A Quantum Chemical and Statistical Study of Phenolic Schiff Bases with Antioxidant Activity against DPPH Free Radical
Abstract
:1. Introduction
2. Material and Methods
2.1. Synthesis and DPPH Free Radical Scavenging Capacity
| Subclass | No. | Hydroxyl Positions | Other Groups | IC50 (μg/mL) |
|---|---|---|---|---|
 | 1 | 2 | - | 0.90 ± 0.045 |
| 2 | 3 | - | 1.1 ± 0.05 | |
| 3 | 4 | - | 0.65 ± 0.045 | |
| 4 | 2, 3 | - | 0.22 ± 0.045 | |
| 5 | 2, 4 | - | 0.34 ± 0.045 | |
| 6 | 2, 5 | - | 0.92 ± 0.045 | |
| 7 | 3, 4 | - | 0.20 ± 0.045 | |
| 8 | 3, 5 | - | 0.91 ± 0.0045 | |
| 9 | 2, 4, 6 | - | 0.35 ± 0.045 | |
| 10 | 3, 4, 5 | - | 0.30 ± 0.045 | |
| 11 | - | 3-OCH3 | >>2 | |
| 12 | - | 4-OCH3 | >>2 | |
| 13 | - | 3,4-diOCH3 | >>2 | |
| 14 | - | 3,5-diOCH3 | >>2 | |
| 15 | 2 | 4- OCH3 | 0.50 ± 0.071 | |
| 16 | 2 | 5-OCH3 | 1.01 ± 0.045 | |
| 17 | 3 | 4- OCH3 | 0.8 ± 0.002 | |
| 18 | 4 | 3-Br | 0.22 ± 0.0045 | |
| 19 | - | 3-Br, 4-Cl | >>2 | |
| 20 | - | 4-F | >>2 | |
| 21 | - | 4-Cl | >>2 | |
| 22 | 3 | 2-I, 4-OCH3 | 1.63 ± 0.21 | |
| 23 | - | 4-COOCH3 | >>2 | |
| 24 | - | 4-NO2 | >>2 | |
| 25 | - | - | >>2 | |
 | 26 | - | - | >>2 |
 | 27 | - | - | >>2 |
 | 28 | - | - | >>2 |
 | 29 | - | - | >>2 |
 | 30 | - | - | >>2 |
2.2. Theoretical Details
3. Results and Discussion
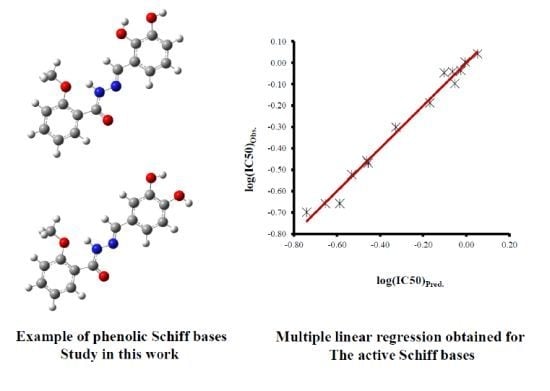
3.1. Simple and Multiple Linear Regressions
| SBs | BDE | BDEd | IPd | IP | EA | χ | η | S | ω | α | μ | A | V | M | logP | nOH | SD | ΔG | log(1/IC50) | Activity |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 83.8 | 88.70 | 6.50 | 6.75 | 2.65 | 4.70 | 4.10 | 0.12 | 2.70 | 307.81 | 10.12 | 346.83 | 368.85 | 270 | 2.87 | 1 | 0.27 | 7.5 | −0.05 | Active |
| 2 | 87.1 | 112.10 | 7.10 | 6.87 | 2.67 | 4.77 | 4.20 | 0.12 | 2.71 | 305.92 | 8.13 | 349.44 | 369.86 | 270 | 2.87 | 1 | 0.37 | 10.8 | 0.04 | Active |
| 3 | 83.2 | 86.10 | 6.40 | 6.62 | 2.54 | 4.58 | 4.07 | 0.12 | 2.58 | 311.99 | 7.65 | 350.17 | 371.21 | 270 | 2.87 | 1 | 0.32 | 6.8 | −0.19 | Active |
| 4 | 77.5 | 76.70 | 6.50 | 6.76 | 2.64 | 4.70 | 4.12 | 0.12 | 2.68 | 314.16 | 9.14 | 356.43 | 379.80 | 286 | 2.58 | 2 | 0.31 | 1.5 | −0.66 | Active |
| 5 | 82.2 | 94.10 | 6.40 | 6.50 | 2.52 | 4.51 | 3.97 | 0.13 | 2.56 | 320.20 | 12.12 | 358.17 | 381.33 | 286 | 2.58 | 2 | 0.25 | 5.7 | −0.47 | Active |
| 6 | 78.8 | 68.80 | 6.20 | 6.52 | 2.69 | 4.60 | 3.84 | 0.13 | 2.76 | 315.42 | 11.00 | 359.45 | 382.83 | 286 | 2.58 | 2 | 0.31 | 2.3 | −0.04 | Active |
