Glycolysis Aids in Human Lens Epithelial Cells’ Adaptation to Hypoxia
Abstract
:1. Introduction
2. Materials and Methods
2.1. Human Lens Epithelial (HLE) Cell Cultures and Treatment
2.2. Light Microscopy
2.3. Cell Counting Kit-8 (CCK8) Assay
2.4. Liquid Chromatography-Tandem Mass Spectrometry (LC-MS/MS) Proteomics
2.5. Western Blotting Analysis
2.6. Quantitative Real-Time Polymerase Chain Reaction Analysis
2.7. Immunofluorescence
2.8. Reactive Oxygen Species (ROS) Measurement
2.9. Determination of Apoptosis
2.10. Statistical Analysis
3. Results
3.1. Hypoxia Treatment Does Not Alter the Cell Morphology and Viability of HLE Cells
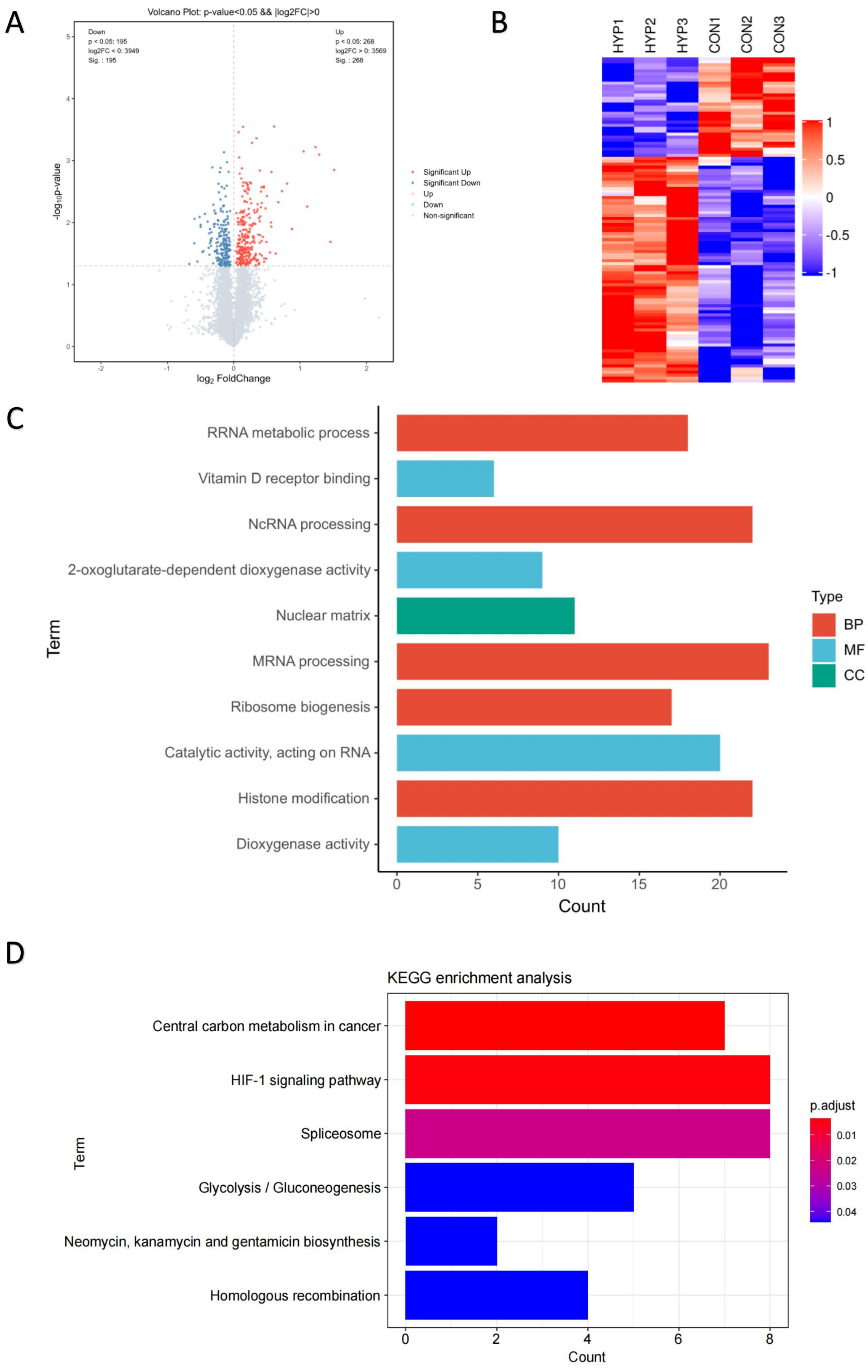
3.2. Proteomic Profiles of HLE Cells under Hypoxia and Normoxia Conditions
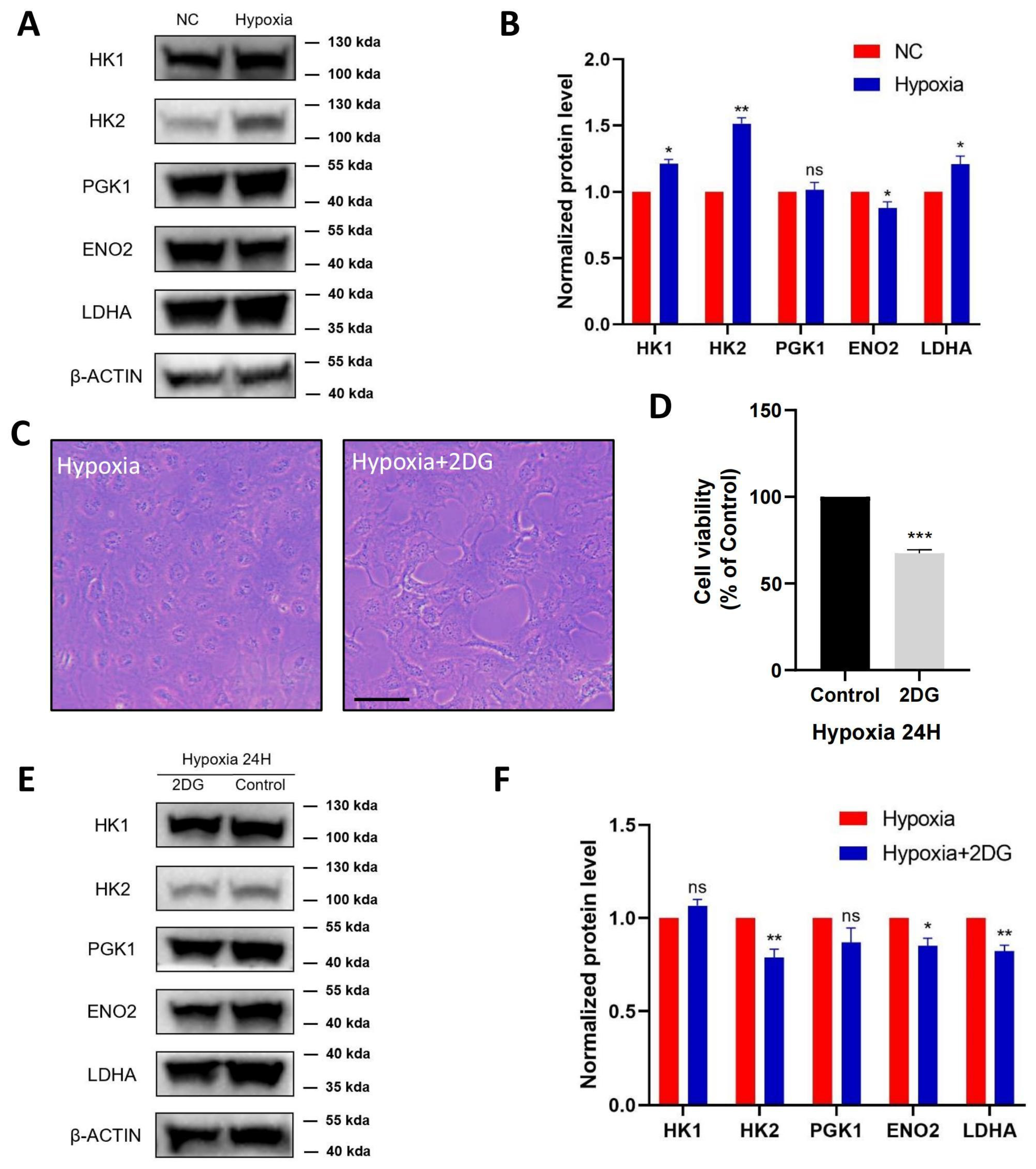
3.3. Validation of Glycolysis Pathway
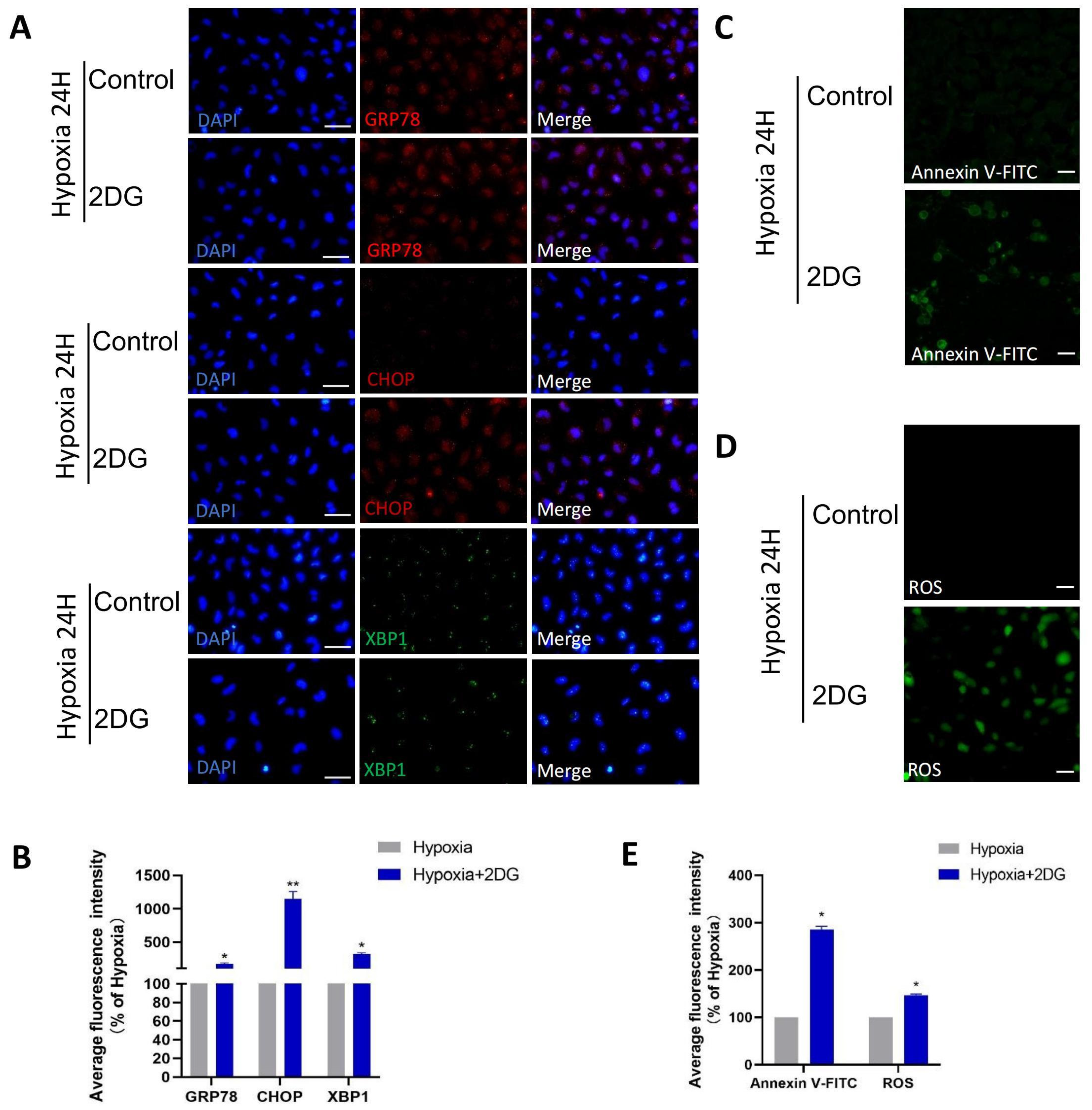
3.4. Inhibition of Glycolysis Induced ROS, ER Stress, and Apoptosis
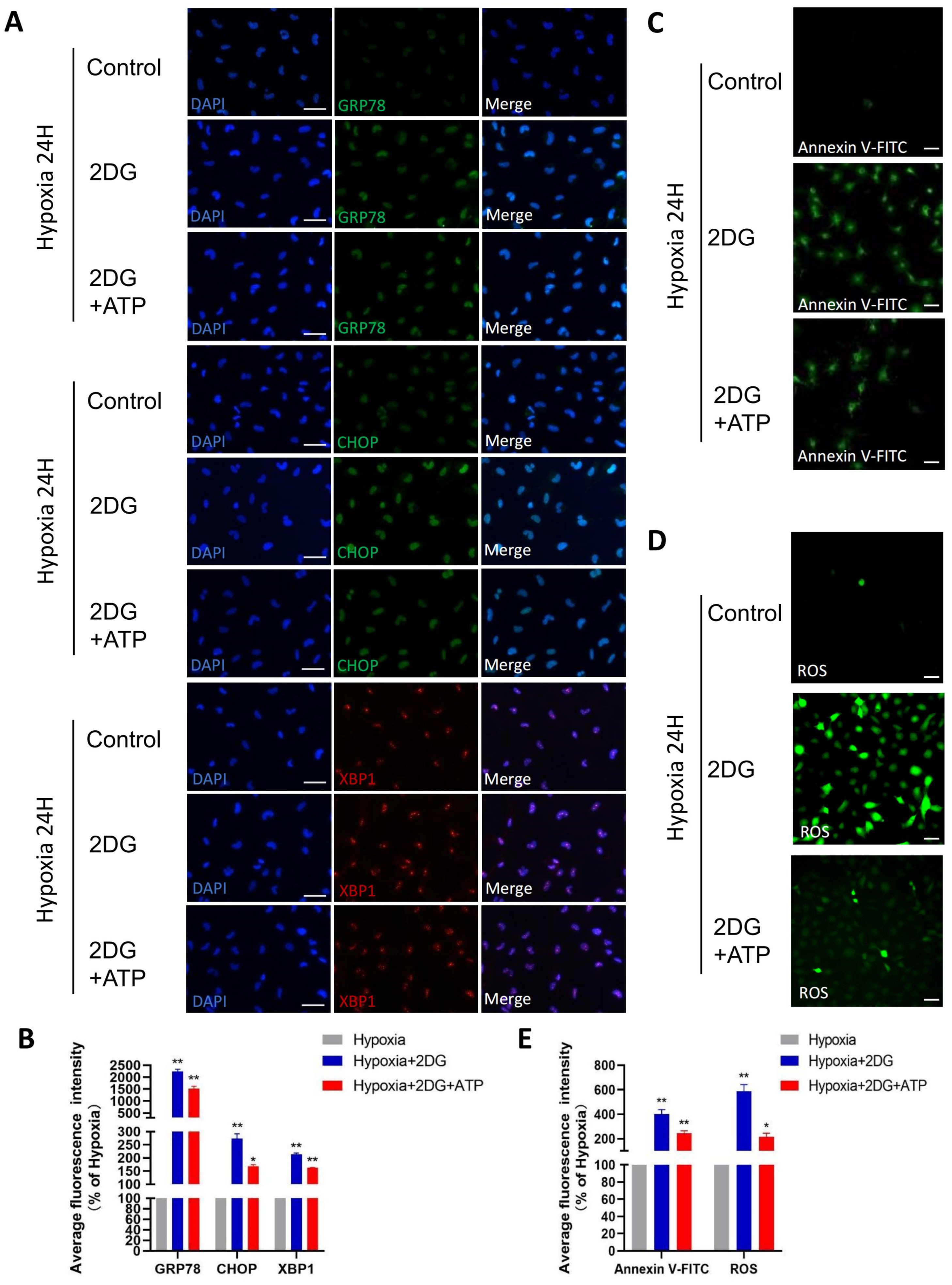
3.5. Replenishing ATP Did Not Rescue Damage Caused by Glycolysis Inhibition in HLE Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Beebe, D.C. Maintaining transparency: A review of the developmental physiology and pathophysiology of two avascular tissues. Semin. Cell Dev. Biol. 2008, 19, 125–133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nahomi, R.B.; Nagaraj, R.H. The role of HIF-1α in the TGF-β2-mediated epithelial-to-mesenchymal transition of human lens epithelial cells. J. Cell. Biochem. 2018, 119, 6814–6827. [Google Scholar] [CrossRef] [PubMed]
- McNulty, R.; Wang, H.; Mathias, R.T.; Ortwerth, B.J.; Truscott, R.J.; Bassnett, S. Regulation of tissue oxygen levels in the mammalian lens. J. Physiol. 2004, 559 Pt 3, 883–898. [Google Scholar] [CrossRef] [PubMed]
- Beebe, D.C.; Shui, Y.B.; Siegfried, C.J.; Holekamp, N.M.; Bai, F. Preserve the (intraocular) environment: The importance of maintaining normal oxygen gradients in the eye. Jpn. J. Ophthalmol. 2014, 58, 225–231. [Google Scholar] [CrossRef]
