Human Coronavirus Cell Receptors Provide Challenging Therapeutic Targets
Abstract
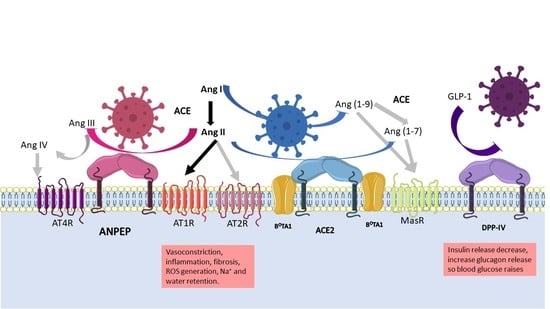
:1. Introduction
2. Human Protein Receptors for Coronaviruses Are Proteases
2.1. ANPEP
2.2. DPP-IV
2.3. ACE2
3. Structural Dissimilarity among the Human Coronaviruses
4. Role of Proteases in Physiological Processes
4.1. Role of the Proteases in Digestion
4.2. Role of the Proteases in Angiogenesis
4.3. Role of the Proteases in the RAAS System
4.4. Role of the Proteases in Metabolism
4.5. Role of the Proteases in the Respiratory System
5. Pathogenesis of the Viruses
6. Treatments for Coronavirus Infection
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cui, J.; Li, F.; Shi, Z.-L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019, 17, 181–192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Woo, P.C.Y.; Lau, S.K.P.; Lam, C.S.F.; Lau, C.C.Y.; Tsang, A.K.L.; Lau, J.H.N.; Bai, R.; Teng, J.L.L.; Tsang, C.C.C.; Wang, M.; et al. Discovery of Seven Novel Mammalian and Avian Coronaviruses in the Genus Deltacoronavirus Supports Bat Coronaviruses as the Gene Source of Alphacoronavirus and Betacoronavirus and Avian Coronaviruses as the Gene Source of Gammacoronavirus and Deltacoronavirus. J. Virol. 2012, 86, 3995–4008. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, X.; Ding, Y.; Du, J.; Fan, Y. 2020 update on human coronaviruses: One health, one world. Med. Nov. Technol. Devices 2020, 8, 100043. [Google Scholar] [CrossRef]
- Zhou, P.; Yang, X.-L.; Wang, X.-G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.-R.; Zhu, Y.; Li, B.; Huang, C.-L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Corman, V.M.; Baldwin, H.J.; Tateno, A.F.; Zerbinati, R.M.; Annan, A.; Owusu, M.; Nkrumah, E.E.; Maganga, G.D.; Oppong, S.; Adu-Sarkodie, Y.; et al. Evidence for an Ancestral Association of Human Coronavirus 229E with Bats. J. Virol. 2015, 89, 11858–11870. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tao, Y.; Shi, M.; Chommanard, C.; Queen, K.; Zhang, J.; Markotter, W.; Kuzmin, I.V.; Holmes, E.C.; Tong, S. Surveillance of Bat Coronaviruses in Kenya Identifies Relatives of Human Coronaviruses NL63 and 229E and Their Recombination History. J. Virol. 2017, 91, e01953-16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kapikian, A.Z. The coronaviruses. Dev. Biol. Stand 1975, 28, 42–64. [Google Scholar] [PubMed]
- Corman, V.M.; Muth, D.; Niemeyer, D.; Drosten, C. Hosts and Sources of Endemic Human Coronaviruses. Adv. Virus Res. 2018, 100, 163–188. [Google Scholar] [CrossRef]
- van der Hoek, L. Human coronaviruses: What do they cause? Antivir. Ther. 2007, 12, 651–658. [Google Scholar] [CrossRef]
- Corman, V.M.; Eckerle, I.; Bleicker, T.; Zaki, A.; Landt, O.; Eschbach-Bludau, M.; van Boheemen, S.; Gopal, R.; Ballhause, M.; Bestebroer, T.M.; et al. Detection of a novel human coronavirus by real-time reverse-transcription polymerase chain reaction. Eurosurveillance 2012, 17, 20285. [Google Scholar] [CrossRef]
- Singh, D.; Yi, S.V. On the origin and evolution of SARS-CoV-2. Exp. Mol. Med. 2021, 53, 537–547. [Google Scholar] [CrossRef] [PubMed]
- Li, F. Structure, Function, and Evolution of Coronavirus Spike Proteins. Annu. Rev. Virol. 2016, 3, 237–261. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Millet, J.K.; Whittaker, G.R. Host cell proteases: Critical determinants of coronavirus tropism and pathogenesis. Virus Res. 2015, 202, 120–134. [Google Scholar] [CrossRef]
- López-Cortés, G.I.; Palacios-Pérez, M.; Zamudio, G.S.; Veledíaz, H.F.; Ortega, E.; José, M.V. Neutral evolution test of the spike protein of SARS-CoV-2 and its implications in the binding to ACE2. Sci. Rep. 2021, 11, 18847. [Google Scholar] [CrossRef] [PubMed]
- López-Cortés, G.I.; Palacios-Pérez, M.; Veledíaz, H.F.; Hernández-Aguilar, M.; López-Hernández, G.R.; Zamudio, G.S.; José, M.V. The Spike Protein of SARS-CoV-2 Is Adapting Because of Selective Pressures. Vaccines 2022, 10, 864. [Google Scholar] [CrossRef]
- Jaimes, J.A.; André, N.M.; Chappie, J.S.; Millet, J.K.; Whittaker, G.R. Phylogenetic Analysis and Structural Modeling of SARS-CoV-2 Spike Protein Reveals an Evolutionary Distinct and Proteolytically Sensitive Activation Loop. J. Mol. Biol. 2020, 432, 3309–3325. [Google Scholar] [CrossRef]
- Millet, J.K.; Jaimes, A.J.; Whittaker, G.R. Molecular diversity of coronavirus host cell entry receptors. FEMS Microbiol. Rev. 2020, 45, fuaa057. [Google Scholar] [CrossRef]
- Peng, G.; Sun, D.; Rajashankar, K.R.; Qian, Z.; Holmes, K.V.; Li, F. Crystal structure of mouse coronavirus receptor-binding domain complexed with its murine receptor. Proc. Natl. Acad. Sci. USA 2011, 108, 10696–10701. [Google Scholar] [CrossRef] [Green Version]
- Qi, F.; Qian, S.; Zhang, S.; Zhang, Z. Single cell RNA sequencing of 13 human tissues identify cell types and receptors of human coronaviruses. Biochem. Biophys. Res. Commun. 2020, 526, 135–140. [Google Scholar] [CrossRef]
- Hoffmann, D.; Mereiter, S.; Jin Oh, Y.; Monteil, V.; Elder, E.; Zhu, R.; Canena, D.; Hain, L.; Laurent, E.; Grünwald-Gruber, C.; et al. Identification of lectin receptors for conserved SARS-CoV-2 glycosylation sites. bioRxiv 2021. [Google Scholar] [CrossRef]
- Daly, J.L.; Simonetti, B.; Klein, K.; Chen, K.E.; Williamson, M.K.; Antón-Plágaro, C.; Shoemark, D.K.; Simón-Gracia, L.; Bauer, M.; Hollandi, R.; et al. Neuropilin-1 is a host factor for SARS-CoV-2 infection. Science 2020, 370, 861–865. [Google Scholar] [CrossRef] [PubMed]
- Mina-Osorio, P. The moonlighting enzyme CD13: Old and new functions to target. Trends Mol. Med. 2008, 14, 361–371. [Google Scholar] [CrossRef] [PubMed]
