotene 15,15′-dioxygenase. Biochem. Biophys. Res. Commun. 2000, 271, 334–336. [Google Scholar] [CrossRef] [PubMed]
- Wyss, A.; Wirtz, G.M.; Woggon, W.D.; Brugger, R.; Wyss, M.; Friedlein, A.; Riss, G.; Bachmann, H.; Hunziker, W. Expression pattern and localization of β,β-carotene 15,15′-dioxygenase in different tissues. Biochem. J. 2001, 354, 521–529. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.S.; Oh, D.K. Substrate specificity of a recombinant chicken β-carotene 15,15′-monooxygenase that converts β-carotene into retinal. Biotechnol. Lett. 2009, 31, 403–408. [Google Scholar] [CrossRef]
- Garcia-Lopez, E.; Gonzalez-Gallardo, A.; Antaramian, A.; Gonzalez-Davalos, M.L.; Shimada, A.; Varela-Echavarria, A.; Mora, O. In vitro conversion of β-carotene to retinal in bovine rumen fluid by a recombinant β-carotene-15, 15′-monooxygenase. Int. J. Vitam. Nutr. Res. 2012, 82, 94–103. [Google Scholar] [CrossRef] [PubMed]
- Singh, H.; Cama, H.R. Enzymatic cleavage of carotenoids. Biochim. Biophys. Acta 1974, 370, 49–61. [Google Scholar] [CrossRef]
- Yan, W.; Jang, G.F.; Haeseleer, F.; Esumi, N.; Chang, J.; Kerrigan, M.; Campochiaro, M.; Campochiaro, P.; Palczewski, K.; Zack, D.J. Cloning and characterization of a human β,β-carotene-15,15′-dioxygenase that is highly expressed in the retinal pigment epithelium. Genomics 2001, 72, 193–202. [Google Scholar] [CrossRef]
- dela Sena, C.; Riedl, K.M.; Narayanasamy, S.; Curley, R.W.; Schwartz, S.J.; Harrison, E.H. The human enzyme that converts dietary provitamin A carotenoids to vitamin A is a dioxygenase. J. Biol. Chem. 2014, 289, 13661–13666. [Google Scholar] [CrossRef] [Green Version]
- Kowatz, T.; Babino, D.; Kiser, P.; Palczewski, K.; von Lintig, J. Characterization of human β,β-carotene-15,15′-monooxygenase (BCMO1) as a soluble monomeric enzyme. Arch. Biochem. Biophys. 2013, 539, 214–222. [Google Scholar] [CrossRef] [Green Version]
- Lindqvist, A.; Andersson, S. Biochemical properties of purified recombinant human β-carotene 15,15′-monooxygenase. J. Biol. Chem. 2002, 277, 23942–23948. [Google Scholar] [CrossRef] [Green Version]
- Kelly, M.E.; Ramkumar, S.; Sun, W.; Colon Ortiz, C.; Kiser, P.D.; Golczak, M.; von Lintig, J. The Biochemical Basis of Vitamin A Production from the Asymmetric Carotenoid β-Cryptoxanthin. ACS Chem. Biol. 2018, 13, 2121–2129. [Google Scholar] [CrossRef]
- Kim, Y.S.; Kim, N.H.; Yeom, S.J.; Kim, S.W.; Oh, D.K. In vitro characterization of a recombinant Blh protein from an uncultured marine bacterium as a β-carotene 15,15′-dioxygenase. J. Biol. Chem. 2009, 284, 15781–15793. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Redmond, T.M.; Gentleman, S.; Duncan, T.; Yu, S.; Wiggert, B.; Gantt, E.; Cunningham, F.X., Jr. Identification, expression, and substrate specificity of a mammalian β-carotene 15,15′-dioxygenase. J. Biol. Chem. 2001, 276, 6560–6565. [Google Scholar] [CrossRef] [Green Version]
- Yu, D.; Zhang, R.; Wang, Y.; Zou, D.; Li, T.; Tang, H.; Jiang, L.; Wang, L. Purification of β-carotene 15,15′-monooxygenase from pig intestine and its enzymatic hydrolysis of pigment in soybean oil. Int. J. Food Sci. 2019, 54, 480–489. [Google Scholar] [CrossRef]