- Andley, U.P. The Lens Epithelium: Focus on the Expression and Function of the Alpha-Crystallin Chaperones. Int. J. Biochem. Cell Biol. 2008, 40, 317–323. [Google Scholar] [CrossRef] [Green Version]
- Luo, Z.; Tian, M.; Yang, G.; Tan, Q.; Chen, Y.; Li, G.; Zhang, Q.; Li, Y.; Wan, P.; Wu, J. Hypoxia signaling in human health and diseases: Implications and prospects for therapeutics. Signal Transduct. Target. Ther. 2022, 7, 218. [Google Scholar] [CrossRef]
- DeBerardinis, R.J.; Thompson, C.B. Cellular metabolism and disease: What do metabolic outliers teach us? Cell 2012, 148, 1132–1144. [Google Scholar] [CrossRef] [Green Version]
- Kell, D.B. Systems biology, metabolic modelling and metabolomics in drug discovery and development. Drug Discov. Today 2006, 11, 1085–1092. [Google Scholar] [CrossRef] [Green Version]
- Yu, L.; Chen, X.; Sun, X.; Wang, L.; Chen, S. The Glycolytic Switch in Tumors: How Many Players Are Involved? J. Cancer 2017, 8, 3430–3440. [Google Scholar] [CrossRef]
- Bell, S.M.; Burgess, T.; Lee, J.; Blackburn, D.J.; Allen, S.P.; Mortiboys, H. Peripheral Glycolysis in Neurodegenerative Diseases. Int. J. Mol. Sci. 2020, 21, 8924. [Google Scholar] [CrossRef]
- Guo, X.; Li, H.; Xu, H.; Woo, S.; Dong, H.; Lu, F.; Lange, A.J.; Wu, C. Glycolysis in the control of blood glucose homeostasis. Acta Pharm. Sin. B 2012, 2, 358–367. [Google Scholar] [CrossRef] [Green Version]
- Fuller, G.G.; Kim, J.K. Compartmentalization and metabolic regulation of glycolysis. J. Cell Sci. 2021, 134, jcs258469. [Google Scholar] [CrossRef] [PubMed]
- Winkler, B.S.; Riley, M.V. Relative contributions of epithelial cells and fibers to rabbit lens ATP content and glycolysis. Investig. Opthalmology Vis. Sci. 1991, 32, 2593–2598. [Google Scholar]
- Ebert, B.L.; Firth, J.D.; Ratcliffe, P.J. Hypoxia and mitochondrial inhibitors regulate expression of glucose transporter-1 via distinct cis-acting sequences. J. Biol. Chem. 1995, 270, 29083–29089. [Google Scholar] [CrossRef] [Green Version]