- Gabrilovac, J.; Čupić, B.; Breljak, D.; Zekušić, M.; Boranić, M. Expression of CD13/aminopeptidase N and CD10/neutral endopeptidase on cultured human keratinocytes. Immunol. Lett. 2004, 91, 39–47. [Google Scholar] [CrossRef] [PubMed]
- Look, A.T.; A Ashmun, R.; Shapiro, L.H.; Peiper, S.C. Human myeloid plasma membrane glycoprotein CD13 (gp150) is identical to aminopeptidase N. J. Clin. Investig. 1989, 83, 1299–1307. [Google Scholar] [CrossRef]
- Griffin, J.D.; Ritz, J.; Nadler, L.M.; Schlossman, S.F. Expression of myeloid differentiation antigens on normal and malignant myeloid cells. J. Clin. Investig. 1981, 68, 932–941. [Google Scholar] [CrossRef] [Green Version]
- López-Cortés, G.I.; Díaz-Alvarez, L.; Ortega, E. Leukocyte Membrane Enzymes Play the Cell Adhesion Game. Front. Immunol. 2021, 12, 742292. [Google Scholar] [CrossRef]
- Chen, L.; Lin, Y.-L.; Peng, G.; Li, F. Structural basis for multifunctional roles of mammalian aminopeptidase N. Proc. Natl. Acad. Sci. USA 2012, 109, 17966–17971. [Google Scholar] [CrossRef] [Green Version]
- Tresnan, D.B.; Levis, R.; Holmes, K.V. Feline aminopeptidase N serves as a receptor for feline, canine, porcine, and human coronaviruses in serogroup I. J. Virol. 1996, 70, 8669. [Google Scholar] [CrossRef] [Green Version]
- Danziger, R.S. Aminopeptidase N in arterial hypertension. Heart Fail. Rev. 2008, 13, 293–298. [Google Scholar] [CrossRef]
- Lu, C.; Amin, M.A.; Fox, D.A. CD13/Aminopeptidase N Is a Potential Therapeutic Target for Inflammatory Disorders. J. Immunol. 2020, 204, 3–11. [Google Scholar] [CrossRef]
- Luan, Y.; Xu, W. The Structure and Main Functions of Aminopeptidase N. Curr. Med. Chem. 2007, 14, 639–647. [Google Scholar] [CrossRef] [PubMed]
- Garay-Canales, C.A.; Díaz-Alvarez, L.; Lopez-Cortes, G.I. Novel immunotherapy strategies involving matrix metalloproteinase (MMP) family. In Immunotherapy in Resistant Cancer: From the Lab Bench Work to Its Clinical Perspectives; Morales-Montor, J., Segovia-Mendoza, M., Eds.; Academic Press Inc.: Cambridge, MA, USA, 2020; Volume 2, pp. 227–247. [Google Scholar]
- Gabrilovac, J.; Breljak, D.; Čupić, B. Regulation of aminopeptidase N (EC 3.4.11.2; APN.; CD13) on the HL-60 cell line by TGF-β1. Int. Immunopharmacol. 2008, 8, 613–623. [Google Scholar] [CrossRef] [PubMed]
- Santos, N.; Langner, J.; Herrmann, M.; Riemann, D. Aminopeptidase N/CD13 is directly linked to signal transduction pathways in monocytes. Eur. PMC 2000, 201, 22–32. [Google Scholar] [CrossRef]
- Subramani, J.; Ghosh, M.; Rahman, M.M.; Caromile, L.A.; Gerber, C.; Rezaul, K.; Han, D.K.; Shapiro, L.H. Tyrosine Phosphorylation of CD13 Regulates Inflammatory Cell–Cell Adhesion and Monocyte Trafficking. J. Immunol. 2013, 191, 3905–3912. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghosh, M.; Gerber, C.; Rahman, M.M.; Vernier, K.M.; Pereira, F.E.; Subramani, J.; Caromile, L.A.; Shapiro, L.H. Molecular mechanisms regulating CD13-mediated adhesion. Immunology 2014, 142, 636–647. [Google Scholar] [CrossRef] [Green Version]
- Licona-Limón, I.; Garay-Canales, C.A.; Muñoz-Paleta, O.; Ortega, E. CD13 mediates phagocytosis in human monocytic cells. J. Leukoc. Biol. 2015, 98, 85–98. [Google Scholar] [CrossRef] [PubMed]
- Mina-Osorio, P.; Winnicka, B.; O’Conor, C.; Grant, C.L.; Vogel, L.K.; Rodriguez-Pinto, D.; Holmes, K.V.; Ortega, E.; Shapiro, L.H. CD13 is a novel mediator of monocytic/endothelial cell adhesion. J. Leukoc. Biol. 2008, 84, 448–459. [Google Scholar] [CrossRef] [Green Version]
- Mina-Osorio, P.; Shapiro, L.H.; Ortega, E. CD13 in cell adhesion: Aminopeptidase N (CD13) mediates homotypic aggregation of monocytic cells. J. Leukoc. Biol. 2006, 79, 719–730. [Google Scholar] [CrossRef]
- Pérez-Figueroa, E.; Álvarez-Carrasco, P.; Ortega, E. Crosslinking of membrane CD13 in human neutrophils mediates phagocytosis and production of reactive oxygen species, neutrophil extracellular traps and proinflammatory cytokines. Front. Immunol. 2022, 13, 6681. [Google Scholar] [CrossRef]
- Mendoza-Coronel, E.; Ortega, E. Macrophage Polarization Modulates FcγR- and CD13-Mediated Phagocytosis and Reactive Oxygen Species Production, Independently of Receptor Membrane Expression. Front. Immunol. 2017, 8, 303. [Google Scholar] [CrossRef]
- Cheng, H.C.; Abdel-Ghany, M.; Pauli, B.U. A Novel Consensus Motif in Fibronectin Mediates Dipeptidyl Peptidase IV Adhesion and Metastasis. J. Biol. Chem. 2003, 278, 24600–24607. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klemann, C.; Wagner, L.; Stephan, M.; von Hörsten, S. Cut to the chase: A review of CD26/dipeptidyl peptidase-4’s (DPP4) entanglement in the immune system. Clin. Exp. Immunol. 2016, 185, 1–21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iwata, B.S.; Morimoto, C. CD26/Dipeptidyl Peptidase IV in Context. J. Exp. Med. 1999, 190, 301–305. [Google Scholar] [CrossRef]
- Engel, M.; Hoffmann, T.; Wagner, L.; Wermann, M.; Heiser, U.; Kiefersauer, R.; Huber, R.; Bode, W.; Demuth, H.-U.; Brandstetter, H. The crystal structure of dipeptidyl peptidase IV (CD26) reveals its functional regulation and enzymatic mechanism. Proc. Natl. Acad. Sci. USA 2003, 100, 5063–5068. [Google Scholar] [CrossRef] [Green Version]
- Nistala, R.; Savin, V. Diabetes, hypertension, and chronic kidney disease progression: Role of DPP4. Am. J. Physiol. Physiol. 2017, 312, F661–F670. [Google Scholar] [CrossRef]
- Raha, A.A.; Chakraborty, S.; Henderson, J.; Mukaetova-Ladinska, E.; Zaman, S.; Trowsdale, J.; Raha-Chowdhury, R. Investigation of CD26, a potential SARS-CoV-2 receptor, as a biomarker of age and pathology. Biosci. Rep. 2020, 40, 20203092. [Google Scholar] [CrossRef] [PubMed]
- Abbott, C.A.; McCaughan, G.W.; Baker, E.; Sutherland, G.R. Genomic organization, exact localization, and tissue expression of the human CD26 (dipeptidyl peptidase IV) gene. Immunogenetics 1994, 40, 331–338. [Google Scholar] [CrossRef]
