Study on the Properties and Synergistic Antioxidant Effects of Novel Bifunctional Fusion Proteins Expressed Using the UTuT6 System
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Construction of Fusion Proteins and SOD3-72P Expression Plasmids
2.3. Protein Expression and Purification
2.4. Incorporation of Copper and Zinc into Fusion Proteins
2.5. SDS-PAGE and Western Blot Analysis
2.6. Assay of Enzyme Activities
2.7. Molecular Modeling and Docking
2.8. Effect of Temperature, pH, and Storage Time on Fusion Proteins
2.9. Steady State Kinetics of Fusion Proteins
2.10. Resistance to Inactivation by H2O2
2.11. Hemolysis Assay
2.12. Inhibition of Lipid Peroxidation in Liver
2.13. Inhibition of Yolk Lipoprotein Peroxidation Induced by FeSO4
3. Results
3.1. Preparation and Identification of Fusion Proteins
3.2. The Assay of Enzyme Activities
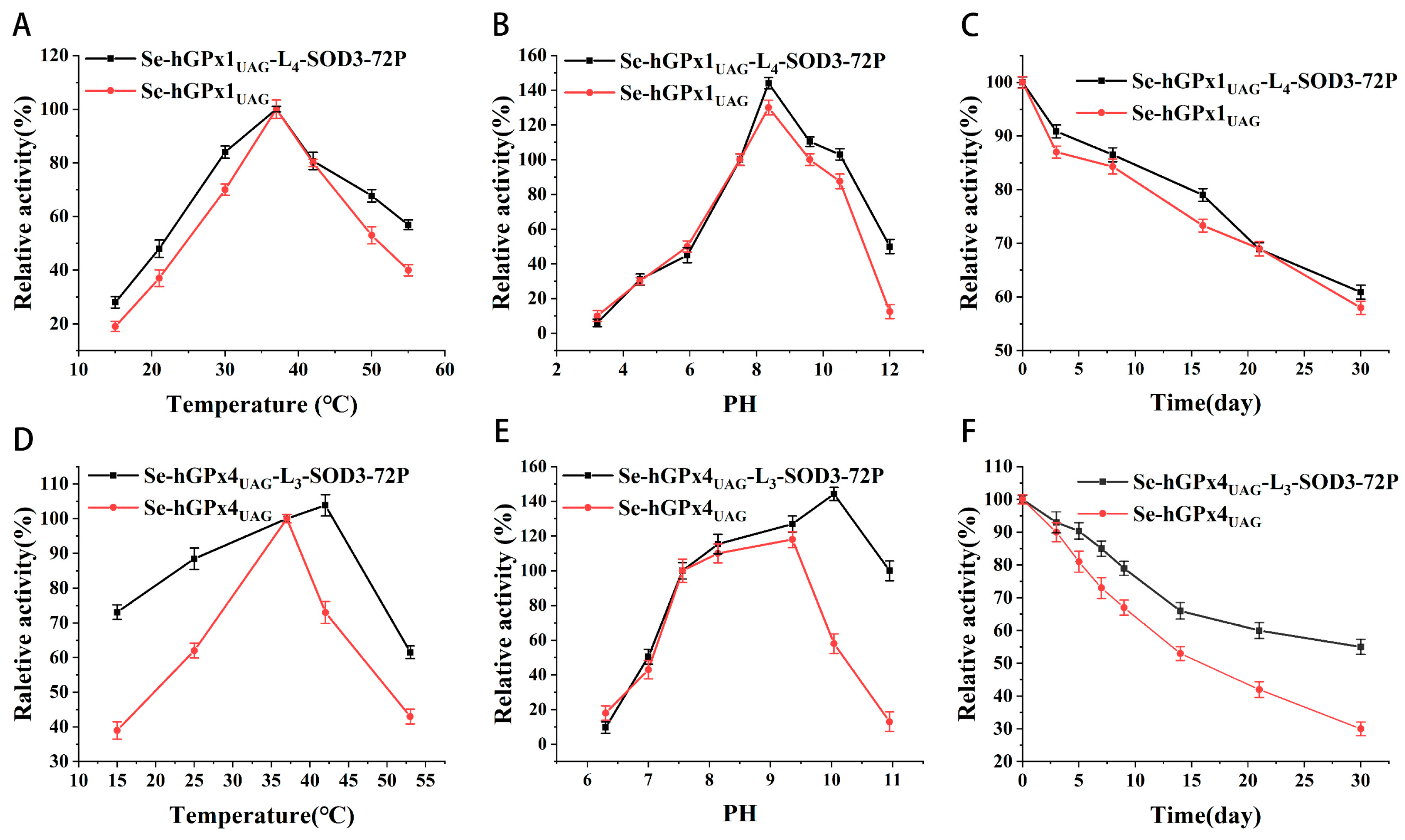
3.3. Effect of Temperature, pH, and Storage Time on Se-hGPx1UAG-L4-SOD3-72P and Se-hGPx4UAG-L3-SOD3-72P
3.4. Steady State Kinetics of Se-hGPx1UAG-L4-SOD3-72P and Se-hGPx4UAG-L3-SOD3-72P
3.5. Determination of the Synergism of Fusion Proteins
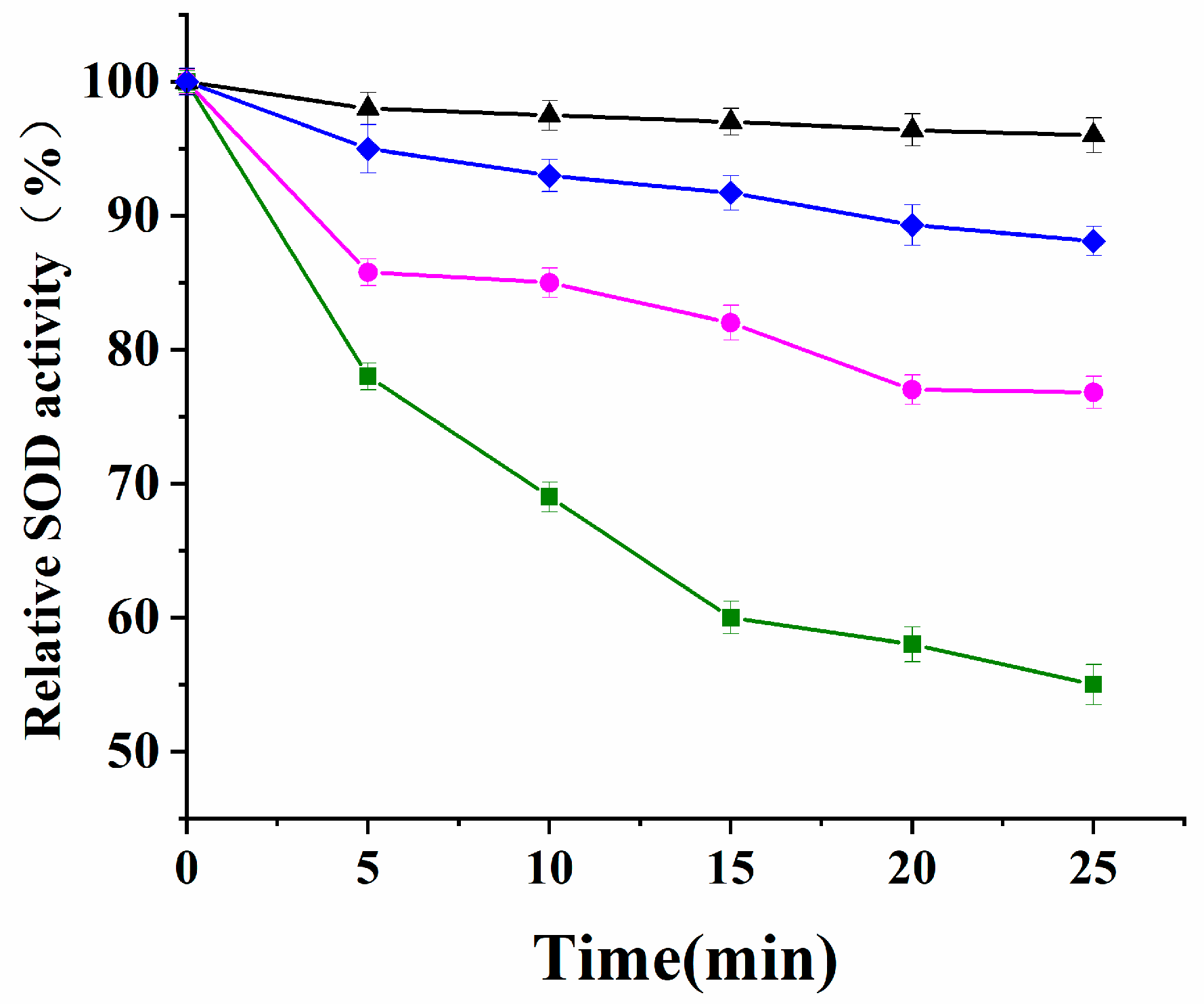
3.5.1. Resistance to Inactivation by H2O2
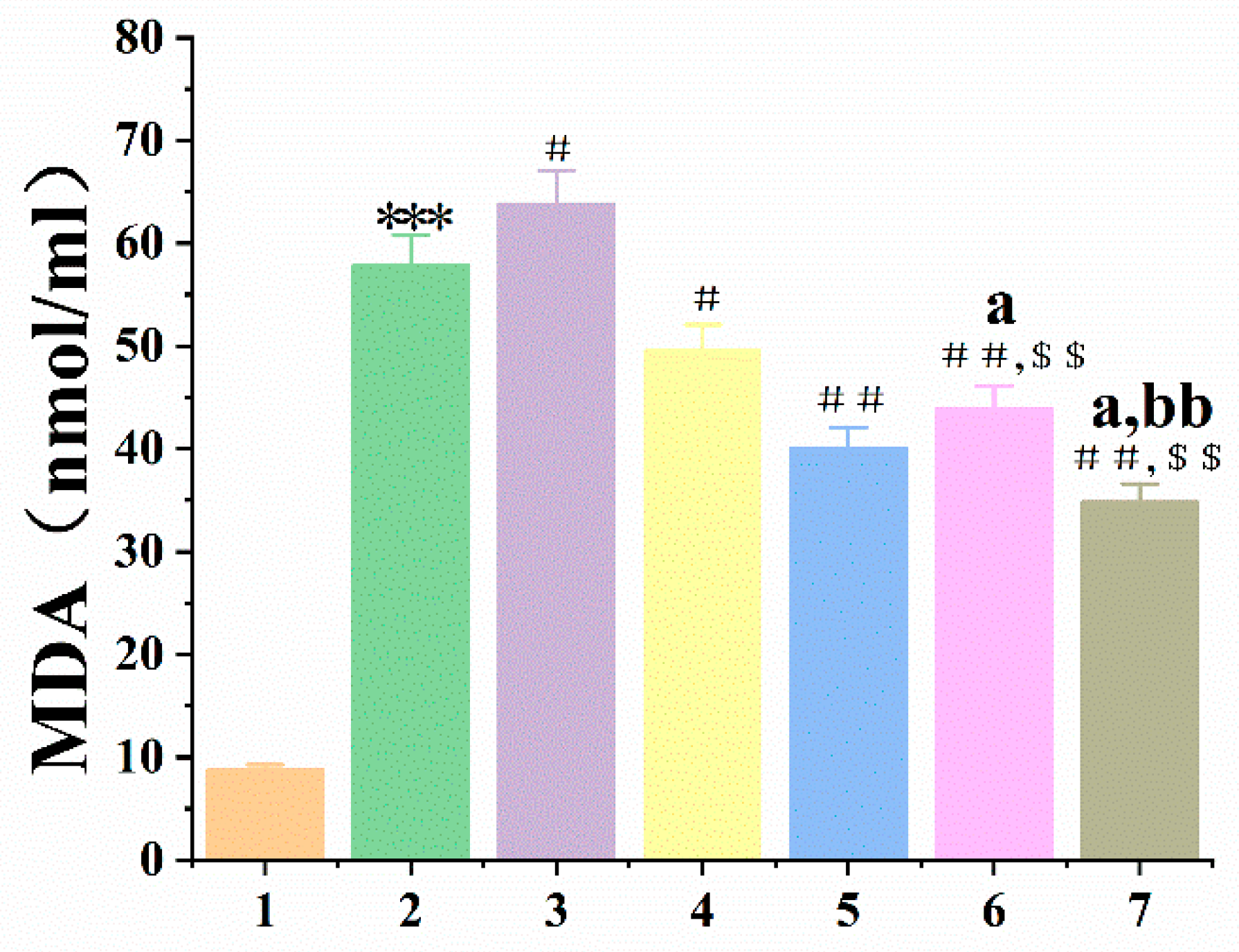
3.5.2. Se-hGPx1UAG-L4-SOD3-72P and Se-hGPx4UAG-L3-SOD3-72P Aid in Protecting RBCs and the Liver against Oxidative Damage
3.5.3. Inhibitory Effect of Se-hGPx1UAG-L4-SOD3-72P and Se-hGPx4UAG-L3-SOD3-72P on Lipid Peroxidation of Yolk Lipoprotein
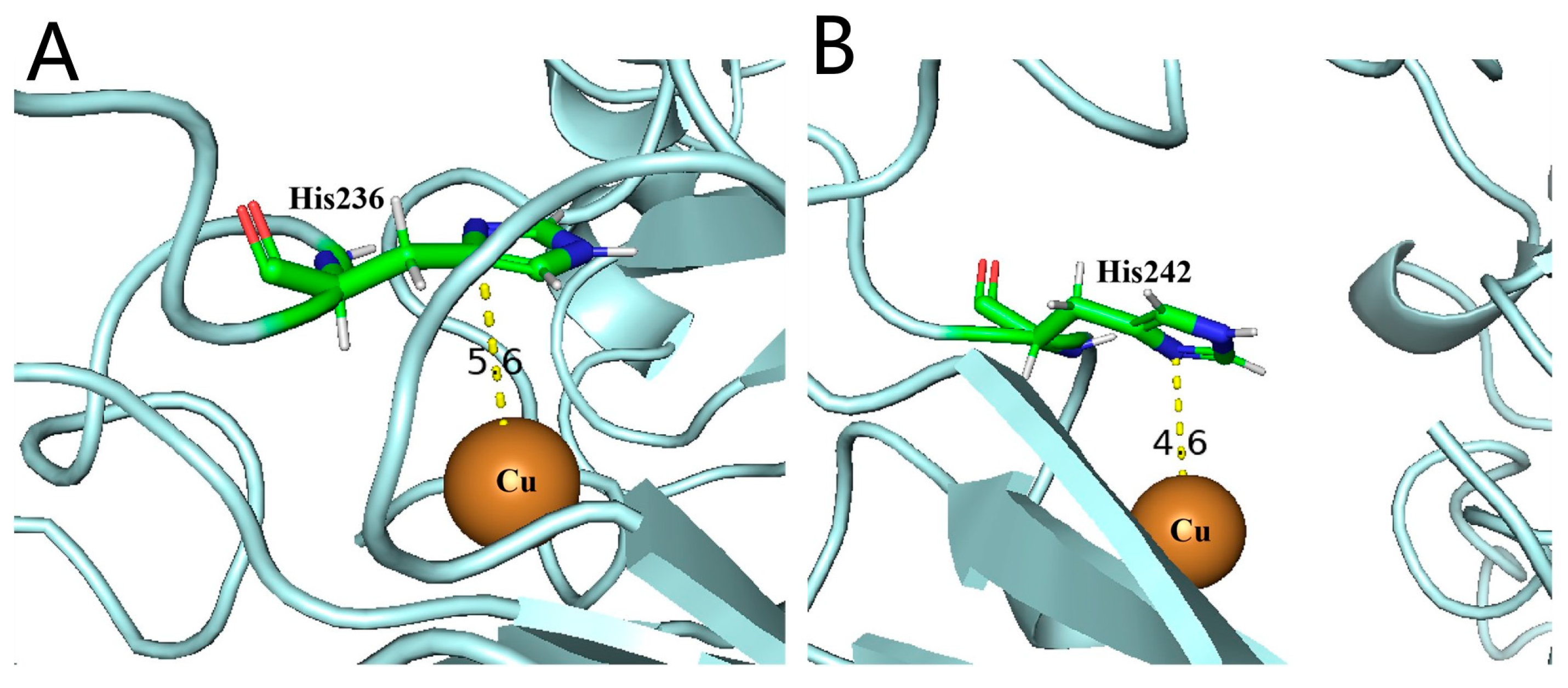
3.6. Exploration of Cys That May Participate in the Formation of Disulfide Bonds