- Iyer, N.V.; Kotch, L.E.; Agani, F.; Leung, S.W.; Laughner, E.; Wenger, R.H.; Gassmann, M.; Gearhart, J.D.; Lawler, A.M.; Yu, A.Y.; et al. Cellular and developmental control of O2 homeostasis by hypoxia-inducible factor 1α. Genes Dev. 1998, 12, 149–162. [Google Scholar] [CrossRef] [Green Version]
- Semenza, G.L.; Roth, P.H.; Fang, H.M.; Wang, G.L. Transcriptional regulation of genes encoding glycolytic enzymes by hypoxia-inducible factor 1. J. Biol. Chem. 1994, 269, 23757–23763. [Google Scholar] [CrossRef]
- Seagroves, T.N.; Ryan, H.E.; Lu, H.; Wouters, B.G.; Knapp, M.; Thibault, P.; Laderoute, K.; Johnson, R.S. Transcription Factor HIF-1 Is a Necessary Mediator of the Pasteur Effect in Mammalian Cells. Mol. Cell. Biol. 2001, 21, 3436–3444. [Google Scholar] [CrossRef] [Green Version]
- Kierans, S.J.; Taylor, C.T. Regulation of glycolysis by the hypoxia-inducible factor (HIF): Implications for cellular physiology. J. Physiol. 2021, 599, 23–37. [Google Scholar] [CrossRef]
- Natua, S.; Ashok, C.; Shukla, S. Hypoxia-induced alternative splicing in human diseases: The pledge, the turn, and the prestige. Cell. Mol. Life Sci. 2021, 78, 2729–2747. [Google Scholar] [CrossRef]
- Patel, A.A.; Steitz, J.A. Splicing double: Insights from the second spliceosome. Nat. Rev. Mol. Cell Biol. 2003, 4, 960–970. [Google Scholar] [CrossRef]
- Kanopka, A. Cell survival: Interplay between hypoxia and pre-mRNA splicing. Exp. Cell Res. 2017, 356, 187–191. [Google Scholar] [CrossRef] [PubMed]
- Nakayama, K.; Kataoka, N. Regulation of Gene Expression under Hypoxia Conditions. Int. J. Mol. Sci. 2019, 20, 3278. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Farina, A.R.; Cappabianca, L.; Sebastiano, M.; Zelli, V.; Guadagni, S.; Mackay, A.R. Hypoxia-induced alternative splicing: The 11th Hallmark of Cancer. J. Exp. Clin. Cancer Res. 2020, 39, 110. [Google Scholar] [CrossRef] [PubMed]
- Frost, J.; Frost, M.; Batie, M.; Jiang, H.; Rocha, S. Roles of HIF and 2-Oxoglutarate-Dependent Dioxygenases in Controlling Gene Expression in Hypoxia. Cancers 2021, 13, 350. [Google Scholar] [CrossRef] [PubMed]
- Webb, J.D.; Coleman, M.L.; Pugh, C.W. Hypoxia, hypoxia-inducible factors (HIF), HIF hydroxylases and oxygen sensing. Cell. Mol. Life Sci. 2009, 66, 3539–3554. [Google Scholar] [CrossRef]