- Salgado, F.J.; Vela, E.; Martın, M.; Franco, R.; Nogueira, M.; Cordero, O.J. Mechanisms of CD26/dipeptidyl peptidase IV cytokine-dependent regulation on human activated lymphocytes. Cytokine 2000, 12, 1136–1141. [Google Scholar] [CrossRef]
- Fan, H.; Tansi, F.L.; Weihofen, W.A.; Böttcher, C.; Hu, J.; Martinez, J.; Saenger, W.; Reutter, W. Molecular mechanism and structural basis of interactions of dipeptidyl peptidase IV with adenosine deaminase and human immunodeficiency virus type-1 transcription transactivator. Eur. J. Cell Biol. 2012, 91, 265–273. [Google Scholar] [CrossRef]
- Morimoto, C.; Schlossman, S.F. The structure and function of CD26 in the T-cell immune response. Immunol. Rev. 1998, 161, 55–70. [Google Scholar] [CrossRef]
- Kameoka, J.; Tanaka, T.; Nojima, Y.; Schlossman, S.F.; Morimoto, C. Direct Association of Adenosine Deaminase with a T Cell Activation Antigen, CD26. Science 1993, 261, 466–469. [Google Scholar] [CrossRef]
- Lambeir, A.-M.; Durinx, C.; Scharpé, S.; De Meester, I. Dipeptidyl-Peptidase IV from Bench to Bedside: An Update on Structural Properties, Functions, and Clinical Aspects of the Enzyme DPP IV. Crit. Rev. Clin. Lab. Sci. 2003, 40, 209–294. [Google Scholar] [CrossRef]
- A Havre, P.; Dang, L.H.; Ohnuma, K.; Iwata, S.; Morimoto, C.; Dang, N.H. CD26 Expression on T-Anaplastic Large Cell Lymphoma (ALCL) Line Karpas 299 is associated with increased expression of Versican and MT1-MMP and enhanced adhesion. BMC Cancer 2013, 13, 517. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gine, S.; Mariño, M.; Mallol, J.; Canela, E.I.; Morimoto, C.; Callebaut, C.; Hovanessian, A.; Casadó, V.; Lluis, C.; Franco, R. Regulation of epithelial and lymphocyte cell adhesion by adenosine deaminase-CD26 interaction. Biochem. J. 2002, 361, 203–209. [Google Scholar] [CrossRef]
- Samavati, L.; Uhal, B.D. ACE2, Much More Than Just a Receptor for SARS-COV-2. Front. Cell. Infect. Microbiol. 2020, 10, 317. [Google Scholar] [CrossRef] [PubMed]
- Gheblawi, M.; Wang, K.; Viveiros, A.; Nguyen, Q.; Zhong, J.-C.; Turner, A.J.; Raizada, M.K.; Grant, M.B.; Oudit, G.Y. Angiotensin-Converting Enzyme 2: SARS-CoV-2 Receptor and Regulator of the Renin-Angiotensin System: Celebrating the 20th Anniversary of the Discovery of ACE2. Circ. Res. 2020, 126, 1456–1474. [Google Scholar] [CrossRef]
- Zhang, Q.; Gefter, J.; Sneddon, W.B.; Mamonova, T.; Friedman, P.A. ACE2 interaction with cytoplasmic PDZ protein enhances SARS-CoV-2 invasion. iScience 2021, 24, 102770. [Google Scholar] [CrossRef]
- Kliche, J.; Kuss, H.; Ali, M.; Ivarsson, Y. Cytoplasmic short linear motifs in ACE2 and integrin β3 link SARS-CoV-2 host cell receptors to mediators of endocytosis and autophagy. Sci. Signal 2021, 14, 1117. [Google Scholar] [CrossRef]
- Hrenak, J.; Simko, F. Renin–angiotensin system: An important player in the pathogenesis of acute respiratory distress syndrome. Int. J. Mol. Sci. 2020, 21, 8038. [Google Scholar] [CrossRef]
- Bourgonje, A.R.; Abdulle, A.E.; Timens, W.; Hillebrands, J.L.; Navis, G.J.; Gordijn, S.J.; Bolling, M.C.; Dijkstra, G.; Voors, A.A.; Osterhaus, A.D.; et al. Angiotensin-converting enzyme 2 (ACE2), SARS-CoV-2 and the pathophysiology of coronavirus disease 2019 (COVID-19). J. Pathol. 2020, 251, 228–248. [Google Scholar] [CrossRef]
- Brandi, M.L. Are sex hormones promising candidates to explain sex disparities in the COVID-19 pandemic? Rev. Endocr. Metab. Disord. 2022, 23, 171–183. [Google Scholar] [CrossRef] [PubMed]
- Purkayastha, A.; Sen, C.; Garcia Jr, G.; Langerman, J.; Shia, D.W.; Meneses, L.K.; Vijayaraj, P.; Durra, A.; Koloff, C.R.; Freund, D.R.; et al. Direct Exposure to SARS-CoV-2 and Cigarette Smoke Increases Infection Severity and Alters the Stem Cell-Derived Airway Repair Response. Cell Stem Cell 2020, 27, 869–875.e4. [Google Scholar] [CrossRef]
- Simõese Silva, A.C.; Silveira, K.D.; Ferreira, A.J.; Teixeira, M.M. ACE2, angiotensin-(1-7) and Mas receptor axis in inflammation and fibrosis. Br. J. Pharmacol. 2013, 169, 477–492. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, C.; Shine, M.; Pyle, A.M.; Zhang, Y. US-align: Universal structure alignments of proteins, nucleic acids, and macromolecular complexes. Nat. Methods 2022, 19, 1109–1115. [Google Scholar] [CrossRef]
- Zhang, Y.; Skolnick, J. TM-align: A protein structure alignment algorithm based on the TM-score. Nucleic Acids Res. 2005, 33, 2302–2309. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, T.; Perlot, T.; Rehman, A.; Trichereau, J.; Ishiguro, H.; Paolino, M.; Sigl, V.; Hanada, T.; Hanada, R.; Lipinski, S.; et al. ACE2 links amino acid malnutrition to microbial ecology and intestinal inflammation. Nature 2012, 487, 477–481. [Google Scholar] [CrossRef]
- Jando, J.; Camargo, S.; Herzog, B.; Verrey, F. Expression and regulation of the neutral amino acid transporter B0AT1 in rat small intestine. PLoS ONE 2017, 12, e0184845. [Google Scholar] [CrossRef] [Green Version]
- Du, Y.; Lu, C.; Morgan, R.L.; Stinson, W.A.; Campbell, P.L.; Cealey, E.; Fu, W.; Lepore, N.J.; Hervoso, J.L.; Cui, H.; et al. Angiogenic and Arthritogenic Properties of the Soluble Form of CD13. J. Immunol. 2019, 203, 360–369. [Google Scholar] [CrossRef]
- Bhagwat, S.V.; Lahdenranta, J.; Giordano, R.; Arap, W.; Pasqualini, R.; Shapiro, L.H. CD13/APN is activated by angiogenic signals and is essential for capillary tube formation. Blood J. Am. Soc. Hematol. 2001, 97, 652–659. [Google Scholar] [CrossRef] [Green Version]
- Fukasawa, K.; Fujii, H.; Saitoh, Y.; Koizumi, K.; Aozuka, Y.; Sekine, K.; Yamada, M.; Saiki, I.; Nishikawa, K. Aminopeptidase N (APN/CD13) is selectively expressed in vascular endothelial cells and plays multiple roles in angiogenesis. Cancer Lett. 2006, 243, 135–143. [Google Scholar] [CrossRef]
