Epithelial Dual Oxidase 2 Shapes the Mucosal Microbiome and Contributes to Inflammatory Susceptibility
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Isolation of Primary Cells
2.3. Isolation and Culture of Intestinal Organoids
2.4. ROS Assay
2.5. Organoid Proliferation Assay
2.6. RNA Isolation and qPCR
2.7. Microbiome Analysis Using 16S Amplicon Sequencing
2.8. Western Blot Analyses
2.9. Histology and Immunostaining
2.10. ELISA
2.11. Data and Code Availability
2.12. Statistics Analysis
3. Results
3.1. Epithelial DUOX2 Functions in Intestinal Homeostasis
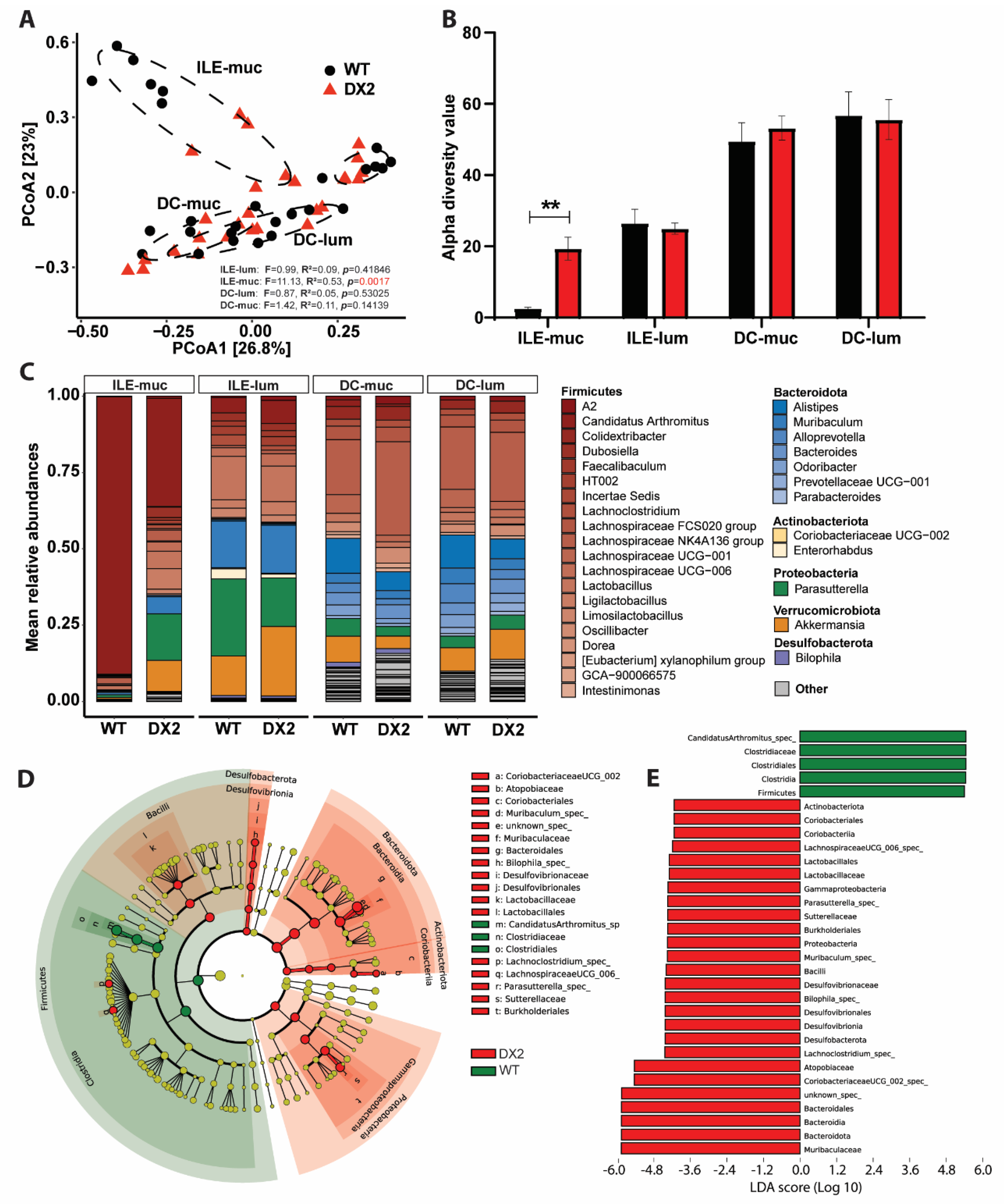
3.2. Deletion of DUOX2 in the Intestinal Epithelium Alters the Mucosal Microbiome
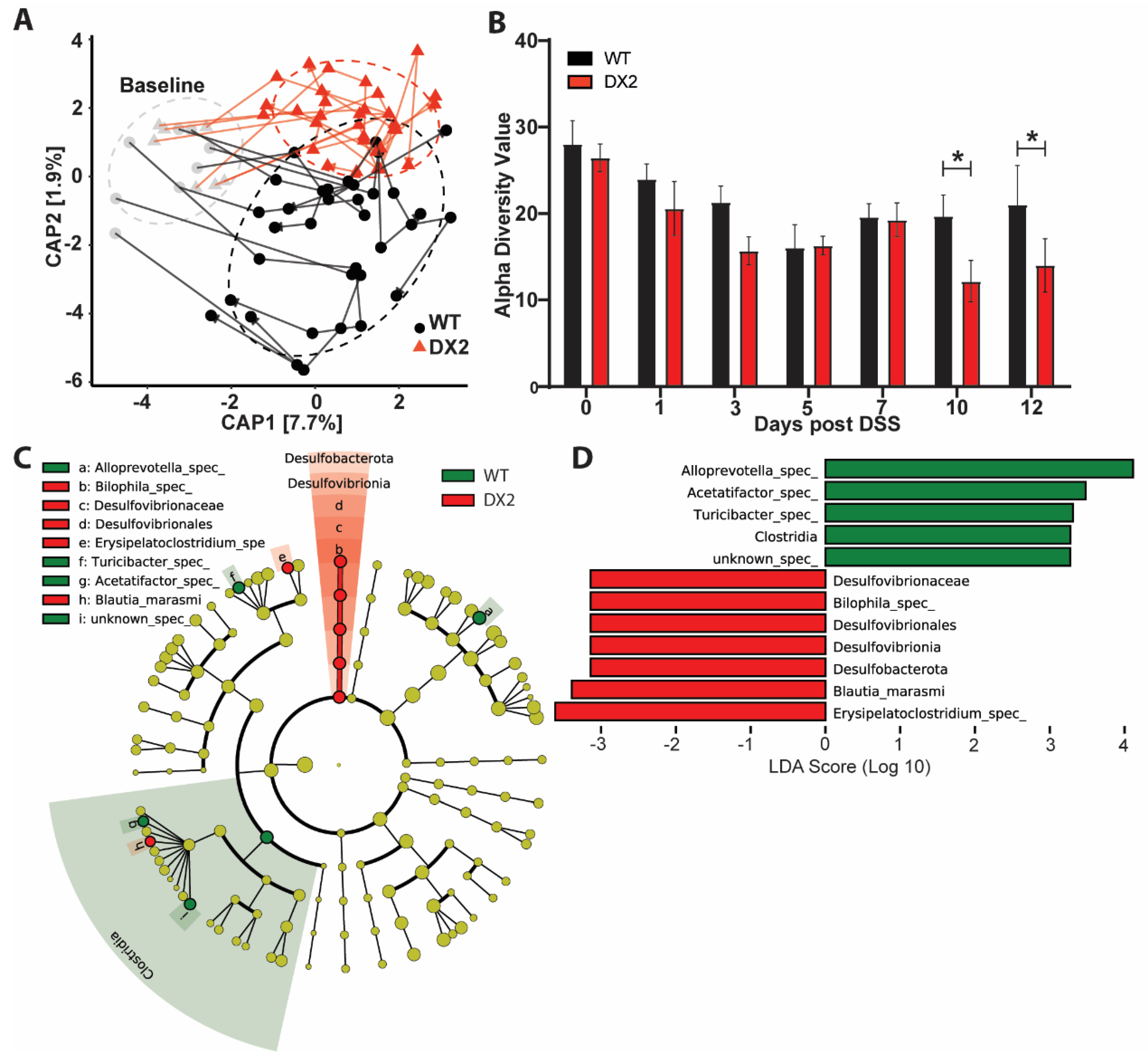
3.3. Duox2∆IEC Mice Are Less Susceptible to DSS Colitis and Develop an Altered Fecal Microbiome
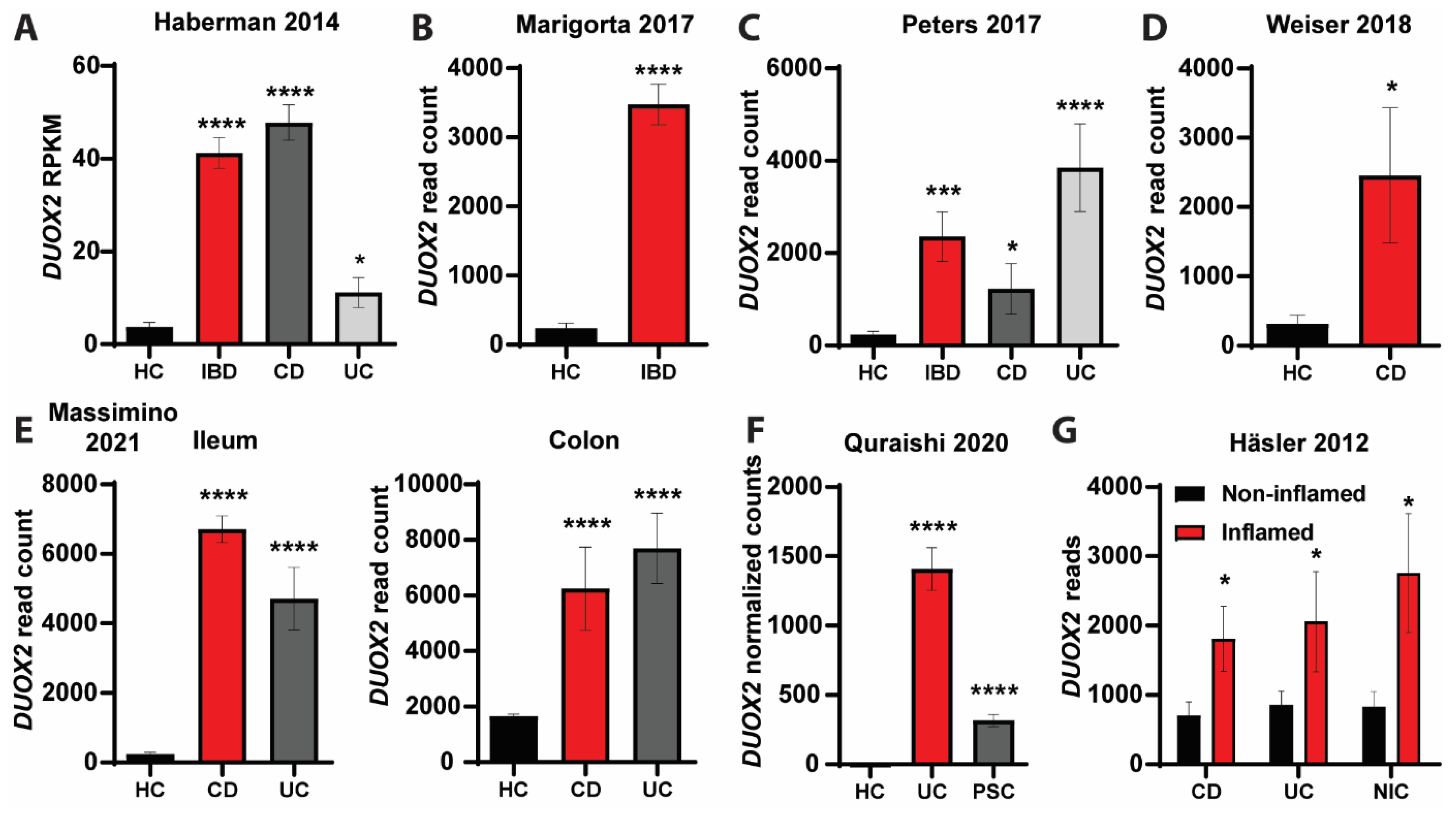
3.4. DUOX2-Microbiome Interactions Are Dysregulated in Patients with Intestinal Inflammation
4. Discussion
4.1. Location Matters—Epithelial DUOX2 Shapes Intestinal Homeostasis
4.2. Reciprocal Interactions between Epithelial DUOX2 and the Intestinal Epithelium Microbiome
4.3. How Does Deletion of Epithelial DUOX2 Protect from Intestinal Inflammation?
4.4. Clinical Relevance of DUOX2
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sommer, F.; Ruhlemann, M.C.; Bang, C.; Hoppner, M.; Rehman, A.; Kaleta, C.; Schmitt-Kopplin, P.; Dempfle, A.; Weidinger, S.; Ellinghaus, E.; et al. Microbiomarkers in Inflammatory Bowel Diseases: Caveats Come with Caviar. Gut 2017, 66, 1734–1738. [Google Scholar] [CrossRef]
- Leto, T.L.; Geiszt, M. Role of Nox Family NADPH Oxidases in Host Defense. Antioxid. Redox Signal 2006, 8, 1549–1561. [Google Scholar] [CrossRef]
- Stenke, E.; Bourke, B.; Knaus, U.G. NAPDH Oxidases in Inflammatory Bowel Disease. Methods Mol. Biol. 2019, 1982, 695–713. [Google Scholar] [CrossRef]
- Aviello, G.; Knaus, U. ROS in Gastrointestinal Inflammation: Rescue Or Sabotage? Br. J. Pharmacol. 2017, 174, 1704–1718. [Google Scholar] [CrossRef]
- Lambeth, J.D. NOX Enzymes and the Biology of Reactive Oxygen. Nat. Rev. Immunol. 2004, 4, 181–189. [Google Scholar] [CrossRef]
- Bokoch, G.M.; Knaus, U.G. NADPH Oxidases: Not Just for Leukocytes Anymore! Trends Biochem. Sci. 2003, 28, 502–508. [Google Scholar] [CrossRef]
