Heterologous Production in the Synechocystis Chassis Suggests the Biosynthetic Pathway of Astaxanthin in Cyanobacteria
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains and Growth Conditions
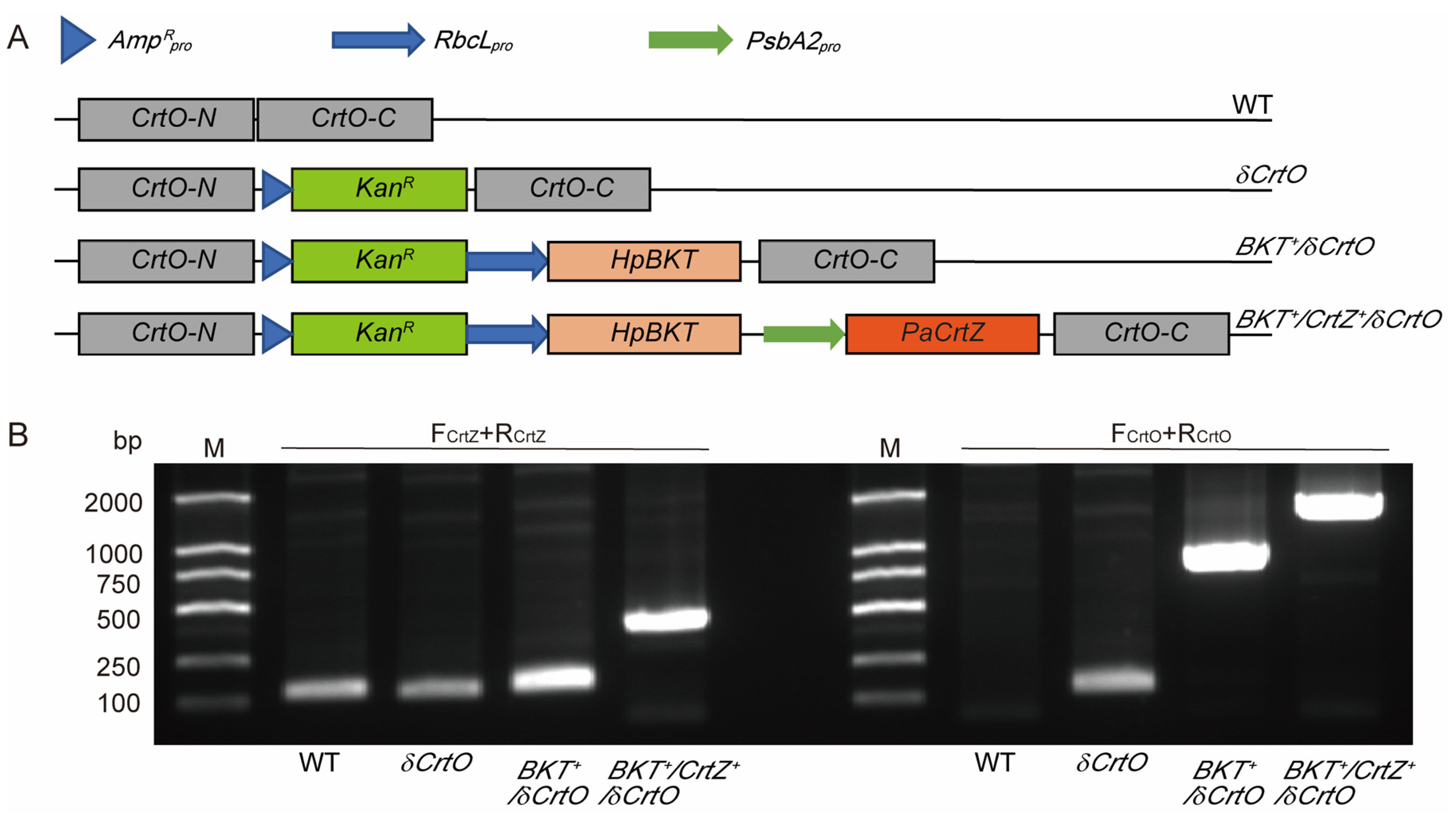
2.2. Gene Cloning and Vector Construction
2.3. Synechocystis Transformation and Mutant Screening
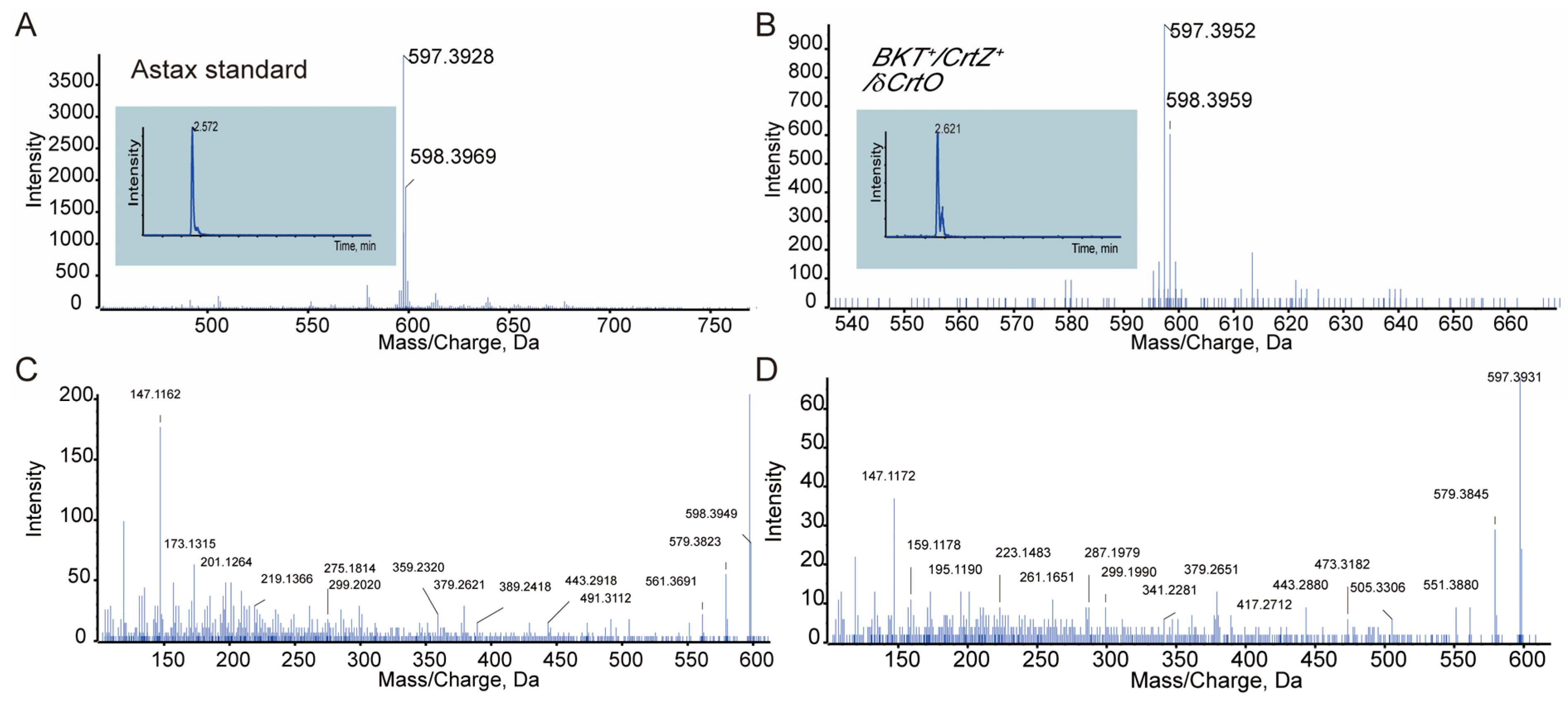
2.4. Carotenoid Extraction and Analysis
2.5. Phylogenetic Analysis
2.6. Statistical Analysis
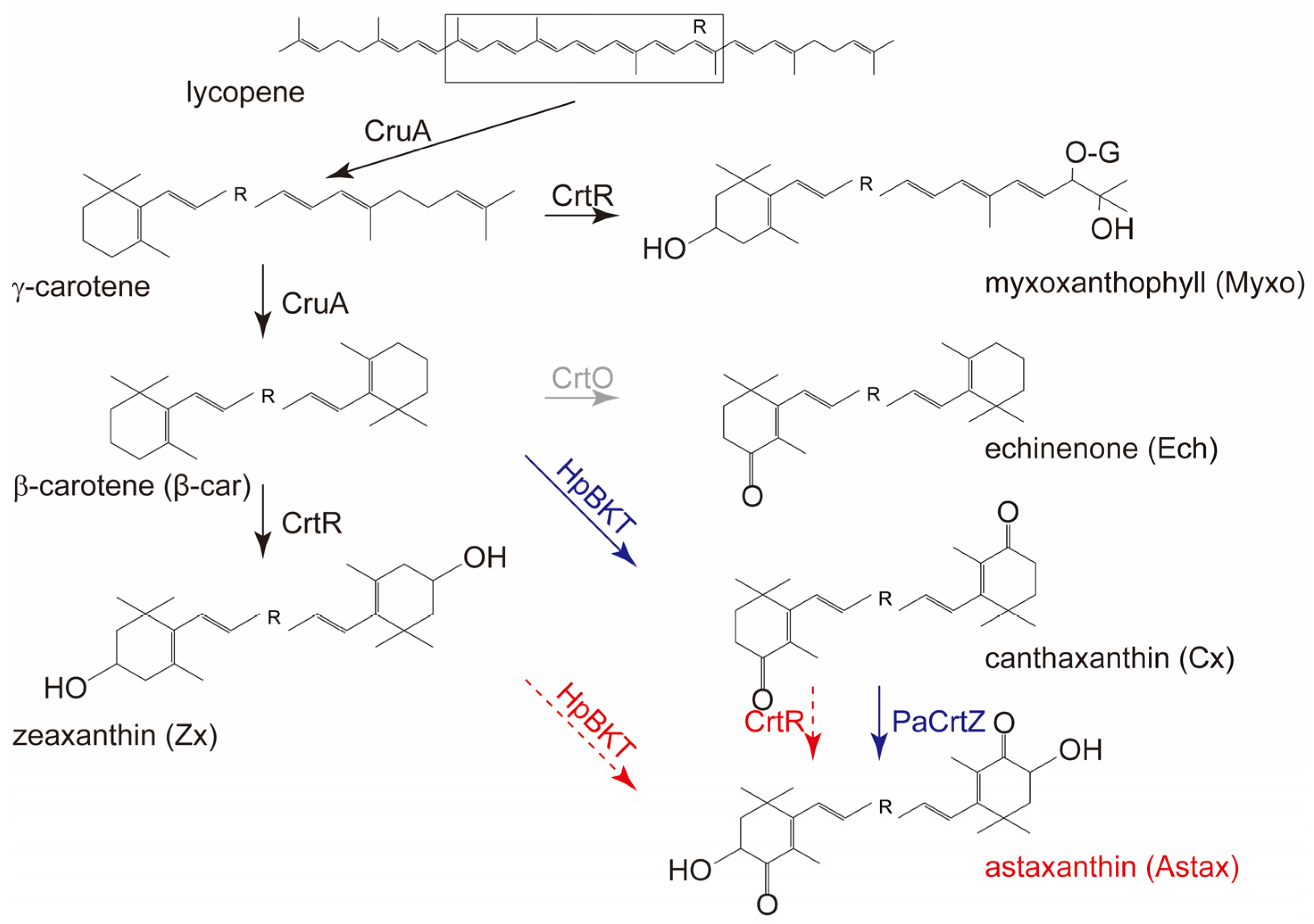
3. Results and Discussion
3.1. Synechocystis Expressing HpBKT Alone Did Not Synthesize Astaxanthin
3.2. PaCrtZ Complemented the Biosynthesis of Astaxanthin in HpBKT-Transformed Cells
3.3. Heterokont CrtZs Might Have Different Catalytic Properties
3.4. Synechocystis Growth Was Not Affected by Astaxanthin Biosynthesis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Abeyrathne, E.; Nam, K.; Huang, X.; Ahn, D.U. Plant- and animal-based antioxidants’ structure, efficacy, mechanisms, and applications: A review. Antioxidants 2022, 11, 1025. [Google Scholar] [CrossRef]
- Sun, T.; Rao, S.; Zhou, X.; Li, L. Plant carotenoids: Recent advances and future perspectives. Mol. Hortic. 2022, 2, 3. [Google Scholar] [CrossRef]
- Mrowicka, M.; Mrowicki, J.; Kucharska, E.; Majsterek, I. Lutein and zeaxanthin and their roles in age-related macular degeneration-neurodegenerative disease. Nutrients 2022, 14, 827. [Google Scholar] [CrossRef]
- Kapoor, B.; Gulati, M.; Rani, P.; Kochhar, R.S.; Atanasov, A.G.; Gupta, R.; Sharma, D.; Kapoor, D. Lycopene: Sojourn from kitchen to an effective therapy in Alzheimer’s disease. Biofactors 2023, 49, 208–227. [Google Scholar] [CrossRef]