- Marchetti, C.; Di Carlo, A.; Facchiano, F.; Senatore, C.; De Cristofaro, R.; Luzi, A.; Federici, M.; Romani, M.; Napolitano, M.; Capogrossi, M.C.; et al. High mobility group box 1 is a novel substrate of dipeptidyl peptidase-IV. Diabetologia 2011, 55, 236–244. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saxena, A.; Fish, J.E.; White, M.D.; Yu, S.; Smyth, J.W.; Shaw, R.M.; DiMaio, J.M.; Srivastava, D. Stromal Cell-Derived Factor-1 Alpha is Cardioprotective After Myocardial Infarction. Circulation 2008, 117, 2224. [Google Scholar] [CrossRef] [PubMed]
- Hinsley, E.E.; de Oliveira, C.E.; Hunt, S.; Coletta, R.D.; Lambert, D.W. Angiotensin 1-7 inhibits angiotensin II-stimulated head and neck cancer progression. Eur. J. Oral Sci. 2017, 125, 247–257. [Google Scholar] [CrossRef] [PubMed]
- Kangussu, L.M.; Marzano, L.A.; Souza, C.F.; Dantas, C.C.; Miranda, A.S.; Simoes e Silva, A.C. The Renin-Angiotensin System and the Cerebrovascular Diseases: Experimental and Clinical Evidence. Protein Pept. Lett. 2019, 27, 463–475. [Google Scholar] [CrossRef]
- Meng, Y.; Yu, C.H.; Li, W.; Li, T.; Luo, W.; Huang, S.; Wu, P.S.; Cai, S.X.; Li, X. Angiotensin-converting enzyme 2/angiotensin-(1-7)/mas axis protects against lung fibrosis by inhibiting the MAPK/NF-κB pathway. Am. J. Respir. Cell Mol. Biol. 2014, 50, 723–736. [Google Scholar] [CrossRef]
- Passos-Silva, D.G.; Verano-Braga, T.; Santos, R.A.S. Angiotensin-(1–7): Beyond the cardio-renal actions. Clin. Sci. 2012, 124, 443–456. [Google Scholar] [CrossRef] [Green Version]
- Zhao, W.; Zhao, T.; Chen, Y.; Sun, Y. Angiotensin 1-7 Promotes Cardiac Angiogenesis Following Infarction. Curr. Vasc. Pharmacol. 2015, 13, 37–42. [Google Scholar] [CrossRef]
- Yu, C.; Tang, W.; Wang, Y.; Shen, Q.; Wang, B.; Cai, C.; Meng, X.; Zou, F. Downregulation of ACE2/Ang-(1-7)/Mas axis promotes breast cancer metastasis by enhancing store-operated calcium entry. Cancer Lett. 2016, 376, 268–277. [Google Scholar] [CrossRef]
- Yang, T.; Xu, C. Physiology and Pathophysiology of the Intrarenal Renin-Angiotensin System: An Update. J. Am. Soc. Nephrol. 2017, 28, 1040–1049. [Google Scholar] [CrossRef] [Green Version]
- Tsujimoto, M.; Goto, Y.; Maruyama, M.; Hattori, A. Biochemical and enzymatic properties of the M1 family of aminopeptidases involved in the regulation of blood pressure. Heart Fail. Rev. 2007, 13, 285–291. [Google Scholar] [CrossRef]
- Chaudhary, M. Anti-Hypertensive Potential and Epigenetics of Angiotensin II type 2 Receptor (AT2R). Curr. Hypertens. Rev. 2021, 17, 176–180. [Google Scholar] [CrossRef] [PubMed]
- Aroor, A.; Zuberek, M.; Duta, C.; Meuth, A.; Sowers, J.R.; Whaley-Connell, A.; Nistala, R. Angiotensin II Stimulation of DPP4 Activity Regulates Megalin in the Proximal Tubules. Int. J. Mol. Sci. 2016, 17, 780. [Google Scholar] [CrossRef] [Green Version]
- Valencia, I.; Peiró, C.; Lorenzo, Ó.; Sánchez-Ferrer, C.F.; Eckel, J.; Romacho, T. DPP4 and ACE2 in Diabetes and COVID-19: Therapeutic Targets for Cardiovascular Complications? Front. Pharmacol. 2020, 11, 1161. [Google Scholar] [CrossRef] [PubMed]
- Ikushima, H.; Munakata, Y.; Ishii, T.; Iwata, S.; Terashima, M.; Tanaka, H.; Schlossman, S.F.; Morimoto, C. Internalization of CD26 by mannose 6-phosphate/insulin-like growth factor II receptor contributes to T cell activation. Proc. Natl. Acad. Sci. USA 2000, 97, 8439–8444. [Google Scholar] [CrossRef] [Green Version]
- Bindom, S.M.; Lazartigues, E. The sweeter side of ACE2: Physiological evidence for a role in diabetes. Mol. Cell. Endocrinol. 2009, 302, 193–202. [Google Scholar] [CrossRef]
- Narula, S.; Yusuf, S.; Chong, M.; Ramasundarahettige, C.; Rangarajan, S.; Bangdiwala, I.S.; van Eikels, M.; Leineweber, K.; Wu, A.; Pigeyre, M.; et al. Plasma ACE2 and risk of death or cardiometabolic diseases: A case-cohort analysis. Lancet 2020, 396, 968–976. [Google Scholar] [CrossRef]
- Park, S.E.; Kim, W.J.; Park, S.W.; Park, J.W.; Lee, N.; Park, C.Y.; Youn, B.S. High urinary ACE2 concentrations are associated with severity of glucose intolerance and microalbuminuria. Eur. J. Endocrinol. Eur. Fed. Endocr. Soc. 2013, 168, 203–210. [Google Scholar] [CrossRef] [Green Version]
- Cherney, D.Z.; Xiao, F.; Zimpelmann, J.; Har, R.L.; Lai, V.; Scholey, J.W.; Reich, H.N.; Burns, K.D. Urinary ACE2 in healthy adults and patients with uncomplicated type 1 diabetes. Can. J. Physiol. Pharmacol. 2014, 92, 703–706. [Google Scholar] [CrossRef] [PubMed]
- Hui, K.P.; Ho, J.C.; Cheung, M.C.; Ng, K.C.; Ching, R.H.; Lai, K.L.; Kam, T.T.; Gu, H.; Sit, K.Y.; Hsin, M.K.; et al. SARS-CoV-2 Omicron variant replication in human bronchus and lung ex vivo. Nature 2022, 603, 715–720. [Google Scholar] [CrossRef]
- Gong, T.; Zhang, X.; Peng, Z.; Ye, Y.; Liu, R.; Yang, Y.; Chen, Z.; Zhang, Z.; Hu, H.; Yin, S.; et al. Macrophage-derived exosomal aminopeptidase N aggravates sepsis-induced acute lung injury by regulating necroptosis of lung epithelial cell. Commun. Biol. 2022, 5, 543. [Google Scholar] [CrossRef]
- Devarakonda, C.K.V.; Meredith, E.; Ghosh, M.; Shapiro, L.H. Coronavirus Receptors as Immune Modulators. J. Immunol. 2021, 206, 923–929. [Google Scholar] [CrossRef] [PubMed]
- Takeda, M. Proteolytic activation of SARS-CoV-2 spike protein. Microbiol. Immunol. 2021, 66, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Jackson, C.B.; Farzan, M.; Chen, B.; Choe, H. Mechanisms of SARS-CoV-2 entry into cells. Nat. Rev. Mol. Cell Biol. 2022, 23, 3–20. [Google Scholar] [CrossRef]