| 7 | 76.1 | 75.90 | 6.20 | 6.52 | 2.55 | 4.54 | 3.97 | 0.13 | 2.59 | 317.53 | 6.18 | 359.09 | 382.23 | 286 | 2.58 | 2 | 0.25 | 0.2 | −0.70 | Active |
| 8 | 86.6 | 107.90 | 7.10 | 6.92 | 2.68 | 4.80 | 4.24 | 0.12 | 2.72 | 312.67 | 8.92 | 361.32 | 383.45 | 286 | 2.58 | 2 | 0.34 | 10.3 | −0.04 | Active |
| 9 | 83 | 83.60 | 6.40 | 6.48 | 2.42 | 4.45 | 4.06 | 0.12 | 2.44 | 326.76 | 8.29 | 367.78 | 392.97 | 302 | 2.3 | 3 | 0.23 | 6.6 | −0.46 | Active |
| 10 | 72.3 | 76.60 | 6.20 | 6.51 | 2.57 | 4.54 | 3.94 | 0.13 | 2.62 | 325.00 | 5.73 | 368.00 | 393.67 | 302 | 2.3 | 3 | 0.27 | -2.7 | −0.52 | Active |
| 11 | 97.7 | - | - | 6.86 | 2.65 | 4.75 | 4.21 | 0.12 | 2.68 | 322.98 | 8.68 | 372.50 | 395.67 | 284 | 2.9 | 0 | 0.5 | 20.5 | >0.30 | Inactive |
| 12 | 95.6 | - | - | 6.56 | 2.54 | 4.55 | 4.02 | 0.12 | 2.58 | 330.86 | 7.50 | 373.23 | 397.38 | 284 | 2.9 | 0 | 0.46 | 18.5 | >0.30 | Inactive |
| 13 | 95.60 | - | - | 6.5 | 2.56 | 4.6 | 4 | 0.12 | 2.60 | 353.49 | 7.17 | 400.14 | 430.15 | 314 | 2.65 | 0 | 0.46 | 18.2 | 0.30 | Inactive |
| 14 | 97.7 | - | - | 6.86 | 2.65 | 4.75 | 4.21 | 0.12 | 2.69 | 346.83 | 7.67 | 404.68 | 433.54 | 314 | 2.65 | 0 | 0.5 | 20.4 | >0.30 | Inactive |
| 15 | 84.1 | 86.30 | 6.40 | 6.48 | 2.53 | 4.51 | 3.95 | 0.13 | 2.57 | 338.44 | 8.42 | 381.97 | 408.56 | 314 | 2.61 | 1 | 0.28 | 7.7 | −0.30 | Active |
| 16 | 79.2 | 91.80 | 6.10 | 6.49 | 2.67 | 4.58 | 3.82 | 0.13 | 2.75 | 331.70 | 10.35 | 382.84 | 408.79 | 314 | 2.61 | 1 | 0.31 | 0 | 0.00 | Active |
| 17 | 86 | 100.60 | 6.30 | 6.47 | 2.55 | 4.51 | 3.92 | 0.13 | 2.60 | 336.25 | 6.16 | 381.75 | 408.07 | 300 | 2.61 | 1 | 0.33 | 9.3 | −0.10 | Active |
| 18 | 82.2 | 86.80 | 6.40 | 6.74 | 2.63 | 4.68 | 4.10 | 0.12 | 2.67 | 341.29 | 6.44 | 373.11 | 399.32 | 349 | 3.66 | 1 | 0.26 | 6 | −0.66 | Active |
| 19 | 99 | - | - | 7.04 | 2.85 | 4.95 | 4.19 | 0.12 | 2.92 | 351.56 | 9.76 | 380.01 | 407.79 | 367 | 4.46 | 0 | 0.53 | 21 | >0.30 | Inactive |
| 20 | 97.4 | - | - | 6.93 | 2.66 | 4.79 | 4.26 | 0.12 | 2.70 | 297.87 | 10.32 | 346.15 | 367.00 | 272 | 3.29 | 0 | 0.49 | 20.4 | >0.30 | Inactive |
| 21 | 97.7 | - | - | 6.95 | 2.75 | 4.85 | 4.20 | 0.12 | 2.80 | 321.19 | 10.50 | 358.15 | 380.50 | 289 | 3.67 | 0 | 0.49 | 20.7 | >0.30 | Inactive |
| 22 | 97.80 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | Inactive |
| 23 | 98.6 | - | - | 7.10 | 3.07 | 5.08 | 4.03 | 0.12 | 3.21 | 353.21 | 12.32 | 424.58 | 398.21 | 312 | 2.86 | 0 | 0.45 | 21.3 | >0.30 | Inactive |
| 24 | 99.6 | - | - | 7.25 | 3.83 | 5.54 | 3.41 | 0.15 | 4.50 | 350.66 | 14.25 | 368.39 | 393.25 | 299 | 3.11 | 0 | 0.5 | 21.2 | >0.30 | Inactive |