- Larsson, E.; Tremaroli, V.; Lee, Y.S.; Koren, O.; Nookaew, I.; Fricker, A.; Nielsen, J.; Ley, R.E.; Bäckhed, F. Analysis of Gut Microbial Regulation of Host Gene Expression along the Length of the Gut and Regulation of Gut Microbial Ecology through MyD88. Gut 2012, 61, 1124–1131. [Google Scholar] [CrossRef]
- Sommer, F.; Nookaew, I.; Sommer, N.; Fogelstrand, P.; Bäckhed, F. Site-Specific Programming of the Host Epithelial Transcriptome by the Gut Microbiota. Genome Biol. 2015, 16, 62. [Google Scholar] [CrossRef]
- Grasberger, H.; Gao, J.; Nagao-Kitamoto, H.; Kitamoto, S.; Zhang, M.; Kamada, N.; Eaton, K.A.; El-Zaatari, M.; Shreiner, A.B.; Merchant, J.L.; et al. Increased Expression of DUOX2 Is an Epithelial Response to Mucosal Dysbiosis Required for Immune Homeostasis in Mouse Intestine. Gastroenterology 2015, 149, 1849–1859. [Google Scholar] [CrossRef]
- Sommer, F.; Bäckhed, F. The Gut Microbiota Engages Different Signaling Pathways to Induce Duox2 Expression in the Ileum and Colon Epithelium. Mucosal Immunol. 2015, 8, 372–379. [Google Scholar] [CrossRef]
- Ogboo, B.C.; Grabovyy, U.V.; Maini, A.; Scouten, S.; van der Vliet, A.; Mattevi, A.; Heppner, D.E. Architecture of the NADPH Oxidase Family of Enzymes. Redox Biol. 2022, 52, 102298. [Google Scholar] [CrossRef]
- Grasberger, H.; El–Zaatari, M.; Dang, D.T.; Merchant, J.L. Dual Oxidases Control Release of Hydrogen Peroxide by the Gastric Epithelium to Prevent Helicobacter Felis Infection and Inflammation in Mice. Gastroenterology 2013, 145, 1045–1054. [Google Scholar] [CrossRef]
- Lipinski, S.; Till, A.; Sina, C.; Arlt, A.; Grasberger, H.; Schreiber, S.; Rosenstiel, P. DUOX2-Derived Reactive Oxygen Species Are Effectors of NOD2-Mediated Antibacterial Responses. J. Cell Sci. 2009, 122, 3522–3530. [Google Scholar] [CrossRef]
- Duan, J.; Matute, J.D.; Unger, L.W.; Hanley, T.; Schnell, A.; Lin, X.; Krupka, N.; Griebel, P.; Lambden, C.; Sit, B.; et al. Endoplasmic Reticulum Stress in the Intestinal Epithelium Initiates Purine Metabolite Synthesis and Promotes Th17 Cell Differentiation in the Gut. Immunity 2023, 56, 1115–1131.e9. [Google Scholar] [CrossRef]
- Hayes, P.; Dhillon, S.; O’Neill, K.; Thoeni, C.; Hui, K.Y.; Elkadri, A.; Guo, C.H.; Kovacic, L.; Aviello, G.; Alvarez, L.A.; et al. Defects in Nicotinamide-Adenine Dinucleotide Phosphate Oxidase Genes NOX1 and DUOX2 in Very Early Onset Inflammatory Bowel Disease. Cell Mol. Gastroenterol. Hepatol. 2015, 1, 489–502. [Google Scholar] [CrossRef]