- Lakshman, M.R.; Okoh, C. Enzymatic conversion of all-trans-β-carotene to retinal. Methods Enzymol. 1993, 214, 256–269. [Google Scholar] [PubMed]
- Huang, F.C.; Molnar, P.; Schwab, W. Cloning and functional characterization of carotenoid cleavage dioxygenase 4 genes. J. Exp. Bot. 2009, 60, 3011–3022. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holger, S.; Kurtzer, R.; Eisenreich, W.; Schwab, W. The carotenase AtCCD1 from Arabidopsis thaliana is a dioxygenase. J. Biol. Chem. 2006, 281, 9845–9851. [Google Scholar]
- Dela Sena, C.; Sun, J.; Narayanasamy, S.; Riedl, K.M.; Yuan, Y.; Curley, R.W., Jr.; Schwartz, S.J.; Harrison, E.H. Substrate specificity of purified recombinant chicken β-carotene 9′,10′-oxygenase (BCO2). J. Biol. Chem. 2016, 291, 14609–14619. [Google Scholar] [CrossRef] [Green Version]
- Hu, K.Q.; Liu, C.; Ernst, H.; Krinsky, N.I.; Russell, R.M.; Wang, X.D. The biochemical characterization of ferret carotene-9′,10′-monooxygenase catalyzing cleavage of carotenoids in vitro and in vivo. J. Biol. Chem. 2006, 281, 19327–19338. [Google Scholar] [CrossRef] [Green Version]
- Lindqvist, A.; He, Y.G.; Andersson, S. Cell type-specific expression of β-carotene 9′,10′-monooxygenase in human tissues. J. Histochem. Cytochem. 2005, 53, 1403–1412. [Google Scholar] [CrossRef]
- Bruno, M.; Beyer, P.; Al-Babili, S. The potato carotenoid cleavage dioxygenase 4 catalyzes a single cleavage of β-ionone ring-containing carotenes and non-epoxidated xanthophylls. Arch. Biochem. Biophys. 2015, 572, 126–133. [Google Scholar] [CrossRef]
- Wei, T.; Jia, B.; Huang, S.; Yang, K.; Jia, C.; Mao, D. Purification and characterization of a novel β-carotene-9′,10′-oxygenase from Saccharomyces cerevisiae ULI3. Biotechnol. Lett. 2015, 37, 1993–1998. [Google Scholar] [CrossRef] [Green Version]
- Rubio-Moraga, A.; Rambla, J.L.; Fernandez-de-Carmen, A.; Trapero-Mozos, A.; Ahrazem, O.; Orzaez, D.; Granell, A.; Gomez-Gomez, L. New target carotenoids for CCD4 enzymes are revealed with the characterization of a novel stress-induced carotenoid cleavage dioxygenase gene from Crocus sativus. Plant Mol. Biol. 2014, 86, 555–569. [Google Scholar] [CrossRef] [PubMed]
- Goodman, D.S.; Huang, H.S.; Shiratori, T. Mechanism of the biosynthesis of vitamin A from β-carotene. J. Biol. Chem. 1966, 241, 1929–1932. [Google Scholar] [CrossRef]
- Poliakov, E.; Gentleman, S.; Cunningham, F.X., Jr.; Miller-Ihli, N.J.; Redmond, T.M. Key role of conserved histidines in recombinant mouse β-carotene 15,15′-monooxygenase-1 activity. J. Biol. Chem. 2005, 280, 29217–29223. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kloer, D.P.; Ruch, S.; Al-Babili, S.; Beyer, P.; Schulz, G.E. The structure of a retinal-forming carotenoid oxygenase. Science 2005, 308, 267–269. [Google Scholar] [CrossRef] [Green Version]
- Bornscheuer, U.T.; Huisman, G.W.; Kazlauskas, R.J.; Lutz, S.; Moore, J.C.; Robins, K. Engineering the third wave of biocatalysis. Nature 2012, 485, 185–194. [Google Scholar] [CrossRef]