- Kim, J.; Lee, H.; Yi, S.J.; Kim, K. Gene regulation by histone-modifying enzymes under hypoxia conditions: A focus on histone methylation and acetylation. Exp. Mol. Med. 2022, 54, 878–889. [Google Scholar] [CrossRef] [PubMed]
- Bindra, R.S.; Crosby, M.E.; Glazer, P.M. Regulation of DNA repair in hypoxia cancer cells. Cancer Metastasis Rev. 2007, 26, 249–260. [Google Scholar] [CrossRef] [PubMed]
- Ron, D.; Walter, P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat. Rev. Mol. Cell Biol. 2007, 8, 519–529. [Google Scholar] [CrossRef]
- Sosa, V.; Moliné, T.; Somoza, R.; Paciucci, R.; Kondoh, H.; LLeonart, M.E. Oxidative stress and cancer: An overview. Ageing Res. Rev. 2013, 12, 376–390. [Google Scholar] [CrossRef]
- Cadenas, S. ROS and redox signaling in myocardial ischemia-reperfusion injury and cardioprotection. Free Radic. Biol. Med. 2018, 117, 76–89. [Google Scholar] [CrossRef]
- Green, D.R.; Galluzzi, L.; Kroemer, G. Cell biology. Metabolic control of cell death. Science 2014, 345, 1250256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Galluzzi, L.; Vitale, I.; Aaronson, S.A.; Abrams, J.M.; Adam, D.; Agostinis, P.; Alnemri, E.S.; Altucci, L.; Amelio, I.; Andrews, D.W.; et al. Molecular mechanisms of cell death: Recommendations of the Nomenclature Committee on Cell Death 2018. Cell Death Differ. 2018, 25, 486–541. [Google Scholar] [CrossRef]
- Wolf, B.B.; Green, D.R. Suicidal tendencies: Apoptotic cell death by caspase family proteinases. J. Biol. Chem. 1999, 274, 20049–20052. [Google Scholar] [CrossRef] [Green Version]
- Elanchezhian, R.; Palsamy, P.; Madson, C.J.; Mulhern, M.L.; Lynch, D.W.; Troia, A.M.; Usukura, J.; Shinohara, T. Low glucose under hypoxic conditions induces unfolded protein response and produces reactive oxygen species in lens epithelial cells. Cell Death Dis. 2012, 3, e301. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kinoshita, J.H. Pathways of Glucose Metabolism in the Lens. Investig. Ophthalmol. 1965, 4, 619–628. [Google Scholar]