| 25 | 97.5 | - | - | 6.94 | 2.67 | 4.81 | 4.28 | 0.12 | 2.70 | 299.19 | 9.46 | 338.81 | 358.66 | 254 | 3.15 | 0 | 0.51 | 20.3 | >0.30 | Inactive |
| 26 | 98.4 | - | - | 7.15 | 2.86 | 5.00 | 4.29 | 0.12 | 2.92 | 291.63 | 11.56 | 334.13 | 352.57 | 255 | 2.7 | 0 | 0.57 | 20.8 | >0.30 | Inactive |
| 27 | 98.5 | - | - | 7.11 | 2.80 | 4.96 | 4.31 | 0.12 | 2.85 | 289.70 | 12.62 | 333.11 | 351.97 | 255 | 2.77 | 0 | 0.51 | 21.2 | >0.30 | Inactive |
| 28 | 99.4 | - | - | 7.26 | 2.97 | 5.12 | 4.29 | 0.12 | 3.05 | 289.74 | 11.55 | 333.31 | 352.15 | 255 | 2.77 | 0 | 0.54 | 21.7 | >0.30 | Inactive |
| 29 | 94.9 | - | - | 6.67 | 2.64 | 4.65 | 4.03 | 0.12 | 2.68 | 275.78 | 10.26 | 319.70 | 333.11 | 244 | 1.79 | 0 | 0.45 | 18 | >0.30 | Inactive |
| 30 | 95.1 | - | - | 6.72 | 2.73 | 4.73 | 3.99 | 0.13 | 2.80 | 297.67 | 10.09 | 328.48 | 346.48 | 260 | 2.13 | 0 | 0.44 | 18.2 | >0.30 | Inactive |
| Descriptors | R2 (%) | R2 Adjusted | SD |
|---|---|---|---|
| BDE | 31.18 | 25.45 | 0.2372 |
| BDEd | 28.26 | 22.28 | 0.2422 |
| IPd | 11.47 | 4% | 0.2691 |
| IP | 3.88 | −4% | 0.2804 |
| EA | 20.57 | 13.95 | 0.2549 |
| χ | 9.58 | 2.05 | 0.2719 |
| η | 0.1 | −8.23 | 0.2858 |
| S | 0.28 | −8.03 | 0.2856 |
| ω | 21.98 | 15.48 | 0.2526 |
| α | 8.43 | 0.7 | 0.2738 |
| μ | 13.07 | 5.82 | 0.2666 |
| A | 0.34 | −7.96 | 0.2855 |
| V | 0.91 | −7.34 | 0.2847 |
| M | 14.42 | 7.29 | 0.2645 |
| logP | 0.33 | −7.98 | 0.2855 |
| nOH | 20.46 | 14.03 | 0.2548 |
| SD | 46.35 | 41.88 | 0.2095 |
| ΔG | 21.78 | 15.26 | 0.2529 |

| SBs | log(IC50)Obs. | Equation 1 | Equation 2 | Equation 3 | |||||
|---|---|---|---|---|---|---|---|---|---|
| log(IC50)Pred. | Residual | log(IC50)Pred. | Residual | log(IC50)Pred. | Residual | ||||
| 1 | −0.05 | −0.10 | −0.06 | −0.22 | −0.17 | −0.15 | −0.10 | ||
| 2 | 0.04 | 0.05 | 0.01 | 0.02 | −0.02 | 0.03 | −0.02 | ||
| 3 | −0.19 | −0.17 | 0.02 | −0.28 | −0.09 | −0.42 | −0.23 | ||
| 4 | −0.66 | −0.59 | 0.07 | −0.57 | 0.09 | −0.36 | 0.30 | ||
| 5 | −0.47 | −0.46 | 0.01 | −0.29 | 0.18 | −0.38 | 0.09 | ||
| 6 | −0.04 | −0.02 | 0.01 | 0.02 | 0.06 | −0.14 | −0.10 | ||
| 7 | −0.70 | −0.74 | −0.04 | −0.54 | 0.16 | −0.60 | 0.10 | ||
| 8 | −0.04 | −0.06 | −0.02 | −0.01 | 0.03 | 0.14 | 0.18 | ||
| 9 | −0.46 | −0.46 | −0.01 | −0.43 | 0.03 | −0.47 | −0.01 | ||
| 10 | −0.52 | −0.53 | −0.01 | −0.72 | −0.20 | −0.62 | −0.10 | ||
| 15 | −0.30 | −0.33 | −0.03 | −0.33 | −0.03 | −0.28 | 0.02 | ||
| 16 | 0.00 | −0.01 | −0.01 | −0.04 | −0.04 | −0.16 | −0.16 | ||