- Bornscheuer, U.T. Biocatalysis: Successfully Crossing Boundaries. Angew. Chem. Int. Ed. Engl. 2016, 55, 4372–4373. [Google Scholar] [CrossRef] [Green Version]
- Sheldon, R.A.; Brady, D. Broadening the Scope of Biocatalysis in Sustainable Organic Synthesis. ChemSusChem 2019, 12, 2859–2881. [Google Scholar] [CrossRef]
- Wu, S.; Snajdrova, R.; Moore, J.C.; Baldenius, K.; Bornscheuer, U.T. Biocatalysis: Enzymatic Synthesis for Industrial Applications. Angew. Chem. Int. Ed. Engl. 2021, 60, 88–119. [Google Scholar] [CrossRef]
- Kim, Y.S.; Kim, N.H.; Kim, H.J.; Lee, J.K.; Kim, S.W.; Oh, D.K. Effective production of retinal from β-carotene using recombinant mouse β-carotene 15,15′-monooxygenase. Appl. Microbiol. Biotechnol. 2007, 76, 1339–1345. [Google Scholar] [CrossRef]
- Kim, N.H.; Kim, Y.S.; Kim, H.J.; Oh, D.K. Optimized formation of detergent micelles of β-carotene and retinal production using recombinant human β,β-carotene 15,15′-monooxygenase. Biotechnol. Prog. 2008, 24, 227–231. [Google Scholar] [CrossRef] [PubMed]
- von Lintig, J.; Vogt, K. Filling the gap in vitamin A research. Molecular identification of an enzyme cleaving β-carotene to retinal. J. Biol. Chem. 2000, 275, 11915–11920. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.H.; Choi, J.G.; Kim, Y.S.; Kim, K.R.; Kim, S.W.; Oh, D.K. Enhancement of retinal production by supplementing the surfactant Span 80 using metabolically engineered Escherichia coli. J. Biosci. Bioeng. 2012, 113, 461–466. [Google Scholar] [CrossRef] [PubMed]
- Jang, H.J.; Yoon, S.H.; Ryu, H.K.; Kim, J.H.; Wang, C.L.; Kim, J.Y.; Oh, D.K.; Kim, S.W. Retinoid production using metabolically engineered Escherichia coli with a two-phase culture system. Microb. Cell. Fact. 2011, 10, 59. [Google Scholar] [CrossRef] [Green Version]
- Jang, H.J.; Ha, B.K.; Zhou, J.; Ahn, J.; Yoon, S.H.; Kim, S.W. Selective retinol production by modulating the composition of retinoids from metabolically engineered E. coli. Biotechnol. Bioeng. 2015, 112, 1604–1612. [Google Scholar] [CrossRef]
- Choi, B.H.; Hwang, H.J.; Lee, J.E.; Oh, S.H.; Hwang, J.S.; Lee, B.Y.; Lee, P.C. Microbial production of retinyl palmitate and its application as a cosmeceutical. Antioxidants 2020, 9, 1130. [Google Scholar] [CrossRef]
- Zhang, C.; Chen, X.; Lindley, N.D.; Too, H.P. A “plug-n-play” modular metabolic system for the production of apocarotenoids. Biotechnol. Bioeng. 2018, 115, 174–183. [Google Scholar] [CrossRef]
- Hu, Q.; Zhang, T.; Yu, H.; Ye, L. Selective biosynthesis of retinol in S. cerevisiae. Bioresour. Bioprocess. 2022, 9, 22. [Google Scholar] [CrossRef]
- Sun, L.; Kwak, S.; Jin, Y.S. Vitamin A production by engineered Saccharomyces cerevisiae from xylose via two-phase in situ extraction. ACS Synth. Biol. 2019, 8, 2131–2140. [Google Scholar] [CrossRef]
- Daruwalla, A.; Zhang, J.; Lee, H.J.; Khadka, N.; Farquhar, E.R.; Shi, W.; von Lintig, J.; Kiser, P.D. Structural basis for carotenoid cleavage by an archaeal carotenoid dioxygenase. Proc. Natl. Acad. Sci. USA 2020, 117, 19914–19925. [Google Scholar] [CrossRef]