- Gupta, A.; Madhavan, M.V.; Sehgal, K.; Nair, N.; Mahajan, S.; Sehrawat, T.S.; Bikdeli, B.; Ahluwalia, N.; Ausiello, J.C.; Wan, E.Y.; et al. Extrapulmonary manifestations of COVID-19. Nat. Med. 2020, 26, 1017–1032. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Channappanavar, R.; Kanneganti, T.-D. Coronaviruses: Innate Immunity, Inflammasome Activation, Inflammatory Cell Death, and Cytokines. Trends Immunol. 2020, 41, 1083–1099. [Google Scholar] [CrossRef]
- Channappanavar, R.; Fehr, A.R.; Zheng, J.; Wohlford-Lenane, C.; Abrahante, J.E.; Mack, M.; Sompallae, R.; McCray, P.B.; Meyerholz, D.K.; Perlman, S. IFN-I response timing relative to virus replication determines MERS coronavirus infection outcomes. J. Clin. Investig. 2019, 129, 3625–3639. [Google Scholar] [CrossRef]
- Cervantes-Barragan, L.; Züst, R.; Weber, F.; Spiegel, M.; Lang, K.S.; Akira, S.; Thiel, V.; Ludewig, B. Control of coronavirus infection through plasmacytoid dendritic-cell–derived type I interferon. Blood 2006, 109, 1131–1137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shah, V.K.; Firmal, P.; Alam, A.; Ganguly, D.; Chattopadhyay, S. Overview of Immune Response During SARS-CoV-2 Infection: Lessons From the Past. Front Immunol 2020, 11, 1949. [Google Scholar] [CrossRef]
- Arunachalam, P.S.; Wimmers, F.; Mok, C.K.P.; Perera, R.A.P.M.; Scott, M.; Hagan, T.; Sigal, N.; Feng, Y.; Bristow, L.; Tsang, O.T.-Y.; et al. Systems biological assessment of immunity to mild versus severe COVID-19 infection in humans. Science 2020, 369, 1210–1220. [Google Scholar] [CrossRef]
- Lee, J.S.; Park, S.; Jeong, H.W.; Ahn, J.Y.; Choi, S.J.; Lee, H.; Choi, B.; Nam, S.K.; Sa, M.; Kwon, J.-S.; et al. Immunophenotyping of COVID-19 and influenza highlights the role of type I interferons in development of severe COVID-19. Sci. Immunol. 2020, 5, 1554. [Google Scholar] [CrossRef]
- Hadjadj, J.; Yatim, N.; Barnabei, L.; Corneau, A.; Boussier, J.; Smith, N.; Péré, H.; Charbit, B.; Bondet, V.; Chenevier-Gobeaux, C.; et al. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science 2020, 369, 718–724. [Google Scholar] [CrossRef] [PubMed]
- Channappanavar, R.; Fehr, A.R.; Vijay, R.; Mack, M.; Zhao, J.; Meyerholz, D.K.; Perlman, S. Dysregulated Type I Interferon and Inflammatory Monocyte-Macrophage Responses Cause Lethal Pneumonia in SARS-CoV-Infected Mice. Cell Host Microbe 2016, 19, 181–193. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tay, M.Z.; Poh, C.M.; Rénia, L.; Macary, P.A.; Ng, L.F.P. The trinity of COVID-19: Immunity, inflammation and intervention. Nat. Rev. Immunol. 2020, 20, 363–374. [Google Scholar] [CrossRef]
- Al-Kuraishy, H.M.; Al-Gareeb, A.I.; Al-Maiahy, T.J.; Alexiou, A.; Mukerjee, N.; Batiha, G.E.-S. An insight into the placental growth factor (PlGf)/angii axis in Covid-19: A detrimental intersection. Biotechnol. Genet. Eng. Rev. 2022, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Riemann, D.; Hansen, G.H.; NIELS-Christiansen, L.L.; Thorsen, E.; Immerdal, L.; Santos, A.N.; Kehlen, A.; Langner, J.; Danielsen, E.M. Caveolae/lipid rafts in fibroblast-like synoviocytes: Ectopeptidase-rich membrane microdomains. Biochem. J. 2001, 354, 47–55. [Google Scholar] [CrossRef] [PubMed]
- Mehrabadi, M.E.; Hemmati, R.; Tashakor, A.; Homaei, A.; Yousefzadeh, M.; Hemati, K.; Hosseinkhani, S. Induced dysregulation of ACE2 by SARS-CoV-2 plays a key role in COVID-19 severity. Biomed. Pharmacother. 2021, 137, 11363. [Google Scholar] [CrossRef] [PubMed]
- Norooznezhad, A.H.; Mansouri, K. Endothelial cell dysfunction, coagulation, and angiogenesis in coronavirus disease 2019 (COVID-19). Microvasc. Res. 2021, 137, 104188. [Google Scholar] [CrossRef] [PubMed]
- Pelle, M.C.; Zaffina, I.; Lucà, S.; Forte, V.; Trapanese, V.; Melina, M.; Giofrè, F.; Arturi, F. Endothelial Dysfunction in COVID-19: Potential Mechanisms and Possible Therapeutic Options. Life 2022, 12, 1605. [Google Scholar] [CrossRef]
- Brand, J.M.V.D.; Smits, S.L.; Haagmans, B.L. Pathogenesis of Middle East respiratory syndrome coronavirus. J. Pathol. 2014, 235, 175–184. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.; Chi, J.; Lv, W.; Wang, Y. Obesity and diabetes as high-risk factors for severe coronavirus disease 2019 (COVID-19). Diabetes Metab. Res. Rev. 2021, 37, e3377. [Google Scholar] [CrossRef]
- Frydrych, L.M.; Bian, G.; O’Lone, D.E.; Ward, P.A.; Delano, M.J. Obesity and type 2 diabetes mellitus drive immune dysfunction, infection development, and sepsis mortality. J. Leukoc. Biol. 2018, 104, 525–534. [Google Scholar] [CrossRef] [PubMed]
- Barron, E.; Bakhai, C.; Kar, P.; Weaver, A.; Bradley, D.; Ismail, H.; Knighton, P.; Holman, N.; Khunti, K.; Sattar, N.; et al. Associations of type 1 and type 2 diabetes with COVID-19-related mortality in England: A whole-population study. Lancet Diabetes Endocrinol. 2020, 8, 813–822. [Google Scholar] [CrossRef]
- Booth, C.M. Clinical Features and Short-term Outcomes of 144 Patients With SARS in the Greater Toronto Area. JAMA 2003, 289, 2801–2809. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferreira, A.J.; Santos, R.A.; Bradford, C.N.; Mecca, A.P.; Sumners, C.; Katovich, M.J.; Raizada, M.K. Therapeutic Implications of the Vasoprotective Axis of the Renin-Angiotensin System in Cardiovascular Diseases. Hypertension 2010, 55, 207–213. [Google Scholar] [CrossRef] [PubMed]
- Santos, P.C.J.L.; Krieger, J.E.; Pereira, A.C. Renin^|^ndash;Angiotensin System, Hypertension, and Chronic Kidney Disease: Pharmacogenetic Implications. J. Pharmacol. Sci. 2012, 120, 77–88. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frühbeck, G.; Catalán, V.; Valentí, V.; Moncada, R.; Gómez-Ambrosi, J.; Becerril, S.; Silva, C.; Portincasa, P.; Escalada, J.; Rodríguez, A. FNDC4 and FNDC5 reduce SARS-CoV-2 entry points and spike glycoprotein S1-induced pyroptosis, apoptosis, and necroptosis in human adipocytes. Cell Mol. Immunol. 2021, 18, 2457–2459. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Colón, G.J.; Ratnasiri, K.; Chen, H.; Jiang, S.; Zanley, E.; Rustagi, A.; Verma, R.; Chen, H.; Andrews, J.R.; Mertz, K.D.; et al. SARS-CoV-2 infection drives an inflammatory response in human adipose tissue through infection of adipocytes and macrophages. Sci. Transl. Med. 2022, 14, eabm9151. [Google Scholar] [CrossRef]