- Crupi, P.; Faienza, M.F.; Naeem, M.Y.; Corbo, F.; Clodoveo, M.L.; Muraglia, M. Overview of the potential beneficial effects of carotenoids on consumer health and well-being. Antioxidants 2023, 12, 1069. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Li, L. Carotenoid metabolism: Biosynthesis, regulation, and beyond. J. Integr. Plant Biol. 2008, 50, 778–785. [Google Scholar] [CrossRef]
- Alcaíno, J.; Baeza, M.; Cifuentes, V. Carotenoid distribution in nature. In Carotenoids in Nature: Biosynthesis, Regulation and Function; Stange, C., Ed.; Springer: Cham, Switzerland, 2016; Volume 79, pp. 3–33. [Google Scholar]
- Wang, Q.; Cao, T.-J.; Zheng, H.; Zhou, C.-F.; Wang, Z.; Wang, R.; Lu, S. Manipulation of carotenoid metabolic flux by lycopene cyclization in ripening red pepper (Capsicum annuum var. conoides) fruits. J. Agric. Food Chem. 2019, 67, 4300–4310. [Google Scholar] [CrossRef]
- Demmig-Adams, B.; Adams, W.W., III. The role of xanthophyll cycle carotenoids in the protection of photosynthesis. Trends Plant Sci. 1996, 1, 21–26. [Google Scholar] [CrossRef]
- Schubert, N.; García-Mendoza, E.; Pacheco-Ruiz, I. Carotenoid composition of marine red algae. J. Phycol. 2006, 42, 1208–1216. [Google Scholar] [CrossRef]
- Takaichi, S. Carotenoids in algae: Distributions, biosyntheses and functions. Mar. Drugs 2011, 9, 1101–1118. [Google Scholar] [CrossRef]
- Ha, S.-H.; Kim, J.-B.; Park, J.-S.; Lee, S.-W.; Cho, K.-J. A comparison of the carotenoid accumulation in Capsicum varieties that show different ripening colours: Deletion of the capsanthin-capsorubin synthase gene is not a prerequisite for the formation of a yellow pepper. J. Exp. Bot. 2007, 58, 3135–3144. [Google Scholar] [CrossRef]
- Aflalo, C.; Meshulam, Y.; Zarka, A.; Boussiba, S. On the relative efficiency of two- vs. one-stage production of astaxanthin by the green alga Haematococcus pluvialis. Biotechnol. Bioeng. 2007, 98, 300–305. [Google Scholar] [CrossRef]