- Eaton, J.W. Is the lens canned? Free Radic. Biol. Med. 1991, 11, 207–213. [Google Scholar] [CrossRef] [PubMed]
- Shui, Y.B.; Beebe, D.C. Age-dependent control of lens growth by hypoxia. Investig. Opthalmology Vis. Sci. 2008, 49, 1023–1029. [Google Scholar] [CrossRef] [PubMed]
- Periyasamy, P.; Shinohara, T. Age-related cataracts: Role of unfolded protein response, Ca2+ mobilization, epigenetic DNA modifications, and loss of Nrf2/Keap1 dependent cytoprotection. Prog. Retin Eye Res. 2017, 60, 1–19. [Google Scholar] [CrossRef]
- Zheng, X.Y.; Xu, J.; Chen, X.I.; Li, W.; Wang, T.Y. Attenuation of oxygen fluctuation-induced endoplasmic reticulum stress in human lens epithelial cells. Exp. Ther. Med. 2015, 10, 1883–1887. [Google Scholar] [CrossRef] [Green Version]
- Druker, J.; Wilson, J.W.; Child, F.; Shakir, D.; Fasanya, T.; Rocha, S. Role of Hypoxia in the Control of the Cell Cycle. Int. J. Mol. Sci. 2021, 22, 4874. [Google Scholar] [CrossRef]
- Muenchau, S.; Deutsch, R.; de Castro, I.J.; Hielscher, T.; Heber, N.; Niesler, B.; Lusic, M.; Stanifer, M.L.; Boulant, S. Hypoxia Environment Promotes Barrier Formation in Human Intestinal Epithelial Cells through Regulation of MicroRNA 320a Expression. Mol. Cell. Biol. 2019, 39, e00553-18. [Google Scholar] [CrossRef] [Green Version]
- Hockwin, O. Age changes of lens metabolism. Altern Entwicklung. Aging Dev. 1971, 1, 95–129. [Google Scholar]
- van Heyningen, R.; Linklater, J. The metabolism of the bovine lens in air and nitrogen. Exp. Eye Res. 1975, 20, 393–396. [Google Scholar] [CrossRef] [PubMed]
- Majewski, N.; Nogueira, V.; Bhaskar, P.; Coy, P.E.; Skeen, J.E.; Gottlob, K.; Chandel, N.S.; Thompson, C.B.; Robey, R.; Hay, N. Hexokinase-mitochondria interaction mediated by Akt is required to inhibit apoptosis in the presence or absence of Bax and Bak. Mol Cell 2004, 16, 819–830. [Google Scholar] [CrossRef]
- Pastorino, J.G.; Shulga, N.; Hoek, J.B. Mitochondrial binding of hexokinase II inhibits Bax-induced cytochrome c release and apoptosis. J. Biol. Chem. 2002, 277, 7610–7618. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heneberg, P. Redox Regulation of Hexokinases. Antioxid. Redox Signal. 2019, 30, 415–442. [Google Scholar] [CrossRef] [PubMed]