- Andersen, K.G.; Rambaut, A.; Lipkin, W.I.; Holmes, E.C.; Garry, R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020, 26, 450–452. [Google Scholar] [CrossRef] [Green Version]
- Kassir, R. Risk of COVID-19 for patients with obesity. Obes. Rev. 2020, 21, e13034. [Google Scholar] [CrossRef] [Green Version]
- Boni, M.F.; Lemey, P.; Jiang, X.; Lam, T.T.Y.; Perry, B.W.; Castoe, T.A.; Rambaut, A.; Robertson, D.L. Evolutionary origins of the SARS-CoV-2 sarbecovirus lineage responsible for the COVID-19 pandemic. Nat. Microbiol. 2020, 5, 1408–1417. [Google Scholar] [CrossRef]
- Ryan, P.M.D.; Caplice, N.M. Is Adipose Tissue a Reservoir for Viral Spread, Immune Activation, and Cytokine Amplification in Coronavirus Disease 2019? Obesity (Silver Spring) 2020, 28, 1191–1194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shinozaki, K.; Ayajiki, K.; Nishio, Y.; Sugaya, T.; Kashiwagi, A.; Okamura, T. Evidence for a Causal Role of the Renin-Angiotensin System in Vascular Dysfunction Associated With Insulin Resistance. Hypertension 2004, 43, 255–262. [Google Scholar] [CrossRef] [PubMed]
- Engeli, S.; Negrel, R.; Sharma, A.M. Physiology and Pathophysiology of the Adipose Tissue Renin-Angiotensin System. Hypertension 2000, 35, 1270–1277. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rathod, M.A.; Rogers, P.M.; Vangipuram, S.D.; McAllister, E.J.; Dhurandhar, N.V. Adipogenic Cascade Can Be Induced Without Adipogenic Media by a Human Adenovirus. Obesity 2009, 17, 657–664. [Google Scholar] [CrossRef] [Green Version]
- Iyer, A.; Fairlie, D.P.; Prins, J.B.; Hammock, B.D.; Brown, L. Inflammatory lipid mediators in adipocyte function and obesity. Nat. Rev. Endocrinol. 2010, 6, 71–82. [Google Scholar] [CrossRef]
- Wu, C.T.; Lidsky, P.V.; Xiao, Y.; Lee, I.T.; Cheng, R.; Nakayama, T.; Jiang, S.; Demeter, J.; Bevacqua, R.J.; Chang, C.A.; et al. SARS-CoV-2 infects human pancreatic β cells and elicits β cell impairment. Cell Metab. 2021, 33, 1565–1576.e5. [Google Scholar] [CrossRef]
- Tang, X.; Uhl, S.; Zhang, T.; Xue, D.; Li, B.; Vandana, J.J.; Acklin, J.A.; Bonnycastle, L.L.; Narisu, N.; Erdos, M.R.; et al. SARS-CoV-2 infection induces beta cell transdifferentiation. Cell Metab. 2021, 33, 1577–1591.e7. [Google Scholar] [CrossRef]
- Popkin, B.M.; Du, S.; Green, W.D.; Beck, M.A.; Algaith, T.; Herbst, C.H.; Alsukait, R.F.; Alluhidan, M.; Alazemi, N.; Shekar, M. Individuals with obesity and COVID-19: A global perspective on the epidemiology and biological relationships. Obes. Rev. 2020, 21, e13128. [Google Scholar] [CrossRef]
- Ana, H.D.A.; de Medeiros, A.F.; Medeiros, I.; de Lima, V.C.; Luz, A.B.; Maciel, B.L.; Passos, T.S. Tamarind ( Tamarindus indica L.) Seed a Candidate Protein Source with Potential for Combating SARS-CoV-2 Infection in Obesity. Drug Target Insights 2021, 15, 5–12. [Google Scholar]
- Jia, H.; Neptune, E.; Cui, H. Targeting ACE2 for COVID-19 Therapy: Opportunities and Challenges. Am. J. Respir. Cell Mol. Biol. 2021, 64, 416–425. [Google Scholar] [CrossRef]
- Asai, A.; Konno, M.; Ozaki, M.; Otsuka, C.; Vecchione, A.; Arai, T.; Kitagawa, T.; Ofusa, K.; Yabumoto, M.; Hirotsu, T.; et al. COVID-19 Drug Discovery Using Intensive Approaches. Int. J. Mol. Sci. 2020, 21, 2839. [Google Scholar] [CrossRef] [PubMed]
- Vaduganathan, M.; Vardeny, O.; Michel, T.; McMurray, J.J.; Pfeffer, M.A.; Solomon, S.D. Renin–Angiotensin–Aldosterone System Inhibitors in Patients with COVID-19. N. Engl. J. Med. 2020, 382, 1653–1659. [Google Scholar] [CrossRef]
- Nojomi, M.; Yassin, Z.; Keyvani, H.; Makiani, M.J.; Roham, M.; Laali, A.; Dehghan, N.; Navaei, M.; Ranjbar, M. Effect of Arbidol (Umifenovir) on COVID-19: A randomized controlled trial. BMC Infect. Dis. 2020, 20, 954. [Google Scholar] [CrossRef] [PubMed]
- Pang, X.C.; Zhang, H.X.; Zhang, Z.; Rinkiko, S.; Cui, Y.M.; Zhu, Y.Z. The Two-Way Switch Role of ACE2 in the Treatment of Novel Coronavirus Pneumonia and Underlying Comorbidities. Molecules 2021, 26, 142. [Google Scholar] [CrossRef]
- El Ouafi, Z.; Rhalem, W.; Habib, N.; Azami, A.I.; Sehli, S.; Al Idrissi, N.; Bakkali, F.; Abderrazak, R.; Merzouki, M.; Allali, I.; et al. Molecular Modeling Targeting the ACE2 Receptor with Cannabis sativa’s Active Ingredients for Antiviral Drug Discovery against SARS-CoV-2 Infections. Bioinform. Biol. Insights 2022, 16, 11779322221145380. [Google Scholar] [CrossRef] [PubMed]
- Krishnamurthy, S.; Lockey, R.F.; Kolliputi, N. Soluble ACE2 as a potential therapy for COVID-19. Am. J. Physiol. Physiol. 2021, 320, C279–C281. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.; Benthin, C.; Zeno, B.; Albertson, T.E.; Boyd, J.; Christie, J.; Hall, R.; Poirier, G.; Ronco, J.J.; Tidswell, M.; et al. A pilot clinical trial of recombinant human angiotensin-converting enzyme 2 in acute respiratory distress syndrome. Crit. Care 2017, 21, 234. [Google Scholar] [CrossRef] [Green Version]
- Murça, T.M.; Moraes, P.L.; Capuruço, C.A.; Santos, S.H.; Melo, M.B.; Santos, R.A.; Shenoy, V.; Katovich, M.J.; Raizada, M.K.; Ferreira, A.J. Oral administration of an angiotensin-converting enzyme 2 activator ameliorates diabetes-induced cardiac dysfunction. Regul. Pept. 2012, 177, 107–115. [Google Scholar] [CrossRef] [Green Version]