- Cunningham, F.X., Jr.; Gantt, E. Elucidation of the pathway to astaxanthin in the flowers of Adonis aestivalis. Plant Cell 2011, 23, 3055–3069. [Google Scholar] [CrossRef] [PubMed]
- Boussiba, S.; Vonshak, A. Astaxanthin accumulation in the green alga Haematococcus pluvialis. Plant Cell Physiol. 1991, 32, 1077–1082. [Google Scholar] [CrossRef]
- Yang, Y.; Seo, J.M.; Nguyen, A.; Pham, T.X.; Park, H.J.; Park, Y.; Kim, B.; Bruno, R.S.; Lee, J. Astaxanthin-rich extract from the green alga Haematococcus pluvialis lowers plasma lipid concentrations and enhances antioxidant defense in apolipoprotein E knockout mice. J. Nutr. 2011, 141, 1611–1617. [Google Scholar] [CrossRef] [PubMed]
- Donoso, A.; González-Durán, J.; Muñoz, A.A.; González, P.A.; Agurto-Muñoz, C. Therapeutic uses of natural astaxanthin: An evidence-based review focused on human clinical trials. Pharmacol. Res. 2021, 166, 105479. [Google Scholar] [CrossRef]
- Kim, E.S.; Baek, Y.; Yoo, H.J.; Lee, J.S.; Lee, H.G. Chitosan-tripolyphosphate nanoparticles prepared by ionic gelation improve the antioxidant activities of astaxanthin in the in vitro and in vivo model. Antioxidants 2022, 11, 479. [Google Scholar] [CrossRef]
- Sayuti, N.H.; Muhammad Nawawi, K.N.; Goon, J.A.; Mokhtar, N.M.; Makpol, S.; Tan, J.K. Preventative and terapeutic efects of ataxanthin on NAFLD. Antioxidants 2023, 12, 1552. [Google Scholar] [CrossRef] [PubMed]
- Mann, V.; Harker, M.; Pecker, I.; Hirschberg, J. Metabolic engineering of astaxanthin production in tobacco flowers. Nat. Biotechnol. 2000, 18, 888–892. [Google Scholar] [CrossRef] [PubMed]
- Ambati, R.R.; Phang, S.M.; Ravi, S.; Aswathanarayana, R.G. Astaxanthin: Sources, extraction, stability, biological activities and its commercial applications—A review. Mar. Drugs 2014, 12, 128–152. [Google Scholar] [CrossRef]