- Grasberger, H.; Magis, A.T.; Sheng, E.; Conomos, M.P.; Zhang, M.; Garzotto, L.S.; Hou, G.; Bishu, S.; Nagao-Kitamoto, H.; El-Zaatari, M.; et al. DUOX2 Variants Associate with Preclinical Disturbances in Microbiota-Immune Homeostasis and Increased Inflammatory Bowel Disease Risk. J. Clin. Investig. 2021, 131. [Google Scholar] [CrossRef]
- Kyodo, R.; Takeuchi, I.; Narumi, S.; Shimizu, H.; Hata, K.; Yoshioka, T.; Tanase-Nakao, K.; Shimizu, T.; Arai, K. Novel Biallelic Mutations in the DUOX2 Gene Underlying Very Early-Onset Inflammatory Bowel Disease: A Case Report. Clin. Immunol. 2022, 238, 109015. [Google Scholar] [CrossRef]
- Hinrichsen, F.; Hamm, J.; Westermann, M.; Schröder, L.; Shima, K.; Mishra, N.; Walker, A.; Sommer, N.; Klischies, K.; Prasse, D.; et al. Microbial Regulation of Hexokinase 2 Links Mitochondrial Metabolism and Cell Death in Colitis. Cell Metab. 2021, 33, 2355–2366.e8. [Google Scholar] [CrossRef]
- Pan, W.-H.; Sommer, F.; Falk-Paulsen, M.; Ulas, T.; Best, P.; Fazio, A.; Kachroo, P.; Luzius, A.; Jentzsch, M.; Rehman, A.; et al. Exposure to the Gut Microbiota Drives Distinct Methylome and Transcriptome Changes in Intestinal Epithelial Cells during Postnatal Development. Genome Med. 2018, 10, 27. [Google Scholar] [CrossRef]
- Sato, T.; Vries, R.G.; Snippert, H.J.; van de Wetering, M.; Barker, N.; Stange, D.E.; van Es, J.H.; Abo, A.; Kujala, P.; Peters, P.J.; et al. Single Lgr5 Stem Cells Build Crypt-Villus Structures in Vitro without a Mesenchymal Niche. Nature 2009, 459, 262–265. [Google Scholar] [CrossRef]
- Sato, T.; Stange, D.E.; Ferrante, M.; Vries, R.G.J.; van Es, J.H.; van den Brink, S.; van Houdt, W.J.; Pronk, A.; van Gorp, J.; Siersema, P.D.; et al. Long-Term Expansion of Epithelial Organoids From Human Colon, Adenoma, Adenocarcinoma, and Barrett’s Epithelium. Gastroenterology 2011, 141, 1762–1772. [Google Scholar] [CrossRef] [PubMed]
- Varga, A.; Makszin, L.; Bufa, A.; Sipos, D.; Kása, P.; Pál, S.; Rosenstiel, P.; Sommer, F.; Kocsis, B.; Péterfi, Z. Efficacy of Lyophilised Bacteria-Rich Faecal Sediment and Supernatant with Reduced Bacterial Count for Treating Patients with Clostridioides Difficile Infection—A Novel Method for Capsule Faecal Microbiota Transfer. Front. Cell Infect. Microbiol. 2023, 13, 1041384. [Google Scholar] [CrossRef] [PubMed]