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Guzik, T.J.; Harrison, D.G. Vascular NADPH Oxidases as Drug Targets for Novel Antioxidant Strategies. Drug Discov. Today 2006, 11, 524–533. [Google Scholar] [CrossRef] [PubMed]
- Bogdan, C.; Röllinghoff, M.; Diefenbach, A. Reactive Oxygen and Reactive Nitrogen Intermediates in Innate and Specific Immunity. Curr. Opin. Immunol. 2000, 12, 64–76. [Google Scholar] [CrossRef] [PubMed]
- Flohé, L.; Toppo, S.; Orian, L. The Glutathione Peroxidase Family: Discoveries and Mechanism. Free Radic. Biol. Med. 2022, 187, 113–122. [Google Scholar] [CrossRef] [PubMed]
- Brigelius-Flohé, R.; Maiorino, M. Glutathione Peroxidases. Biochim. Biophys. Acta (BBA) General. Subj. 2013, 1830, 3289–3303. [Google Scholar] [CrossRef] [PubMed]
- Cui, C.; Yang, F.; Li, Q. Post-Translational Modification of GPX4 Is a Promising Target for Treating Ferroptosis-Related Diseases. Front. Mol. Biosci. 2022, 9, 901565. [Google Scholar] [CrossRef]
- Ursini, F.; Bosello Travain, V.; Cozza, G.; Miotto, G.; Roveri, A.; Toppo, S.; Maiorino, M. A White Paper on Phospholipid Hydroperoxide Glutathione Peroxidase (GPx4) Forty Years Later. Free Radic. Biol. Med. 2022, 188, 117–133. [Google Scholar] [CrossRef]
- Hatfield, D.; Diamond, A. UGA: A Split Personality in the Universal Genetic Code. Trends Genet. 1993, 9, 69–70. [Google Scholar] [CrossRef]
- Chambers, I.; Frampton, J.; Goldfarb, P.; Affara, N.; McBain, W.; Harrison, P.R. The Structure of the Mouse Glutathione Peroxidase Gene: The Selenocysteine in the Active Site Is Encoded by the ‘Termination’ Codon, TGA. EMBO J. 1986, 5, 1221–1227. [Google Scholar] [CrossRef]