- Kathiresan, S.; Chandrashekar, A.; Ravishankar, G.A.; Sarada, R. Regulation of astaxanthin and its intermediates through cloning and genetic transformation of β-carotene ketolase in Haematococcus pluvialis. J. Biotechnol. 2015, 196–197, 33–41. [Google Scholar] [CrossRef]
- Misawa, N.; Satomi, Y.; Kondo, K.; Yokoyama, A.; Kajiwara, S.; Saito, T.; Ohtani, T.; Miki, W. Structure and functional analysis of a marine bacterial carotenoid biosynthesis gene cluster and astaxanthin biosynthetic pathway proposed at the gene level. J. Bacteriol. 1995, 177, 6575–6584. [Google Scholar] [CrossRef] [PubMed]
- Kajiwara, S.; Kakizono, T.; Saito, T.; Kondo, K.; Ohtani, T.; Nishio, N.; Nagai, S.; Misawa, N. Isolation and functional identification of a novel cDNA for astaxanthin biosynthesis from Haematococcus pluvialis, and astaxanthin synthesis in Escherichia coli. Plant Mol. Biol. 1995, 29, 343–352. [Google Scholar] [CrossRef] [PubMed]
- Lotan, T.; Hirschberg, J. Cloning and expression in Escherichia coli of the gene encoding β-C-4-oxygenase, that converts β-carotene to the ketocarotenoid canthaxanthin in Haematococcus pluvialis. FEBS Lett. 1995, 364, 125–128. [Google Scholar] [CrossRef]
- Galarza, J.I.; Gimpel, J.A.; Rojas, V.; Arredondo-Vega, B.O.; Henríquez, V. Over-accumulation of astaxanthin in Haematococcus pluvialis through chloroplast genetic engineering. Algal Res. 2018, 31, 291–297. [Google Scholar] [CrossRef]
- Cao, T.-J.; Wang, L.-J.; Huang, X.-Q.; Deng, Y.-Y.; Yang, L.-E.; Lu, S. Manipulation of Synechocystis sp. PCC 6803 as a platform for functional identification of genes involved in carotenoid metabolism. Plant Biotechnol. J. 2020, 18, 605–607. [Google Scholar] [CrossRef] [PubMed]