- Sommer, F.; Adam, N.; Johansson, M.E.V.; Xia, L.; Hansson, G.C.; Bäckhed, F. Altered Mucus Glycosylation in Core 1 O-Glycan-Deficient Mice Affects Microbiota Composition and Intestinal Architecture. PLoS ONE 2014, 9, e85254. [Google Scholar] [CrossRef]
- Fadrosh, D.W.; Ma, B.; Gajer, P.; Sengamalay, N.; Ott, S.; Brotman, R.M.; Ravel, J. An Improved Dual-Indexing Approach for Multiplexed 16S RRNA Gene Sequencing on the Illumina MiSeq Platform. Microbiome 2014, 2, 6. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Huntley, J.; Fierer, N.; Owens, S.M.; Betley, J.; Fraser, L.; Bauer, M.; et al. Ultra-High-Throughput Microbial Community Analysis on the Illumina HiSeq and MiSeq Platforms. ISME J. 2012, 6, 1621–1624. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-Resolution Sample Inference from Illumina Amplicon Data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA Ribosomal RNA Gene Database Project: Improved Data Processing and Web-Based Tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S. Phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef]
- Dixon, P. VEGAN, a Package of R Functions for Community Ecology. J. Veg. Sci. 2003, 14, 927–930. [Google Scholar] [CrossRef]
- Mallick, H.; Rahnavard, A.; McIver, L.J.; Ma, S.; Zhang, Y.; Nguyen, L.H.; Tickle, T.L.; Weingart, G.; Ren, B.; Schwager, E.H.; et al. Multivariable Association Discovery in Population-Scale Meta-Omics Studies. PLoS Comput. Biol. 2021, 17, e1009442. [Google Scholar] [CrossRef]
- Kumar, M.S.; Slud, E.V.; Okrah, K.; Hicks, S.C.; Hannenhalli, S.; Corrada Bravo, H. Analysis and Correction of Compositional Bias in Sparse Sequencing Count Data. BMC Genom. 2018, 19, 799. [Google Scholar] [CrossRef] [PubMed]
- Lloyd-Price, J.; Arze, C.; Ananthakrishnan, A.N.; Schirmer, M.; Avila-Pacheco, J.; Poon, T.W.; Andrews, E.; Ajami, N.J.; Bonham, K.S.; Brislawn, C.J.; et al. Multi-Omics of the Gut Microbial Ecosystem in Inflammatory Bowel Diseases. Nature 2019, 569, 655–662. [Google Scholar] [CrossRef] [PubMed]
- Siegmund, B.; Lehr, H.-A.; Fantuzzi, G.; Dinarello, C.A. IL-1β-Converting Enzyme (Caspase-1) in Intestinal Inflammation. Proc. Natl. Acad. Sci. USA 2001, 98, 13249–13254. [Google Scholar] [CrossRef] [PubMed]
- Czarnewski, P.; Parigi, S.M.; Sorini, C.; Diaz, O.E.; Das, S.; Gagliani, N.; Villablanca, E.J. Conserved Transcriptomic Profile between Mouse and Human Colitis Allows Unsupervised Patient Stratification. Nat. Commun. 2019, 10, 2892. [Google Scholar] [CrossRef] [PubMed]