- Aldag, C.; Bröcker, M.J.; Hohn, M.J.; Prat, L.; Hammond, G.; Plummer, A.; Söll, D. Rewiring Translation for Elongation Factor Tu-Dependent Selenocysteine Incorporation. Angew. Chem. Int. Ed. 2013, 52, 1441–1445. [Google Scholar] [CrossRef]
- Fan, Z.; Song, J.; Guan, T.; Lv, X.; Wei, J. Efficient Expression of Glutathione Peroxidase with Chimeric TRNA in Amber-Less Escherichia Coli. ACS Synth. Biol. 2018, 7, 249–257. [Google Scholar] [CrossRef]
- Zelko, I.N.; Mariani, T.J.; Folz, R.J. Superoxide Dismutase Multigene Family: A Comparison of the CuZn-SOD (SOD1), Mn-SOD (SOD2), and EC-SOD (SOD3) Gene Structures, Evolution, and Expression. Free. Radic. Biol. Med. 2002, 33, 337–349. [Google Scholar] [CrossRef] [PubMed]
- Kwon, M.J.; Kim, B.H.; Lee, Y.S.; Kim, T.Y. Role of Superoxide Dismutase 3 in Skin Inflammation. J. Dermatol. Sci. 2012, 67, 81–87. [Google Scholar] [CrossRef] [PubMed]
- Antonyuk, S.V.; Strange, R.W.; Marklund, S.L.; Hasnain, S.S. The Structure of Human Extracellular Copper-Zinc Superoxide Dismutase at 1.7 Å Resolution: Insights into Heparin and Collagen Binding. J. Mol. Biol. 2009, 388, 310–326. [Google Scholar] [CrossRef] [PubMed]
- Yan, F.; Yan, G.; Lv, S.; Shen, N.; Mu, Y.; Chen, T.; Gong, P.; Xu, Y.; Lv, L.; Liu, J.; et al. A Novel 65-Mer Peptide Imitates the Synergism of Superoxide Dismutase and Glutathione Peroxidase. Int. J. Biochem. Cell Biol. 2011, 43, 1802–1811. [Google Scholar] [CrossRef]