- Diao, J.; Song, X.; Zhang, L.; Cui, J.; Chen, L.; Zhang, W. Tailoring cyanobacteria as a new platform for highly efficient synthesis of astaxanthin. Metab. Eng. 2020, 61, 275–287. [Google Scholar] [CrossRef]
- Hasunuma, T.; Takaki, A.; Matsuda, M.; Kato, Y.; Vavricka, C.J.; Kondo, A. Single-stage astaxanthin production enhances the nonmevalonate pathway and photosynthetic central metabolism in Synechococcus sp. PCC 7002. ACS Synth. Biol 2019, 8, 2701–2709. [Google Scholar] [CrossRef] [PubMed]
- Cao, T.-J.; Huang, X.-Q.; Qu, Y.-Y.; Zhuang, Z.; Deng, Y.-Y.; Lu, S. Cloning and functional characterization of a lycopene β-cyclase from macrophytic red alga Bangia fuscopurpurea. Mar. Drugs 2017, 15, 116. [Google Scholar] [CrossRef]
- Furubayashi, M.; Kubo, A.; Takemura, M.; Otani, Y.; Maoka, T.; Terada, Y.; Yaoi, K.; Ohdan, K.; Misawa, N.; Mitani, Y. Capsanthin production in Escherichia coli by overexpression of capsanthin/capsorubin synthase from Capsicum annuum. J. Agric. Food Chem. 2021, 69, 5076–5085. [Google Scholar] [CrossRef] [PubMed]
- Bongirwar, R.; Shukla, P. Metabolic sink engineering in cyanobacteria: Perspectives and applications. Bioresour. Technol. 2023, 379, 128974. [Google Scholar] [CrossRef] [PubMed]
- Summerfield, T.C.; Crawford, T.S.; Young, R.D.; Chua, J.P.; Macdonald, R.L.; Sherman, L.A.; Eaton-Rye, J.J. Environmental pH affects photoautotrophic growth of Synechocystis sp. PCC 6803 strains carrying mutations in the lumenal proteins of PSII. Plant Cell Physiol. 2013, 54, 859–874. [Google Scholar] [CrossRef]
- Huang, H.-H.; Camsund, D.; Lindblad, P.; Heidorn, T. Design and characterization of molecular tools for a synthetic biology approach towards developing cyanobacterial biotechnology. Nucleic Acids Res. 2010, 38, 2577–2593. [Google Scholar] [CrossRef] [PubMed]