- Häsler, R.; Feng, Z.; Bäckdahl, L.; Spehlmann, M.E.; Franke, A.; Teschendorff, A.; Rakyan, V.K.; Down, T.A.; Wilson, G.A.; Feber, A.; et al. A Functional Methylome Map of Ulcerative Colitis. Genome Res. 2012, 22, 2130–2137. [Google Scholar] [CrossRef]
- Quraishi, M.N.; Acharjee, A.; Beggs, A.D.; Horniblow, R.; Tselepis, C.; Gkoutos, G.; Ghosh, S.; Rossiter, A.E.; Loman, N.; van Schaik, W.; et al. A Pilot Integrative Analysis of Colonic Gene Expression, Gut Microbiota, and Immune Infiltration in Primary Sclerosing Cholangitis-Inflammatory Bowel Disease: Association of Disease With Bile Acid Pathways. J. Crohns Colitis 2020, 14, 935–947. [Google Scholar] [CrossRef]
- Massimino, L.; Lamparelli, L.A.; Houshyar, Y.; D’Alessio, S.; Peyrin-Biroulet, L.; Vetrano, S.; Danese, S.; Ungaro, F. The Inflammatory Bowel Disease Transcriptome and Metatranscriptome Meta-Analysis (IBD TaMMA) Framework. Nat. Comput. Sci. 2021, 1, 511–515. [Google Scholar] [CrossRef]
- Weiser, M.; Simon, J.M.; Kochar, B.; Tovar, A.; Israel, J.W.; Robinson, A.; Gipson, G.R.; Schaner, M.S.; Herfarth, H.H.; Sartor, R.B.; et al. Molecular Classification of Crohn’s Disease Reveals Two Clinically Relevant Subtypes. Gut 2018, 67, 36–42. [Google Scholar] [CrossRef]
- Peters, L.A.; Perrigoue, J.; Mortha, A.; Iuga, A.; Song, W.; Neiman, E.M.; Llewellyn, S.R.; Di Narzo, A.; Kidd, B.A.; Telesco, S.E.; et al. A Functional Genomics Predictive Network Model Identifies Regulators of Inflammatory Bowel Disease. Nat. Genet. 2017, 49, 1437–1449. [Google Scholar] [CrossRef]
- Marigorta, U.M.; Denson, L.A.; Hyams, J.S.; Mondal, K.; Prince, J.; Walters, T.D.; Griffiths, A.; Noe, J.D.; Crandall, W.V.; Rosh, J.R.; et al. Transcriptional Risk Scores Link GWAS to EQTLs and Predict Complications in Crohn’s Disease. Nat. Genet. 2017, 49, 1517–1521. [Google Scholar] [CrossRef]
- Haberman, Y.; Tickle, T.L.; Dexheimer, P.J.; Kim, M.-O.; Tang, D.; Karns, R.; Baldassano, R.N.; Noe, J.D.; Rosh, J.; Markowitz, J.; et al. Pediatric Crohn Disease Patients Exhibit Specific Ileal Transcriptome and Microbiome Signature. J. Clin. Investig. 2014, 124, 3617–3633. [Google Scholar] [CrossRef] [PubMed]
- The Integrative Human Microbiome Project. Nature 2019, 569, 641–648. [CrossRef] [PubMed]