- Lajoie, M.J.; Rovner, A.J.; Goodman, D.B.; Aerni, H.R.; Haimovich, A.D.; Kuznetsov, G.; Mercer, J.A.; Wang, H.H.; Carr, P.A.; Mosberg, J.A.; et al. Genomically Recoded Organisms Expand Biological Functions. Science 2013, 342, 357–360. [Google Scholar] [CrossRef]
- Rhodius, V.A.; Segall-Shapiro, T.H.; Sharon, B.D.; Ghodasara, A.; Orlova, E.; Tabakh, H.; Burkhardt, D.H.; Clancy, K.; Peterson, T.C.; Gross, C.A.; et al. Design of Orthogonal Genetic Switches Based on a Crosstalk Map of Σs, Anti-Σs, and Promoters. Mol. Syst. Biol. 2013, 9, 702. [Google Scholar] [CrossRef]
- Wilson, S.R.; Zucker, P.A.; Huang, R.R.C.; Spector, A. Development of Synthetic Compounds with Glutathione Peroxidase Activity. J. Am. Chem. Soc. 1989, 111, 5936–5939. [Google Scholar] [CrossRef]
- Guan, T.; Song, J.; Wang, Y.; Guo, L.; Yuan, L.; Zhao, Y.; Gao, Y.; Lin, L.; Wang, Y.; Wei, J. Expression and Characterization of Recombinant Bifunctional Enzymes with Glutathione Peroxidase and Superoxide Dismutase Activities. Free Radic. Biol. Med. 2017, 110, 188–195. [Google Scholar] [CrossRef]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J.E. The Phyre2 Web Portal for Protein Modeling, Prediction and Analysis. Nat. Protoc. 2015, 10, 845–858. [Google Scholar] [CrossRef]
- Zhang, J.; Liang, Y.; Zhang, Y. Atomic-Level Protein Structure Refinement Using Fragment-Guided Molecular Dynamics Conformation Sampling. Structure 2011, 19, 1784–1795. [Google Scholar] [CrossRef]
- Zhang, W.; Bell, E.W.; Yin, M.; Zhang, Y. EDock: Blind Protein-Ligand Docking by Replica-Exchange Monte Carlo Simulation. J. Cheminform. 2020, 12, 37. [Google Scholar] [CrossRef]