- Koch, M.; Bruckmoser, J.; Scholl, J.; Hauf, W.; Rieger, B.; Forchhammer, K. Maximizing PHB content in Synechocystis sp. PCC 6803: A new metabolic engineering strategy based on the regulator PirC. Microb. Cell Factories 2020, 19, 231. [Google Scholar] [CrossRef]
- Schwarzkopf, M.; Yoo, Y.C.; Hückelhoven, R.; Park, Y.M.; Proels, R.K. Cyanobacterial Phytochrome2 regulates the heterotrophic metabolism and has a function in the heat and high-light stress response. Plant Physiol. 2014, 164, 2157–2166. [Google Scholar] [CrossRef]
- Xiong, W.; Shen, G.; Bryant, D.A. Synechocystis sp. PCC 6803 CruA (sll0147) encodes lycopene cyclase and requires bound chlorophyll a for activity. Photosynth. Res. 2017, 131, 267–280. [Google Scholar] [CrossRef]
- Song, S.; Jin, R.; Chen, Y.; He, S.; Li, K.; Tang, Q.; Wang, Q.; Wang, L.; Kong, M.; Dudareva, N.; et al. The functional evolution of architecturally different plant geranyl diphosphate synthases from geranylgeranyl diphosphate synthase. Plant Cell 2023, 35, 2293–2315. [Google Scholar] [CrossRef]
- Zhou, P.; Ye, L.; Xie, W.; Lv, X.; Yu, H. Highly efficient biosynthesis of astaxanthin in Saccharomyces cerevisiae by integration and tuning of algal crtZ and bkt. Appl. Microbiol. Biotechnol. 2015, 99, 8419–8428. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Trifinopoulos, J.; Nguyen, L.T.; von Haeseler, A.; Minh, B.Q. W-IQ-TREE: A fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 2016, 44, W232–W235. [Google Scholar] [CrossRef]
- Harker, M.; Hirschberg, J. Biosynthesis of ketocarotenoids in transgenic cyanobacteria expressing the algal gene for β-C-4-oxygenase, crtO. FEBS Lett. 1997, 404, 129–134. [Google Scholar] [CrossRef] [PubMed]