- Burgueño, J.F.; Fritsch, J.; González, E.E.; Landau, K.S.; Santander, A.M.; Fernández, I.; Hazime, H.; Davies, J.M.; Santaolalla, R.; Phillips, M.C.; et al. Epithelial TLR4 Signaling Activates DUOX2 to Induce Microbiota-Driven Tumorigenesis. Gastroenterology 2021, 160, 797–808.e6. [Google Scholar] [CrossRef] [PubMed]
- Grasberger, H.; De Deken, X.; Mayo, O.B.; Raad, H.; Weiss, M.; Liao, X.-H.; Refetoff, S. Mice Deficient in Dual Oxidase Maturation Factors Are Severely Hypothyroid. Mol. Endocrinol. 2012, 26, 481–492. [Google Scholar] [CrossRef]
- Hazime, H.; Ducasa, G.M.; Santander, A.M.; Brito, N.; González, E.E.; Ban, Y.; Kaunitz, J.; Akiba, Y.; Fernández, I.; Burgueño, J.F.; et al. Intestinal Epithelial Inactivity of Dual Oxidase 2 Results in Microbiome-Mediated Metabolic Syndrome. Cell Mol. Gastroenterol. Hepatol. 2023, 16, 557–572. [Google Scholar] [CrossRef]
- Juan, C.A.; Pérez de la Lastra, J.M.; Plou, F.J.; Pérez-Lebeña, E. The Chemistry of Reactive Oxygen Species (ROS) Revisited: Outlining Their Role in Biological Macromolecules (DNA, Lipids and Proteins) and Induced Pathologies. Int. J. Mol. Sci. 2021, 22, 4642. [Google Scholar] [CrossRef]
- Schnupf, P.; Gaboriau-Routhiau, V.; Gros, M.; Friedman, R.; Moya-Nilges, M.; Nigro, G.; Cerf-Bensussan, N.; Sansonetti, P.J. Growth and Host Interaction of Mouse Segmented Filamentous Bacteria in Vitro. Nature 2015, 520, 99–103. [Google Scholar] [CrossRef]
- Kuwahara, T.; Ogura, Y.; Oshima, K.; Kurokawa, K.; Ooka, T.; Hirakawa, H.; Itoh, T.; Nakayama-Imaohji, H.; Ichimura, M.; Itoh, K.; et al. The Lifestyle of the Segmented Filamentous Bacterium: A Non-Culturable Gut-Associated Immunostimulating Microbe Inferred by Whole-Genome Sequencing. DNA Res. 2011, 18, 291–303. [Google Scholar] [CrossRef]
- He, G.; Shankar, R.A.; Chzhan, M.; Samouilov, A.; Kuppusamy, P.; Zweier, J.L. Noninvasive Measurement of Anatomic Structure and Intraluminal Oxygenation in the Gastrointestinal Tract of Living Mice with Spatial and Spectral EPR Imaging. Proc. Natl. Acad. Sci. USA 1999, 96, 4586–4591. [Google Scholar] [CrossRef]
- Loh, G.; Blaut, M. Role of Commensal Gut Bacteria in Inflammatory Bowel Diseases. Gut Microbes 2012, 3, 544–555. [Google Scholar] [CrossRef]
- Jin, S.; Zhao, D.; Cai, C.; Song, D.; Shen, J.; Xu, A.; Qiao, Y.; Ran, Z.; Zheng, Q. Low-Dose Penicillin Exposure in Early Life Decreases Th17 and the Susceptibility to DSS Colitis in Mice through Gut Microbiota Modification. Sci. Rep. 2017, 7, 43662. [Google Scholar] [CrossRef] [PubMed]