- Wang, S.; Zhao, Y.; Yang, J.; Liu, S.; Ni, W.; Bai, X.; Yang, Z.; Zhao, D.; Liu, M. Ginseng Polysaccharide Attenuates Red Blood Cells Oxidative Stress Injury by Regulating Red Blood Cells Glycolysis and Liver Gluconeogenesis. J. Ethnopharmacol. 2023, 300, 115716. [Google Scholar] [CrossRef]
- Dorochow, E.; Gurke, R.; Rischke, S.; Geisslinger, G.; Hahnefeld, L. Effects of Different Storage Conditions on Lipid Stability in Mice Tissue Homogenates. Metabolites 2023, 13, 504. [Google Scholar] [CrossRef]
- Xu, Y.; Zhou, Y.; Yin, R.; Wang, C.; Chu, H.; Wang, J. A Novel 76-Mer Peptide Mimic with the Synergism of Superoxide Dismutase and Glutathione Peroxidase. In Vitro Cell Dev. Biol. Anim. 2018, 54, 335–345. [Google Scholar] [CrossRef] [PubMed]
- Toppo, S.; Flohé, L.; Ursini, F.; Vanin, S.; Maiorino, M. Catalytic Mechanisms and Specificities of Glutathione Peroxidases: Variations of a Basic Scheme. Biochim. Biophys. Acta Gen. Subj. 2009, 1790, 1486–1500. [Google Scholar] [CrossRef] [PubMed]
- Aboelella, N.S.; Brandle, C.; Kim, T.; Ding, Z.-C.; Zhou, G. Oxidative Stress in the Tumor Microenvironment and Its Relevance to Cancer Immunotherapy. Cancers 2021, 13, 986. [Google Scholar] [CrossRef] [PubMed]
- García-Sánchez, A.; Miranda-Díaz, A.G.; Cardona-Muñoz, E.G. The Role of Oxidative Stress in Physiopathology and Pharmacological Treatment with Pro- and Antioxidant Properties in Chronic Diseases. Oxid. Med. Cell Longev. 2020, 2020, 2082145. [Google Scholar] [CrossRef]