- Jeknić, Z.; Morré, J.T.; Jeknić, S.; Jevremović, S.; Subotić, A.; Chen, T.H.H. Cloning and functional characterization of a gene for capsanthin-capsorubin synthase from tiger lily (Lilium lancifolium Thunb. ‘Splendens’). Plant Cell Physiol. 2012, 53, 1899–1912. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Concepcion, M.; Avalos, J.; Bonet, M.L.; Boronat, A.; Gomez-Gomez, L.; Hornero-Mendez, D.; Limon, M.C.; Meléndez-Martínez, A.J.; Olmedilla-Alonso, B.; Palou, A.; et al. A global perspective on carotenoids: Metabolism, biotechnology, and benefits for nutrition and health. Prog. Lipid Res. 2018, 70, 62–93. [Google Scholar] [CrossRef]
- Keeling, P.J. The endosymbiotic origin, diversification and fate of plastids. Philos. Trans. R. Soc. B Biol. Sci. 2010, 365, 729–748. [Google Scholar] [CrossRef] [PubMed]


| SyneCrtR | PaCrtZ | HpCrtZ | HfCrtZ | CtCrtZ | |
|---|---|---|---|---|---|
| SyneCrtR | 100 | ||||
| PaCrtZ | 9.7 | 100 | |||
| HpCrtZ | 12.4 | 38.4 | 100 | ||
| HfCrtZ | 15.9 | 26 | 26.7 | 100 | |
| CtCrtZ | 12.4 | 19.5 | 25.6 | 34.6 | 100 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liang, H.; Chen, H.; Liu, X.; Wang, Z.; Li, P.; Lu, S. Heterologous Production in the Synechocystis Chassis Suggests the Biosynthetic Pathway of Astaxanthin in Cyanobacteria. Antioxidants 2023, 12, 1826. https://doi.org/10.3390/antiox12101826
Liang H, Chen H, Liu X, Wang Z, Li P, Lu S. Heterologous Production in the Synechocystis Chassis Suggests the Biosynthetic Pathway of Astaxanthin in Cyanobacteria. Antioxidants. 2023; 12(10):1826. https://doi.org/10.3390/antiox12101826
Chicago/Turabian StyleLiang, Hanyu, Hongjuan Chen, Xinya Liu, Zihan Wang, Pengfu Li, and Shan Lu. 2023. "Heterologous Production in the Synechocystis Chassis Suggests the Biosynthetic Pathway of Astaxanthin in Cyanobacteria" Antioxidants 12, no. 10: 1826. https://doi.org/10.3390/antiox12101826