| 17 | −0.10 | −0.05 | 0.04 | −0.13 | −0.04 | −0.14 | −0.05 | ||
| 18 | −0.66 | −0.65 | 0.00 | −0.62 | 0.03 | −0.59 | 0.07 | ||
3.2. Principal Component Analysis (PCA)
| Component | Eigenvalue | Variance (%) | Cumulated Variance (%) |
|---|---|---|---|
| PC1 | 2.306 | 76.88 | 76.88 |
| PC2 | 0.6261 | 20.87 | 97.75 |

| Variable | PC1 | PC2 |
|---|---|---|
| BDE | 0.6276 | 0.3014 |
| μ | 0.4724 | −0.8803 |
| logP | 0.6189 | 0.3663 |
3.3. Hierarchical Cluster Analysis (HCA)

4. Conclusions
Acknowledgments
References
- Vicini, P.; Geronikaki, A.; Incerti, M.; Busonera, B.; Poni, G.; Cabras, C.A.; La Colla, P. Synthesis and biological evaluation of benzo[d]isothiazole, benzothiazole and thiazole Schiff bases. Bioorg. Med. Chem. 2003, 11, 4785–4789. [Google Scholar] [CrossRef]
- Tarafder, M.; Kasbollah, A.; Saravanan, N.; Crouse, K.A.; Ali, A.M. S-methyldithiocarbazate and its Schiff bases: Evaluation of bondings and biological properties. J. Biochem. Mol. Biol. Biophys 2002, 6, 85–91. [Google Scholar] [CrossRef]
- Solmaz, R.; Altunbaş, E.; Kardaş, G. Adsorption and corrosion inhibition effect of 2-((5-mercapto-1, 3, 4-thiadiazol-2-ylimino) methyl) phenol Schiff base on mild steel. Mater. Chem. Phys. 2011, 125, 796–801. [Google Scholar] [CrossRef]
- Khan, K.M.; Ahmad, A.; Ambreen, N.; Amyn, A.; Perveen, S.; Khan, S.A.; Choudhary, M.I. Schiff Bases of 3-Formylchromones as Antibacterial, Antifungal, and Phytotoxic Agents (Supplementry Table). Lett. Drug. Des. Discov. 2009, 6, 363–373. [Google Scholar]
- Chohan, Z.H.; Pervez, H.; Rauf, A.; Khan, K.M.; Supuran, C.T. Antibacterial cobalt (II), copper (II), nickel (II) and zinc (II) complexes of mercaptothiadiazole-derived furanyl, thienyl, pyrrolyl, salicylyl and pyridinyl Schiff bases. J. Enzyme Inhib. Med. Chem. 2006, 21, 193–201. [Google Scholar] [CrossRef]
- Chohan, Z.H.; Pervez, H.; Rauf, A.; Khan, K.M.; Maharvi, G.M.; Supuran, C.T. Antibacterial and Antifugal Mono-and Di-substituted Symmetrical and Unsymmetrical Triazine-derived Schiff-bases and their Transition Metal Complexes. J. Enzyme Inhib. Med. Chem. 2004, 19, 161–168. [Google Scholar] [CrossRef]
- Kabeer, A.S.; Baseer, M.; Mote, N. Synthesis and antimicrobial activity of some Schiff bases from benzothiazoles. Asian J. Chem. 2001, 13, 496–500. [Google Scholar]
- Chohan, Z.H.; Arif, M.; Shafiq, Z.; Yaqub, M.; Supuran, C.T. In vitro antibacterial, antifungal & cytotoxic activity of some isonicotinoylhydrazide Schiff’s bases and their cobalt (II), copper (II), nickel (II) and zinc (II) complexes. J. Enzyme Inhib. Med. Chem. 2006, 21, 95–103. [Google Scholar] [CrossRef]