- Poliakov, E.; Gentleman, S.; Chander, P.; Cunningham, F.X., Jr.; Grigorenko, B.L.; Nemuhin, A.V.; Redmond, T.M. Biochemical evidence for the tyrosine involvement in cationic intermediate stabilization in mouse β-carotene 15, 15′-monooxygenase. BMC Biochem. 2009, 10, 31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoon, S.H.; Kim, J.E.; Lee, S.H.; Park, H.M.; Choi, M.S.; Kim, J.Y.; Lee, S.H.; Shin, Y.C.; Keasling, J.D.; Kim, S.W. Engineering the lycopene synthetic pathway in E. coli by comparison of the carotenoid genes of Pantoea agglomerans and Pantoea ananatis. Appl. Microbiol. Biotechnol. 2007, 74, 131–139. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.H.; Park, H.M.; Kim, J.E.; Lee, S.H.; Choi, M.S.; Kim, J.Y.; Oh, D.K.; Keasling, J.D.; Kim, S.W. Increased β-carotene production in recombinant Escherichia coli harboring an engineered isoprenoid precursor pathway with mevalonate addition. Biotechnol. Prog. 2007, 23, 599–605. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Wang, Y.; Tang, Q.; Kong, W.; Chung, W.J.; Lu, T. MEP pathway-mediated isopentenol production in metabolically engineered Escherichia coli. Microb. Cell Fact. 2014, 13, 135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, C.; Seow, V.Y.; Chen, X.; Too, H.P. Multidimensional heuristic process for high-yield production of astaxanthin and fragrance molecules in Escherichia coli. Nat. Commun. 2018, 9, 1858. [Google Scholar] [CrossRef] [Green Version]
- Chatzivasileiou, A.O.; Ward, V.; Edgar, S.M.; Stephanopoulos, G. Two-step pathway for isoprenoid synthesis. Proc. Natl. Acad. Sci. USA 2019, 116, 506–511. [Google Scholar] [CrossRef] [Green Version]
- Kim, M.J.; Noh, M.H.; Woo, S.; Lim, H.G.; Jung, G.Y. Enhanced lycopene production in Escherichia coli by expression of two MEP pathway enzymes from Vibrio sp. Dhg. Catalysts 2019, 9, 1003. [Google Scholar] [CrossRef] [Green Version]
- Ferrando, J.; Solomon, L.A. Recent progress using de novo design to study protein structure, design and binding interactions. Life 2021, 11, 225. [Google Scholar] [CrossRef]
- Zhang, T.; Wu, Z.F.; Chen, H.; Wu, Q.; Tang, Z.Z.; Gou, J.B.; Wang, L.H.; Hao, W.W.; Wang, C.M.; Li, C.M. Progress in strategies for sequence diversity library creation for directed evolution. Afr. J. Biotechnol. 2010, 9, 9277–9285. [Google Scholar]
- Grayson, K.J.; Anderson, J.L.R. Designed for life: Biocompatible de novo designed proteins and components. J. R. Soc. Interface 2018, 15, 20180472. [Google Scholar] [CrossRef]
- Tippmann, S.; Anfelt, J.; David, F.; Rand, J.M.; Siewers, V.; Uhlen, M.; Nielsen, J.; Hudson, E.P. Affibody scaffolds improve sesquiterpene production in Saccharomyces cerevisiae. ACS Synth. Biol. 2017, 6, 19–28. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Tian, L.Q.; Tang, S.S.; Xie, C.J.; Xu, J.L.; Jiang, L. Design and tailoring of an artificial DNA scaffolding system for efficient lycopene synthesis using zinc-finger-guided assembly. J. Ind. Microbiol. Biot. 2020, 47, 209–222. [Google Scholar] [CrossRef] [PubMed]