- Iljazovic, A.; Roy, U.; Gálvez, E.J.C.; Lesker, T.R.; Zhao, B.; Gronow, A.; Amend, L.; Will, S.E.; Hofmann, J.D.; Pils, M.C.; et al. Perturbation of the Gut Microbiome by Prevotella Spp. Enhances Host Susceptibility to Mucosal Inflammation. Mucosal Immunol. 2021, 14, 113–124. [Google Scholar] [CrossRef] [PubMed]
- Khattab, M.S.A.; El Tawab, A.M.A.; Fouad, M.T. Isolation and Characterization of Anaerobic Bacteria from Frozen Rumen Liquid and Its Potential Characterizations. Int. J. Dairy Sci. 2016, 12, 47–51. [Google Scholar] [CrossRef]
- Kalyana Chakravarthy, S.; Jayasudha, R.; Sai Prashanthi, G.; Ali, M.H.; Sharma, S.; Tyagi, M.; Shivaji, S. Dysbiosis in the Gut Bacterial Microbiome of Patients with Uveitis, an Inflammatory Disease of the Eye. Indian. J. Microbiol. 2018, 58, 457–469. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Wang, W.; Zhou, R.; Ng, S.C.; Li, J.; Huang, M.; Zhou, F.; Wang, X.; Shen, B.A.; Kamm, M.; et al. Characteristics of Fecal and Mucosa-Associated Microbiota in Chinese Patients With Inflammatory Bowel Disease. Medicine 2014, 93, e51. [Google Scholar] [CrossRef] [PubMed]
- Srinivas, U.S.; Tan, B.W.Q.; Vellayappan, B.A.; Jeyasekharan, A.D. ROS and the DNA Damage Response in Cancer. Redox Biol. 2019, 25, 101084. [Google Scholar] [CrossRef]
- Williams, R.; Yao, H.; Peng, F.; Yang, Y.; Bethel-Brown, C.; Buch, S. Cooperative Induction of CXCL10 Involves NADPH Oxidase: Implications for HIV Dementia. Glia 2010, 58, 611–621. [Google Scholar] [CrossRef]
- You, N.; Li, J.; Huang, X.; Wu, K.; Tang, Y.; Wang, L.; Li, H.; Mi, N.; Zheng, L. COMMD7 Activates CXCL10 Production by Regulating NF-ΚB and the Production of Reactive Oxygen Species. Mol. Med. Rep. 2018, 17, 6784–6788. [Google Scholar] [CrossRef]
- Mancini, N.L.; Rajeev, S.; Jayme, T.S.; Wang, A.; Keita, Å.V.; Workentine, M.L.; Hamed, S.; Söderholm, J.D.; Lopes, F.; Shutt, T.E.; et al. Crohn’s Disease Pathobiont Adherent-Invasive E Coli Disrupts Epithelial Mitochondrial Networks with Implications for Gut Permeability. Cell Mol. Gastroenterol. Hepatol. 2021, 11, 551–571. [Google Scholar] [CrossRef]
- Feng, Q.; Liang, S.; Jia, H.; Stadlmayr, A.; Tang, L.; Lan, Z.; Zhang, D.; Xia, H.; Xu, X.; Jie, Z.; et al. Gut Microbiome Development along the Colorectal Adenoma–Carcinoma Sequence. Nat. Commun. 2015, 6, 6528. [Google Scholar] [CrossRef]