- Santiago, A.P.; Chaves, E.A.; Oliveira, M.F.; Galina, A. Reactive oxygen species generation is modulated by mitochondrial kinases: Correlation with mitochondrial antioxidant peroxidases in rat tissues. Biochimie 2008, 90, 1566–1577. [Google Scholar] [CrossRef] [PubMed]
- Filomeni, G.; De Zio, D.; Cecconi, F. Oxidative stress and autophagy: The clash between damage and metabolic needs. Cell Death Differ. 2015, 22, 377–388. [Google Scholar] [CrossRef] [Green Version]
- Valvona, C.J.; Fillmore, H.L.; Nunn, P.B.; Pilkington, G.J. The Regulation and Function of Lactate Dehydrogenase A: Therapeutic Potential in Brain Tumor. Brain Pathol. 2016, 26, 3–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, W.; Wang, C.; Hu, X.; Lian, Y.; Ding, C.; Ming, L. Inhibition of LDHA suppresses cell proliferation and increases mitochondrial apoptosis via the JNK signaling pathway in cervical cancer cells. Oncol. Rep. 2022, 47, 77. [Google Scholar] [CrossRef]
- Urbańska, K.; Orzechowski, A. Unappreciated Role of LDHA and LDHB to Control Apoptosis and Autophagy in Tumor Cells. Int. J. Mol. Sci. 2019, 20, 2085. [Google Scholar] [CrossRef] [Green Version]
- Sun, C.; Liu, M.; Zhang, W.; Wang, S.; Qian, G.; Wang, M.; Zhang, G. Overexpression of enolase 2 is associated with worsened prognosis and increased glycikolysis in papillary renal cell carcinoma. J. Cell. Physiol. 2021, 236, 3821–3831. [Google Scholar] [CrossRef] [PubMed]
- Pan, J.; Jin, Y.; Xu, X.; Wei, W.; Pan, H. Integrated Analysis of the Role of Enolase 2 in Clear Cell Renal Cell Carcinoma. Dis. Markers 2022, 2022, 6539203. [Google Scholar] [CrossRef] [PubMed]
- Leung, S.W.S.; Shi, Y. The glycolytic process in endothelial cells and its implications. Acta Pharmacol. Sin. 2022, 43, 251–259. [Google Scholar] [CrossRef] [PubMed]
- Magnuson, D.K.; Maier, R.V.; Pohlman, T.H. Protein kinase C: A potential pathway of endothelial cell activation by endotoxin, tumor necrosis factor, and interleukin-1. Surgery 1989, 106, 216–223. [Google Scholar]