- Hatlem, D.; Trunk, T.; Linke, D.; Leo, J.C. Catching a SPY: Using the SpyCatcher-SpyTag and related systems for labeling and localizing bacterial proteins. Int. J. Mol. Sci. 2019, 20, 2119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Keeble, A.H.; Howarth, M. Power to the protein: Enhancing and combining activities using the Spy toolbox. Chem. Sci. 2020, 11, 7281–7291. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Yan, P.; Li, Y.; Liu, X.; Wang, Z.; Chen, T.; Zhao, X. Enhancing β-Carotene Production in Escherichia coli by Perturbing Central Carbon Metabolism and Improving the NADPH Supply. Front. Bioeng. Biotechnol. 2020, 8, 585. [Google Scholar] [CrossRef]
- Jung, J.; Lim, J.H.; Kim, S.Y.; Im, D.K.; Seok, J.Y.; Lee, S.J.V.; Oh, M.K.; Jung, G.Y. Precise precursor rebalancing for isoprenoids production by fine control of gapA expression in Escherichia coli. Metab. Eng. 2016, 38, 401–408. [Google Scholar] [CrossRef]




| Type /Organism | Accession No. | Optimum Temperature (°C) | Optimum pH | Molecular Weight (kDa) | Associated Form | Metal Ion | References | |
|---|---|---|---|---|---|---|---|---|
| Subunit | Native | |||||||
| BCO1 | ||||||||
| Caenorhabditis elegansR | Q9U2E4 | 37 | 7.5 | – | – | dimer | – | [21] |
| ChickenR | Q9I993 | – | – | 60 | – | – | – | [22] |
| ChickenR | 37 | 8.0 | 60 | ≈50 | monomer | – | [23] | |
| ChickenR | 37 | 8.0 | 60 | 240 | tetramer | Fe2+ | [24] | |
| ChickenR | 37 | – | 64 | – | – | – | [25] | |
| Guinea pigN | – | 37 | 8.5 | – | – | – | Fe2+ | [26] |
| HumanR | Q9HAY6 | 37 | 6.5 | 65 | – | monomer | Fe2+ | [27] |
| HumanR | 37 | 8.0 | – | – | – | – | [28] | |
| HumanR | 37 | 8.0 | – | – | – | – | [8] | |
| HumanR | 28 | 7.6 | – | – | – | – | [29] | |
| HumanR | 37 | 7.5–8.0 | ≈64 | ≈230 | tetramer | Fe2+ | [30] | |
| HumanR | 30 | 7.0 | – | – | dimer | Fe2+ | [31] | |
| Uncultured marine bacteriumR | Q4PNI0 | 40 | 8.0 | 32 | 64 | dimer | Fe2+ | [32] |
| MouseR | Q9JJS6 | 37 | 8.0 | 65 | – | – | Fe2+ | [33] |
| PigN | – | 37 | 8.0 | 156 | – | – | – | [34] |
| RatN | – | – | 7.5–8.0 | 62 | 62 | monomer | – | [35] |
| RatN | – | 37 | 8.0 | – | – | – | – | [20] |
| BCO2 | ||||||||
| AppleR | A9Z0V8 | 30 | ≈7.4 | – | – | – | – | [36] |
| Arabidopsis thalianaR | O65572 | 30 | ≈7.4 | – | – | – | – | [37] |
| ChickenR | E1C8E0 | 37 | 8.0 | 64 | – | – | Fe2+ | [38] |
| ChrysanthemumR | B0FLM7 | 30 | ≈7.4 | – | – | – | – | [36] |
| FerretR | Q6QT07 | 37 | 8.5 | ≈64 | – | – | Fe2+ | [39] |
| HumanR | Q9BYV7 | – | – | 64 | – | – | – | [40] |
| HumanR | 37 | 8.0 | 58 | 240 | tetramer | Fe2+ | [15] | |
| MouseR | Q99NF1 | 37 | 8.0 | – | – | – | – | [11] |
| MouseR | 30 | 7.0 | – | – | dimer | Fe2+ | [31] | |
| OsmanthusR | B0FLM9 | 30 | ≈7.4 | – | – | – | – | [36] |
| PotatoN | – | 28 | 7.8 | – | – | – | Fe2+ | [41] |
| RoseR | B0FLM8 | 30 | ≈7.4 | – | – | – | – | [36] |
| Saccharomyces cerevisiaeN | – | 45 | 8.0 | 50 | – | – | Ca2+ | [42] |
| SaffronR | A0SE36 | 30 | – | – | – | – | – | [43] |
| Type /Organism | Expression | Substrate | Specific Activity (nmol/min/mg) | kcat/Km (min−1 mM−1) | References |
|---|---|---|---|---|---|
| BCO1 | |||||
| Chicken | pBAD system in E. coli cellsP | β-carotene | 6.8 | [25] | |
| Chicken | SFV system in BHK cellsH | β-carotene | 0.04 | [22] | |