- Guo, Z.; Xing, R.; Liu, S.; Zhong, Z.; Ji, X.; Wang, L.; Li, P. Antifungal properties of Schiff bases of chitosan, N-substituted chitosan and quaternized chitosan. Carbohydr. Res. 2007, 342, 1329–1332. [Google Scholar] [CrossRef]
- Taha, M.; Baharudin, M.S.; Ismail, N.H.; Khan, K.M.; Jaafar, F.M.; Siddiqui, S.; Iqbal Choudhary, M. Synthesis of 2-methoxybenzoylhydrazone and evaluation of their antileishmanial activity. Bioorg. Med. Chem. Lett. 2013, 23, 3463–3466. [Google Scholar] [CrossRef]
- Mohammed Khan, K.; Taha, M.; Naz, F.; Siddiqui, S.; Ali, S.; Rahim, F.; Perveen, S.; Iqbal Choudhary, M. Acylhydrazide Schiff Bases: DPPH Radical and Superoxide Anion Scavengers. Med. Chem. 2012, 8, 705–710. [Google Scholar] [CrossRef]
- Mohammed Khan, K.; Shah, Z.; Uddin Ahmad, V.; Khan, M.; Taha, M.; Rahim, F.; Ali, S.; Ambreen, N.; Perveen, S.; Iqbal Choudhary, M. 2,4,6-Trichlorophenylhydrazine Schiff bases as DPPH radical and super oxide anion scavengers. Med. Chem. 2012, 8, 452–461. [Google Scholar] [CrossRef]
- Anouar, E.; Calliste, C.; Kosinova, P.; di Meo, F.; Duroux, J.; Champavier, Y.; Marakchi, K.; Trouillas, P. Free radical scavenging properties of guaiacol oligomers: A combined experimental and quantum study of the guaiacyl-moiety role. J. Phys. Chem. A 2009, 113, 13881–13891. [Google Scholar] [CrossRef]
- Trouillas, P.; Marsal, P.; Siri, D.; Lazzaroni, R.; Duroux, J.-L. A DFT study of the reactivity of OH groups in quercetin and taxifolin antioxidants: The specificity of the 3-OH site. Food Chem. 2006, 97, 679–688. [Google Scholar] [CrossRef]
- Kozlowski, D.; Trouillas, P.; Calliste, C.; Marsal, P.; Lazzaroni, R.; Duroux, J.-L. Density Functional Theory Study of the Conformational, Electronic, and Antioxidant Properties of Natural Chalcones. J. Phys. Chem. A 2007, 111, 1138–1145. [Google Scholar] [CrossRef]
- Leopoldini, M.; Russo, N.; Toscano, M. The molecular basis of working mechanism of natural polyphenolic antioxidants. Food Chem. 2011, 125, 288–306. [Google Scholar] [CrossRef]
- Rice-Evans, C.; Miller, N.J.; Paganga, G. Structure-antioxidant activity relationships of flavonoids and phenolic acids. Free Radic. BioMed. 1996, 20, 933–956. [Google Scholar] [CrossRef]
- Rastija, V.; Medić-Šarić, M. QSAR study of antioxidant activity of wine polyphenols. Eur. J. Med. Chem. 2009, 44, 400–408. [Google Scholar] [CrossRef]