- Iwanaga, N.; Cooper, L.; Rong, L.; Maness, N.J.; Beddingfield, B.; Qin, Z.; Crabtree, J.; Tripp, R.A.; Yang, H.; Blair, R.; et al. ACE2-IgG1 fusions with improved in vitro and in vivo activity against SARS-CoV-2. iScience 2022, 25, 103670. [Google Scholar] [CrossRef]
- de Ligt, M.; Hesselink, M.K.; Jorgensen, J.; Hoebers, N.; Blaak, E.E.; Goossens, G.H. Resveratrol supplementation reduces ACE2 expression in human adipose tissue. Adipocyte 2021, 10, 408–411. [Google Scholar] [CrossRef]
- Jardine, M.J.; Kotwal, S.S.; Bassi, A.; Hockham, C.; Jones, M.; Wilcox, A.; Pollock, C.; Burrell, L.M.; McGree, J.; Rathore, V.; et al. Angiotensin receptor blockers for the treatment of covid-19: Pragmatic, adaptive, multicentre, phase 3, randomised controlled trial. BMJ 2022, 379, e072175. [Google Scholar] [CrossRef]
- Kotfis, K.; Lechowicz, K.; Drożdżal, S.; Niedźwiedzka-Rystwej, P.; Wojdacz, T.K.; Grywalska, E.; Biernawska, J.; Wiśniewska, M. COVID-19—The potential beneficial therapeutic effects of spironolactone during SARS-CoV-2 infection. Pharmaceuticals 2021, 14, 71. [Google Scholar] [CrossRef]
- Saeedi Saravi, S.S.; Beer, J.H. Apelin-potential therapy for COVID-19? J. Mol. Cell Cardiol. 2020, 145, 84–87. [Google Scholar] [CrossRef]
- Valdez-Cruz, N.A.; Garcia-Hernandez, E.; Espitia, C.; Cobos-Marín, L.; Altamirano, C.; Bando-Campos, C.G.; Cofas-Vargas, L.F.; Coronado-Aceves, E.W.; Gonzalez-Hernandez, R.A.; Hernandez-Peralta, P.; et al. Integrative overview of antibodies against SARS-CoV-2 and their possible applications in COVID-19 prophylaxis and treatment. Microb. Cell Fact. 2021, 20, 88. [Google Scholar] [CrossRef]
- Cascella, M.; Rajnik, M.; Dulebohn, S.C.; di Napoli, R. Features, Evaluation, and Treatment of Coronavirus (COVID-19); StatPearls Publishing: Treasure Island, FL, USA, 2022. [Google Scholar]
- Berry, J.D.; Hay, K.; Rini, J.M.; Yu, M.; Wang, L.; Plummer, F.A.; Corbett, C.R.; Andonov, A. Neutralizing epitopes of the SARS-CoV S-protein cluster independent of repertoire, antigen structure or mAb technology. MAbs 2010, 2, 53–66. [Google Scholar] [CrossRef] [Green Version]
- Du, L.; Yang, Y.; Zhou, Y.; Lu, L.; Li, F.; Jiang, S. MERS-CoV spike protein: A key target for antivirals. Expert Opin. Ther. Targets 2017, 21, 131–143. [Google Scholar] [CrossRef] [Green Version]
- Wang, P.; Casner, R.G.; Nair, M.S.; Yu, J.; Guo, Y.; Wang, M.; Chan, J.F.W.; Cerutti, G.; Iketani, S.; Liu, L.; et al. A monoclonal antibody that neutralizes SARS-CoV-2 variants, SARS-CoV, and other sarbecoviruses. Emerg. Microbes Infect. 2022, 11, 147–157. [Google Scholar] [CrossRef]
- Akbar, R.; Bashour, H.; Rawat, P.; Robert, P.A.; Smorodina, E.; Cotet, T.-S.; Flem-Karlsen, K.; Frank, R.; Mehta, B.B.; Vu, M.H.; et al. Progress and challenges for the machine learning-based design of fit-for-purpose monoclonal antibodies. Mabs 2022, 14. [Google Scholar] [CrossRef]
- Pang, J.; Liu, M.; Ling, W.; Jin, T. Friend or foe? ACE2 inhibitors and GLP-1R agonists in COVID-19 treatment. Obes. Med. 2021, 22, 100312. [Google Scholar] [CrossRef]
- Haber, P.K.; Ye, M.; Wysocki, J.; Maier, C.; Haque, S.K.; Batlle, D. Angiotensin-converting enzyme 2-independent action of presumed angiotensin-converting enzyme 2 activators: Studies in vivo, ex vivo, and in vitro. Hypertension 2014, 63, 774–782. [Google Scholar] [CrossRef] [Green Version]
- Shapira, T.; Monreal, I.A.; Dion, S.P.; Buchholz, D.W.; Imbiakha, B.; Olmstead, A.D.; Jager, M.; Désilets, A.; Gao, G.; Martins, M.; et al. A TMPRSS2 inhibitor acts as a pan-SARS-CoV-2 prophylactic and therapeutic. Nature 2022, 605, 340–348. [Google Scholar] [CrossRef]
- Fraser, B.J.; Beldar, S.; Seitova, A.; Hutchinson, A.; Mannar, D.; Li, Y.; Kwon, D.; Tan, R.; Wilson, R.P.; Leopold, K.; et al. Structure and activity of human TMPRSS2 protease implicated in SARS-CoV-2 activation. Nat. Chem. Biol. 2022, 18, 963–971. [Google Scholar] [CrossRef]
- Howell, R.; Clarke, M.A.; Reuschl, A.K.; Chen, T.; Abbott-Imboden, S.; Singer, M.; Lowe, D.M.; Bennett, C.L.; Chain, B.; Jolly, C.; et al. Executable network of SARS-CoV-2-host interaction predicts drug combination treatments. Npj Digit. Med. 2022, 5, 18. [Google Scholar] [CrossRef]
- Leach, D.A.; Mohr, A.; Giotis, E.S.; Cil, E.; Isac, A.M.; Yates, L.L.; Barclay, W.S.; Zwacka, R.M.; Bevan, C.L.; Brooke, G.N. The antiandrogen enzalutamide downregulates TMPRSS2 and reduces cellular entry of SARS-CoV-2 in human lung cells. Nat. Commun. 2021, 12, 4068. [Google Scholar] [CrossRef]
- Chen, Y.; Lear, T.B.; Evankovich, J.W.; Larsen, M.B.; Lin, B.; Alfaras, I.; Kennerdell, J.R.; Salminen, L.; Camarco, D.P.; Lockwood, K.C.; et al. A high-throughput screen for TMPRSS2 expression identifies FDA-approved compounds that can limit SARS-CoV-2 entry. Nat. Commun. 2021, 12, 3907. [Google Scholar] [CrossRef]
- Meng, B.; Abdullahi, A.; Ferreira, I.A.; Goonawardane, N.; Saito, A.; Kimura, I.; Yamasoba, D.; Gerber, P.P.; Fatihi, S.; Rathore, S.; et al. Altered TMPRSS2 usage by SARS-CoV-2 Omicron impacts infectivity and fusogenicity. Nature 2022, 603, 706–714. [Google Scholar] [CrossRef]
- Zhao, M.M.; Zhu, Y.; Zhang, L.; Zhong, G.; Tai, L.; Liu, S.; Yin, G.; Lu, J.; He, Q.; Li, M.J.; et al. Novel cleavage sites identified in SARS-CoV-2 spike protein reveal mechanism for cathepsin L-facilitated viral infection and treatment strategies. Cell Discov. 2022, 8, 53. [Google Scholar] [CrossRef]
- Zhao, M.M.; Yang, W.L.; Yang, F.Y.; Zhang, L.; Huang, W.J.; Hou, W.; Fan, C.F.; Jin, R.H.; Feng, Y.M.; Wang, Y.C.; et al. Cathepsin L plays a key role in SARS-CoV-2 infection in humans and humanized mice and is a promising target for new drug development. Signal Transduct. Target. Ther. 2021, 6, 134. [Google Scholar] [CrossRef]
- Zadeh, N.M.; Asl, N.S.M.; Forouharnejad, K.; Ghadimi, K.; Parsa, S.; Mohammadi, S.; Omidi, A. Mechanism and adverse effects of COVID-19 drugs: A basic review. Int. J. Physiol. Pathophysiol. Pharmacol. 2021, 13, 102. [Google Scholar]