- Beltran, C.; Pardo, R.; Bou-Teen, D.; Ruiz-Meana, M.; Villena, J.A.; Ferreira-González, I.; Barba, I. Enhancing Glycolysis Protects against Ischemia-Reperfusion Injury by Reducing ROS Production. Metabolites 2020, 10, 132. [Google Scholar] [CrossRef] [Green Version]
- Jian, S.L.; Chen, W.W.; Su, Y.C.; Su, Y.W.; Chuang, T.H.; Hsu, S.C.; Huang, L.R. Glycolysis regulates the expansion of myeloid-derived suppressor cells in tumor-bearing hosts through prevention of ROS-mediated apoptosis. Cell Death Dis. 2017, 8, e2779. [Google Scholar] [CrossRef] [Green Version]
- Yang, H.; Ping, X.; Cui, Y.; Zheng, S.; Shentu, X. Role of Rapamycin and 3-MA in oxidative damage of HLECs caused by two doses of UVB radiation. AOPR 2023, 3, 15–22. [Google Scholar] [CrossRef]
- Chen, Y.; Mehta, G.; Vasiliou, V. Antioxidant defenses in the ocular surface. Ocul. Surf. 2009, 7, 176–185. [Google Scholar] [CrossRef] [Green Version]
- Holekamp, N.M.; Shui, Y.B.; Beebe, D.C. Vitrectomy surgery increases oxygen exposure to the lens: A possible mechanism for nuclear cataract formation. Am. J. Ophthalmol. 2005, 139, 302–310. [Google Scholar] [CrossRef]
- Harocopos, G.J.; Shui, Y.B.; McKinnon, M.; Holekamp, N.M.; Gordon, M.O.; Beebe, D.C. Importance of vitreous liquefaction in age-related cataract. Investig. Opthalmology Vis. Sci. 2004, 45, 77–85. [Google Scholar] [CrossRef]
- Patil, N.; Howe, O.; Cahill, P.; Byrne, H.J. Monitoring and modelling the dynamics of the cellular glycolysis pathway: A review and future perspectives. Mol. Metab. 2022, 66, 101635. [Google Scholar] [CrossRef] [PubMed]
- Finlay, D.K. Regulation of glucose metabolism in T cells: New insight into the role of Phosphoinositide 3-kinases. Front. Immunol. 2012, 3, 247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arthur, S.A.; Blaydes, J.P.; Houghton, F.D. Glycolysis Regulates Human Embryonic Stem Cell Self-Renewal under Hypoxia through HIF-2α and the Glycolytic Sensors CTBPs. Stem Cell Rep. 2019, 12, 728–742. [Google Scholar] [CrossRef] [Green Version]