- Bergeron, L.M.; Gomez, L.; Whitehead, T.A.; Clark, D.S. Self-Renaturing Enzymes: Design of an Enzyme-Chaperone Chimera as a New Approach to Enzyme Stabilization. Biotechnol. Bioeng. 2009, 102, 1316–1322. [Google Scholar] [CrossRef]
- Wang, Y.; Niu, S.; Li, B.; Yan, Q.; Zhao, Y.; Wang, Z.; Xiong, C.; Wei, J. Improvement of Stability and in Vivo Antioxidant Effect of Human Glutathione Peroxidase Mutant by PEGylation. Int. J. Pharm. 2021, 609, 121152. [Google Scholar] [CrossRef]
- Chen, X.; Zaro, J.L.; Shen, W.C. Fusion Protein Linkers: Property, Design and Functionality. Adv. Drug Deliv. Rev. 2013, 65, 1357–1369. [Google Scholar] [CrossRef]
- Zhang, L.; Chou, C.P.; Moo-Young, M. Disulfide Bond Formation and Its Impact on the Biological Activity and Stability of Recombinant Therapeutic Proteins Produced by Escherichia Coli Expression System. Biotechnol. Adv. 2011, 29, 923–929. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Dai, S.; Chen, X.; Xu, L.; Yan, J.; Yang, M.; Yan, Y. Alteration of Chain-Length Selectivity and Thermostability of Rhizopus Oryzae Lipase via Virtual Saturation Mutagenesis Coupled with Disulfide Bond Design. Appl. Environ. Microbiol. 2023, 89, e01878-22. [Google Scholar] [CrossRef]
- Pernow, J.; Mahdi, A.; Yang, J.; Zhou, Z. Red Blood Cell Dysfunction: A New Player in Cardiovascular Disease. Cardiovasc. Res. 2019, 115, 1596–1605. [Google Scholar] [CrossRef] [PubMed]
- Stolwijk, J.M.; Falls-Hubert, K.C.; Searby, C.C.; Wagner, B.A.; Buettner, G.R. Simultaneous Detection of the Enzyme Activities of GPx1 and GPx4 Guide Optimization of Selenium in Cell Biological Experiments. Redox Biol. 2020, 32, 101518. [Google Scholar] [CrossRef] [PubMed]