- Forni, D.; Cagliani, R.; Clerici, M.; Sironi, M. Molecular Evolution of Human Coronavirus Genomes. Trends Microbiol. 2016, 25, 35–48. [Google Scholar] [CrossRef] [Green Version]
- Heilmann, E.; Costacurta, F.; Moghadasi, S.A.; Ye, C.; Pavan, M.; Bassani, D.; Volland, A.; Ascher, C.; Weiss, A.K.H.; Bante, D.; et al. SARS-CoV-2 3CL pro mutations selected in a VSV-based system confer resistance to nirmatrelvir, ensitrelvir, and GC376. Sci. Transl. Med. 2022, 15. [Google Scholar] [CrossRef] [PubMed]
- Silveira, M.A.D.; De Jong, D.; Berretta, A.A.; dos Santos Galvão, E.B.; Ribeiro, J.C.; Cerqueira-Silva, T.; Amorim, T.C.; da Conceicao, L.F.M.R.; Gomes, M.M.D.; Teixeira, M.B.; et al. Efficacy of Brazilian green propolis (EPP-AF®) as an adjunct treatment for hospitalized COVID-19 patients: A randomized, controlled clinical trial. Biomed. Pharmacother. 2021, 138, 111526. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Fernandez, J.; Ochoa, J.; Ojeda, M.L.; Nogales, F.; Carreras, O.; Díaz-Castro, J. Inflammation and oxidative stress, the links between obesity and COVID-19: A narrative review. J. Physiol. Biochem. 2022, 78, 581–591. [Google Scholar] [CrossRef] [PubMed]



| Coronavirus | Genera | Identified | Most Probable Ancestor Host | Receptor | Ref |
|---|---|---|---|---|---|
| HCoV-229 E | Alphacoronavirus | 1965 | Bats Hipposideros and camelids | ANPEP/CD13 | [1,5,6] |
| HCoV-OC43 | Betacoronavirus | 1967 | Rodents and swine | Sialic acid | [7] |
| SARS-CoV | Betacoronavirus | 2002 | Bat Rhinolophus and civet | ACE2 | [8] |
| HCoV-NL63 | Alphacoronavirus | 2004 | Bats Triaenops | ACE2 | [6] |
| HCoV-HKU1 | Betacoronavirus | 2005 | Rodents | Sialic acid | [8,9] |
| MERS-CoV | Betacoronavirus | 2012 | Bat and camel | DPP IV/CD26 | [10] |
| SARS-CoV-2 | Betacoronavirus | 2019 | Bat Rhinolophus affini and | ACE2 | [11] |
| Drug Family | Drug | Action Mechanism | Coronavirus Treatment | Other Uses | Ref |
|---|---|---|---|---|---|
| ANPEP inhibitors | Ubenimex | Inhibit the catalytic activity of ANPEP. | NR | Cancer treatment in study. | [132] |
| DPP-IV inhibitors | Gliptins | Inhibit the catalytic activity of membrane and soluble DPP-IV. Reduce glucose levels. Suppress T cell proliferation and pro-inflammatory cytokine synthesis. | Experimental models | Diabetes type 2 treatment. Anti-inflammatory drug. | [84] |
| ACE2 inhibitors | MLN-4760 | Inhibit catalytic activity of ACE2. | Experimental models | Treatment for hypertension, cardiovascular diseases, chronic kidney disease, and diabetes mellitus. | [133] |
| ACE2 internalization inhibitor | Arbidol | Interacts with aromatic residues within the viral proteins and the plasma membrane. Suppresses the expression of IL-1β, IL-6, IL-12, and TNF-α. | Experimental models | Treatment for various virus including influenza, Ebola virus and hepatitis C virus. | [133,134] |
| ACE2 viral-binding-site blockers | NAAE (N-[2-aminoethyl]-1 aziridine ethanamine), 6-Prenylapigenin, cannabivarin and Δ8-tetrahydrocannabinolic acid-A | Block the viral docking sites of ACE2 and thus the membrane fusion with the cell membranes. | Experimental and computational models | NR | [133,135,136] |
| ACE2 as decoy molecules | sACE2 (GSK2586881, APN01, dimeric ACE2) | Binds to extracellular viral S proteins, thus neutralizing the virus. Ang II decreases and Ang (1–7) increases. | GSK2586881 and APN01 in Phase II clinical trial | NR | [137,138,139] |
| Chimeric sACE2- IgG Fc fragment | Chimeric protein N terminus of ACE2 with a human IgG Fc fragment at the C-terminus which enhances phagocytosis and complement activation via interaction with Fc receptors. | Experimental models | NR | [140] | |
| Decrease transmembrane ACE2 | PMA (phorbol 12-myristate 13-acetate) | Enhances ADAM17 activity to increase ACE2 shedding. | In vitro study | NR | [133] |
| Ionomycin | Enhances ADAM10 activity to increase ACE2 shedding. | In vitro study | NR | [133] | |
| Resveratrol | Reduce ACE2 expression | Phase I clinical trial | Antioxidant, anticoagulant, anti-inflammatory drug | [141] | |
| Increase ACE2 activity | XNT (1-[(2-dimethylamino) ethylamino]-4-(hydroxymethyl)-7-[(4-methylphenyl) sulfonyl oxy]-9H-xanthen-9-one) | Increase ACE2 activity | Experimental model | NR | [133,139] |
| Diminazene aceturate | Increases ACE2 activity. | Experimental model | Anti-protozoa drugs. | [133] | |
| Olmesartan, losartan, telmisartan, azilsartan | ARB increases the ACE2 expression levels. | Phase III clinical trial | Used to downregulate high blood pressure. | [135,142] | |
| Spironolactone | Increases ACE2 mRNA in macrophages. | Clinical trials not concluded | Treatment for hyperaldosteronism and diuretic drug. | [135,143] | |
| Apelin-13 | Substrate of ACE2. | As hypothesis | Peptide used to treat cardiovascular diseases. | [135,144] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
López-Cortés, G.I.; Palacios-Pérez, M.; Hernández-Aguilar, M.M.; Veledíaz, H.F.; José, M.V. Human Coronavirus Cell Receptors Provide Challenging Therapeutic Targets. Vaccines 2023, 11, 174. https://doi.org/10.3390/vaccines11010174
López-Cortés GI, Palacios-Pérez M, Hernández-Aguilar MM, Veledíaz HF, José MV. Human Coronavirus Cell Receptors Provide Challenging Therapeutic Targets. Vaccines. 2023; 11(1):174. https://doi.org/10.3390/vaccines11010174
Chicago/Turabian StyleLópez-Cortés, Georgina I., Miryam Palacios-Pérez, Margarita M. Hernández-Aguilar, Hannya F. Veledíaz, and Marco V. José. 2023. "Human Coronavirus Cell Receptors Provide Challenging Therapeutic Targets" Vaccines 11, no. 1: 174. https://doi.org/10.3390/vaccines11010174