| Chicken | SFV system in BHK cells | β-carotene | 0.04 | [23] | |
| Chicken | pET system in E. coli cellsH | β-carotene | 0.32 | 64.0 | [24] |
| α-carotene | 1.3 | ||||
| γ-carotene | 0.7 | ||||
| β-cryptoxanthin | 7.1 | ||||
| β-apo-4′-carotenal | 4.9 | ||||
| β-apo-8′-carotenal | 6.0 | ||||
| Guinea pig | Extraction from intestineP | β-carotene | 0.03 | [26] | |
| lutein | 0.02 | ||||
| β-apo-10′-carotenal | 0.02 | ||||
| Human | expression in SF9 cells | β-carotene | 18.5 | [29] | |
| Human | expression in SF9 cells | β-carotene | 93.0 | [30] | |
| β-cryptoxanthin | 19.0 | ||||
| Human | pET system in E. coli cellsH | β-carotene | 2.20 | 6.10 | [8] |
| α-carotene | 1.81 | ||||
| β-cryptoxanthin | 1.43 | ||||
| lycopene | 8.85 | ||||
| β-apo-8′-carotenal | 2.72 | ||||
| Uncultured marine bacterium | pET system in E. coli cellsH | β-carotene | 45.0 | 97.0 | [32] |
| α-carotene | 3.60 | ||||
| γ-carotene | 0.55 | ||||
| β-cryptoxanthin | 28.0 | ||||
| β-apo-4′-carotenal | 4.20 | ||||
| Mouse | pBAD system in E. coli cellsH | β-carotene | 0.39 | [33] | |
| Rat | extraction from intestine | β-carotene | 0.012 | [20] | |
| BCO2 | |||||
| Chicken | pET system in E. coli cellsH | β-carotene | 0.49 | [38] | |
| Human | pET system in E. coli cellsH | β-carotene | 80.0 | [15] | |
| S. cerevisiae | extraction from microbial cells | β-carotene | 380.0 | 77.0 | [42] |
| α-carotene | 49.0 | ||||
| lutein | 10.0 |
| Biocatalyst | Source/Host | Substrate (mg/L) | Product (mg/L) | Reaction Time (h) | Conversion Yield (%) | Productivity (mg/L/h) | References |
|---|---|---|---|---|---|---|---|
| BCO1 | Mouse | 200 | 72 | 2 | 36 | 4.8 | [51] |
| Human | 200 | 98 | 16 | 49 | 6.13 | [52] | |
| Chicken | 5.37 | 3.2 | 16 | 60 | 1.06 | [24] | |
| Fruit fly | 0.17 | 0.13 | 2 | 18 | 0.065 | [53] | |
| Uncultured Marine bacterium | 350 | 181 | 20 | 52 | 9.1 | [14] | |
| Pig | 44 | 14.65 | 20 | 33.3 | 0.73 | [34] | |
| Metabolically engineered cells | E. coli | – | 600 | 33 | – | 18 | [54] |
| E. coli | – | 67 | 72 | – | 0.93 | [55] | |
| E. coli | – | 7 | 72 | – | 0.1 | [56] | |
| E. coli | – | 2.26 | 36 | – | 0.062 | [57] | |
| E. coli | – | 5.1 | 50 | – | 0.102 | [58] | |
| S. cerevisiae | – | 221.37 | 20 | – | 11.065 | [59] | |
| S. cerevisiae | – | 2094 | 160 | – | 13.1 | [60] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shin, K.-C.; Seo, M.-J.; Kim, Y.-S.; Yeom, S.-J. Molecular Properties of β-Carotene Oxygenases and Their Potential in Industrial Production of Vitamin A and Its Derivatives. Antioxidants 2022, 11, 1180. https://doi.org/10.3390/antiox11061180
Shin K-C, Seo M-J, Kim Y-S, Yeom S-J. Molecular Properties of β-Carotene Oxygenases and Their Potential in Industrial Production of Vitamin A and Its Derivatives. Antioxidants. 2022; 11(6):1180. https://doi.org/10.3390/antiox11061180
Chicago/Turabian StyleShin, Kyung-Chul, Min-Ju Seo, Yeong-Su Kim, and Soo-Jin Yeom. 2022. "Molecular Properties of β-Carotene Oxygenases and Their Potential in Industrial Production of Vitamin A and Its Derivatives" Antioxidants 11, no. 6: 1180. https://doi.org/10.3390/antiox11061180