- Zhang, C.C.; Sadek, H.A. Hypoxia and metabolic properties of hematopoietic stem cells. Antioxid. Redox Signal. 2014, 20, 1891–1901. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schröder, M.; Kaufman, R.J. ER stress and the unfolded protein response. Mutat. Res. 2005, 569, 29–63. [Google Scholar] [CrossRef] [PubMed]
- Santos, C.X.; Tanaka, L.Y.; Wosniak, J.; Laurindo, F.R. Mechanisms and implications of reactive oxygen species generation during the unfolded protein response: Roles of endoplasmic reticulum oxidoreductases, mitochondrial electron transport, and NADPH oxidase. Antioxid. Redox Signal. 2009, 11, 2409–2427. [Google Scholar] [CrossRef]
- Pierce, G.B.; Parchment, R.E.; Lewellyn, A.L. Hydrogen peroxide as a mediator of programmed cell death in the blastocyst. Differentiation 1991, 46, 181–186. [Google Scholar] [CrossRef]
- Ma, J.; Chen, T.; Wu, S.; Yang, C.; Bai, M.; Shu, K.; Li, K.; Zhang, G.; Jin, Z.; He, F.; et al. iProX: An integrated proteome resource. Nucleic Acids Res. 2019, 47, D1211–D1217. [Google Scholar] [CrossRef] [Green Version]
- Chen, T.; Ma, J.; Liu, Y.; Chen, Z.; Xiao, N.; Lu, Y.; Fu, Y.; Yang, C.; Li, M.; Wu, S.; et al. iProX in 2021: Connecting proteomics data sharing with big data. Nucleic Acids Res. 2022, 50, D1522–D1527. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, Y.; Ping, X.; Cui, Y.; Yang, H.; Bao, J.; Yin, Q.; Ailifeire, H.; Shentu, X. Glycolysis Aids in Human Lens Epithelial Cells’ Adaptation to Hypoxia. Antioxidants 2023, 12, 1304. https://doi.org/10.3390/antiox12061304
Huang Y, Ping X, Cui Y, Yang H, Bao J, Yin Q, Ailifeire H, Shentu X. Glycolysis Aids in Human Lens Epithelial Cells’ Adaptation to Hypoxia. Antioxidants. 2023; 12(6):1304. https://doi.org/10.3390/antiox12061304
Chicago/Turabian StyleHuang, Yuxin, Xiyuan Ping, Yilei Cui, Hao Yang, Jing Bao, Qichuan Yin, Hailaiti Ailifeire, and Xingchao Shentu. 2023. "Glycolysis Aids in Human Lens Epithelial Cells’ Adaptation to Hypoxia" Antioxidants 12, no. 6: 1304. https://doi.org/10.3390/antiox12061304