- Anouar, E.; Raweh, S.; Bayach, I.; Taha, M.; Baharudin, M.S.; di Meo, F.; Hasan, M.H.; Adam, A.; Ismail, N.H.; Weber, J.-F.F.; et al. Antioxidant properties of phenolic Schiff bases: Structure–activity relationship and mechanism of action. J. Comput. Aid. Mol. Des. 2013, 27, 951–964. [Google Scholar] [CrossRef]
- Weber, K.C.; Honório, K.M.; da Silva, S.; Mercadante, R.; da Silva, A. Selection of quantum chemical descriptors by chemometric methods in the study of antioxidant activity of flavonoid compounds. Int. J. Quant. Chem. 2005, 103, 731–737. [Google Scholar] [CrossRef]
- Iuga, C.; Alvarez-Idaboy, J.R.l.; Russo, N. Antioxidant activity of trans-resveratrol toward hydroxyl and hydroperoxyl radicals: A quantum chemical and computational kinetics study. J. Org. Chem. 2012, 77, 3868–3877. [Google Scholar] [CrossRef]
- Leopoldini, M.; Chiodo, S.G.; Russo, N.; Toscano, M. Detailed investigation of the OH radical quenching by natural antioxidant caffeic acid studied by quantum mechanical models. J. Chem. Theory Comput. 2011, 7, 4218–4233. [Google Scholar] [CrossRef]
- Mayer, J.M.; Hrovat, D.A.; Thomas, J.L.; Borden, W.T. Proton-Coupled Electron Transfer versus Hydrogen Atom Transfer in Benzyl/Toluene, Methoxyl/Methanol, and Phenoxyl/Phenol Self-Exchange Reactions. J. Am. Chem. Soc. 2002, 124, 11142–11147. [Google Scholar] [CrossRef] [PubMed]
- Tishchenko, O.; Truhlar, D.G.; Ceulemans, A.; Nguyen, M.T. A unified perspective on the hydrogen atom transfer and proton-coupled electron transfer mechanisms in terms of topographic features of the ground and excited potential energy surfaces as exemplified by the reaction between phenol and radicals. J. Am. Chem. Soc. 2008, 130, 7000–7010. [Google Scholar] [CrossRef] [PubMed]
- Mendes, A.P.; Borges, R.S.; Neto, A.M.C.; de Macedo, L.G.; da Silva, A.B. The basic antioxidant structure for flavonoid derivatives. J. Mol. Mod. 2012, 18, 4073–4080. [Google Scholar] [CrossRef]
- Sarkar, A.; Middya, T.R.; Jana, A.D. A QSAR study of radical scavenging antioxidant activity of a series of flavonoids using DFT based quantum chemical descriptors—The importance of group frontier electron density. J. Mol. Mod. 2012, 18, 2621–2631. [Google Scholar] [CrossRef]
- Anouar, E.; Shah, S.; Hassan, N.; Moussaoui, N.; Ahmad, R.; Zulkefeli, M.; Weber, J.-F. Antioxidant Activity of Hispidin Oligomers from Medicinal Fungi: A DFT Study. Molecules 2014, 19, 3489–3507. [Google Scholar] [CrossRef]
- HyperChem, release 7.52 for Windows; Hypercube Inc: Gainesville, FL, USA, 2002.