- Li, J.; Sung, C.Y.J.; Lee, N.; Ni, Y.; Pihlajamäki, J.; Panagiotou, G.; El-Nezami, H. Probiotics Modulated Gut Microbiota Suppresses Hepatocellular Carcinoma Growth in Mice. Proc. Natl. Acad. Sci. USA 2016, 113, E1306–E1315. [Google Scholar] [CrossRef]
- Slingerland, A.E.; Schwabkey, Z.; Wiesnoski, D.H.; Jenq, R.R. Clinical Evidence for the Microbiome in Inflammatory Diseases. Front. Immunol. 2017, 8, 400. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Xu, X.; Fu, D.; Gu, Y.; Fan, R.; Yi, H.; He, X.; Wang, C.; Ouyang, B.; Zhao, P.; et al. Butyrate-Producing Eubacterium Rectale Suppresses Lymphomagenesis by Alleviating the TNF-Induced TLR4/MyD88/NF-ΚB Axis. Cell Host Microbe 2022, 30, 1139–1150.e7. [Google Scholar] [CrossRef] [PubMed]
- Faden, H. The Role of Faecalibacterium, Roseburia, and Butyrate in Inflammatory Bowel Disease. Dig. Dis. 2022, 40, 793–795. [Google Scholar] [CrossRef] [PubMed]
- Weber-Stiehl, S.; Järke, L.; Castrillón-Betancur, J.C.; Gilbert, F.; Sommer, F. Mitochondrial Function and Microbial Metabolites as Central Regulators of Intestinal Immune Responses and Cancer. Front. Microbiol. 2022, 13, 919424. [Google Scholar] [CrossRef]
- Massironi, S.; Viganò, C.; Palermo, A.; Pirola, L.; Mulinacci, G.; Allocca, M.; Peyrin-Biroulet, L.; Danese, S. Inflammation and Malnutrition in Inflammatory Bowel Disease. Lancet Gastroenterol. Hepatol. 2023, 8, 579–590. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Castrillón-Betancur, J.C.; López-Agudelo, V.A.; Sommer, N.; Cleeves, S.; Bernardes, J.P.; Weber-Stiehl, S.; Rosenstiel, P.; Sommer, F. Epithelial Dual Oxidase 2 Shapes the Mucosal Microbiome and Contributes to Inflammatory Susceptibility. Antioxidants 2023, 12, 1889. https://doi.org/10.3390/antiox12101889
Castrillón-Betancur JC, López-Agudelo VA, Sommer N, Cleeves S, Bernardes JP, Weber-Stiehl S, Rosenstiel P, Sommer F. Epithelial Dual Oxidase 2 Shapes the Mucosal Microbiome and Contributes to Inflammatory Susceptibility. Antioxidants. 2023; 12(10):1889. https://doi.org/10.3390/antiox12101889
Chicago/Turabian StyleCastrillón-Betancur, Juan Camilo, Víctor Alonso López-Agudelo, Nina Sommer, Sven Cleeves, Joana Pimenta Bernardes, Saskia Weber-Stiehl, Philip Rosenstiel, and Felix Sommer. 2023. "Epithelial Dual Oxidase 2 Shapes the Mucosal Microbiome and Contributes to Inflammatory Susceptibility" Antioxidants 12, no. 10: 1889. https://doi.org/10.3390/antiox12101889