- Qian, S.Y.; Wang, H.P.; Schafer, F.Q.; Buettner, G.R. EPR Detection of Lipid-Derived Free Radicals from PUFA, LDL, and Cell Oxidations. Free Radic. Biol. Med. 2000, 29, 568–579. [Google Scholar] [CrossRef] [PubMed]





| Catalyst | GPx Activity (U/mg) | SOD Activity (U/mg) |
|---|---|---|
| Se-hGPx1UAG | 321 ± 31 | ND |
| Se-hGPx4UAG | 38 ± 2 | ND |
| SOD3-72P Se-Cu/Zn-65P [14,24] | ND 112 | 2411.5 ± 113 1254 |
| Se-hGPx1Ser-L-APSOD [18] | 52.1 ± 4.8 | 101 ± 13 |
| Se-hGPx1UAG-L3-SOD3-72P | 190 ± 29 | 1537 ± 211 |
| Se-hGPx1UAG-L4-SOD3-72P | 199 ± 16 | 2790 ± 125 |
| Se-hGPx4UAG-L3-SOD3-72P | 20.78 ± 3.1 | 2506.3 ± 223 |
| Se-hGPx4UAG-L4-SOD3-72P | 19.2 ± 2.9 | 2403.6 ± 145 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yan, Q.; Wei, J.; Song, J.; Li, M.; Guan, X.; Song, J. Study on the Properties and Synergistic Antioxidant Effects of Novel Bifunctional Fusion Proteins Expressed Using the UTuT6 System. Antioxidants 2023, 12, 1766. https://doi.org/10.3390/antiox12091766
Yan Q, Wei J, Song J, Li M, Guan X, Song J. Study on the Properties and Synergistic Antioxidant Effects of Novel Bifunctional Fusion Proteins Expressed Using the UTuT6 System. Antioxidants. 2023; 12(9):1766. https://doi.org/10.3390/antiox12091766
Chicago/Turabian StyleYan, Qi, Jingyan Wei, Junxia Song, Mengna Li, Xin Guan, and Jian Song. 2023. "Study on the Properties and Synergistic Antioxidant Effects of Novel Bifunctional Fusion Proteins Expressed Using the UTuT6 System" Antioxidants 12, no. 9: 1766. https://doi.org/10.3390/antiox12091766