- Ghose, A.K.; Pritchett, A.; Crippen, G.M. Atomic physicochemical parameters for three dimensional structure directed quantitative structure-activity relationships III: Modeling hydrophobic interactions. J. Comput. Chem. 1988, 9, 80–90. [Google Scholar] [CrossRef]
- Viswanadhan, V.N.; Ghose, A.K.; Revankar, G.R.; Robins, R.K. Atomic physicochemical parameters for three dimensional structure directed quantitative structure-activity relationships. 4. Additional parameters for hydrophobic and dispersive interactions and their application for an automated superposition of certain naturally occurring nucleoside antibiotics. J. Chem. Inf. Comput. Sci. 1989, 29, 163–172. [Google Scholar] [CrossRef]
- Parr, R.G.; Yang, W. Density-Functional Theory of Atoms and Molecules; Oxford University Press: Cary, NC, USA, 1989. [Google Scholar]
- Neshev, N.; Mineva, T. Metal-Ligand Interactions; Springer: Berlin/Heidelberg, Germany, 1996; pp. 361–405. [Google Scholar]
- Grigorov, M.; Weber, J.; Chermette, H.; Tronchet, J.M. Numerical evaluation of the internal orbitally resolved chemical hardness tensor in density functional theory. Inter. J. Quantum Chem. 1997, 61, 551–562. [Google Scholar] [CrossRef]
- Janak, J. Proof that ∂ E/∂ ni = ε in density-functional theory. Phys. Rev. B 1978, 18, 7165–7168. [Google Scholar] [CrossRef]
- De Luca, G.; Sicilia, E.; Russo, N.; Mineva, T. On the hardness evaluation in solvent for neutral and charged systems. J. Am. Chem. Soc. 2002, 124, 1494–1499. [Google Scholar] [CrossRef]
- Mineva, T.; Sicilia, E.; Russo, N. Density-functional approach to hardness evaluation and its use in the study of the maximum hardness principle. J. Am. Chem. Soc. 1998, 120, 9053–9058. [Google Scholar] [CrossRef]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Mennucci, B.; Petersson, G.A.; et al. Gaussian 09, Revision A.02; Gaussian, Inc.: Wallingford, CT, USA, 2009. [Google Scholar]
- Tomasi, J.; Mennucci, B.; Cammi, R. Quantum Mechanical Continuum Solvation Models. Chem. Rev. 2005, 105, 2999–3093. [Google Scholar] [CrossRef]
- Guerra, M.; Amorati, R.; Pedulli, G.F. Water Effect on the OH Dissociation Enthalpy of Para-Substituted Phenols: A DFT Study. J. Org. Chem. 2004, 69, 5460–5467. [Google Scholar] [CrossRef]
- Amić, A.; Marković, Z.; Dimitrić Marković, J.M.; Stepanić, V.; Lučić, B.; Amić, D. Towards an improved prediction of the free radical scavenging potency of flavonoids: The significance of double PCET mechanisms. Food Chem. 2014, 152, 578–585. [Google Scholar] [CrossRef]
- Ishihara, M.; Wakabayashi, H.; Motohashi, N.; Sakagami, H. Quantitative Structure-Cytotoxicity Relationship of Newly Synthesized Tropolones Determined by a Semiempirical Molecular-orbital Method (PM5). Anticancer Res. 2010, 30, 129–133. [Google Scholar] [PubMed]
- Yang, H.-L.; Chen, G.-H.; Li, Y.-Q. A quantum chemical and statistical study of ganoderic acids with cytotoxicity against tumor cell. Eur. J. Med. Chem. 2005, 40, 972–976. [Google Scholar] [CrossRef]
- Johnson, R.A.; Wichern, D.W. Applied Multivariate Statistical Analysis; Prentice Hall: Upper Saddle River, NJ, USA, 2002; Volume 5. [Google Scholar]
- Kowalski, B.; Bender, C. Pattern recognition. Powerful approach to interpreting chemical data. J. Am. Chem. Soc. 1972, 94, 5632–5639. [Google Scholar] [CrossRef]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Anouar, E.H. A Quantum Chemical and Statistical Study of Phenolic Schiff Bases with Antioxidant Activity against DPPH Free Radical. Antioxidants 2014, 3, 309-322. https://doi.org/10.3390/antiox3020309
Anouar EH. A Quantum Chemical and Statistical Study of Phenolic Schiff Bases with Antioxidant Activity against DPPH Free Radical. Antioxidants. 2014; 3(2):309-322. https://doi.org/10.3390/antiox3020309
Chicago/Turabian StyleAnouar, El Hassane. 2014. "A Quantum Chemical and Statistical Study of Phenolic Schiff Bases with Antioxidant Activity against DPPH Free Radical" Antioxidants 3, no. 2: 309-322. https://doi.org/10.3390/antiox3020309