Longitudinal Analysis of Humoral and Cellular Immune Response up to 6 Months after SARS-CoV-2 BA.5/BF.7/XBB Breakthrough Infection and BA.5/BF.7-XBB Reinfection
Abstract
:1. Introduction
2. Materials and Methods
2.1. Serum Samples
2.2. Cell Lines
2.3. Construction and Production of Variant Pseudoviruses
2.4. Pseudovirus Neutralization Assays
2.5. Protein Expression and Purification
2.6. Memory B Cells Detection by Flow Cytometry
2.7. T Cells Detection by Flow Cytometry
2.8. ELISA
2.9. Quantification and Statistical Analysis
3. Results
3.1. COVID-19 Breakthrough Infection Cohorts
3.2. Anti-Spike and RBD Specific Antibody Magnitude and Durability Elicited by Breakthrough Infection
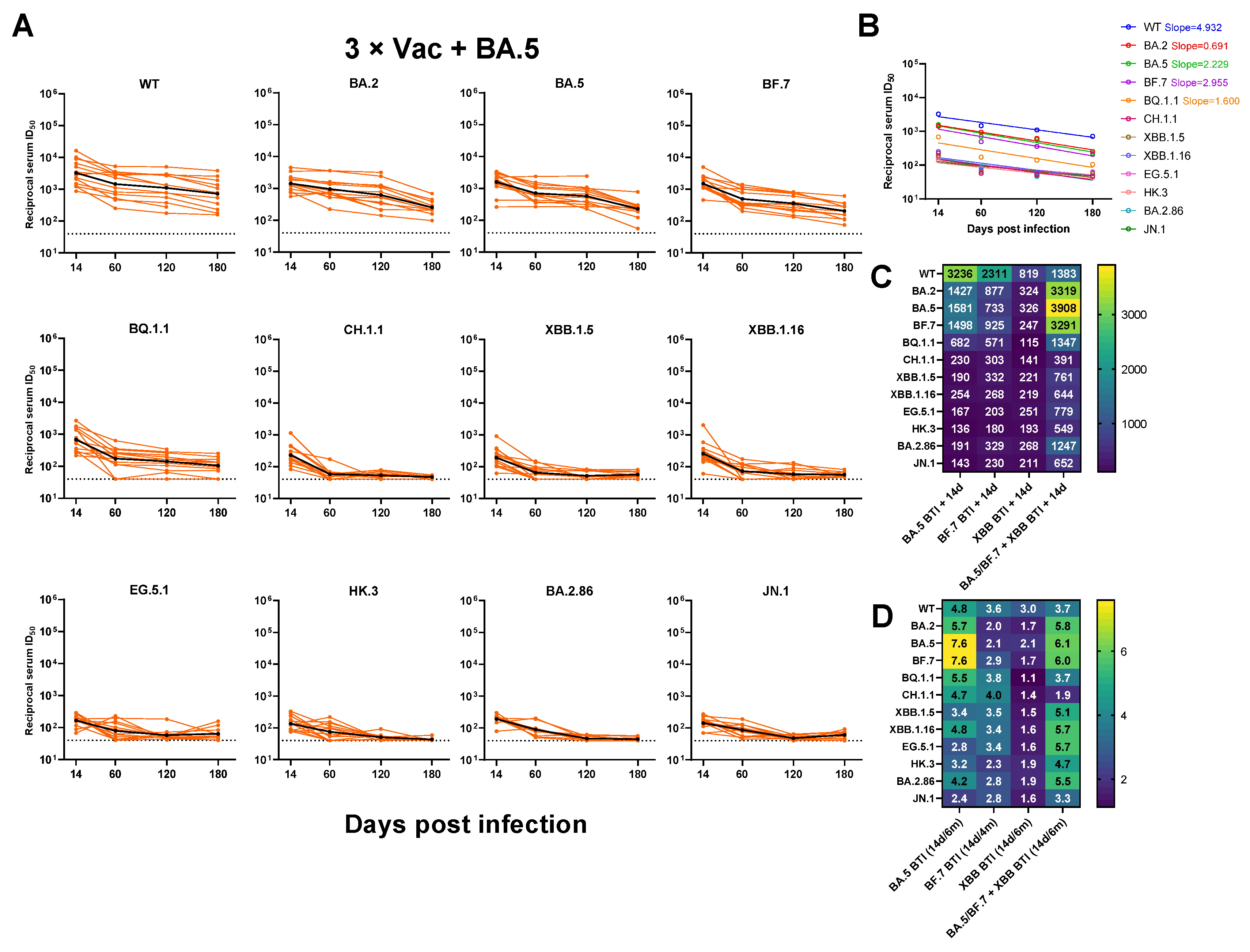
3.3. Neutralizing Antibody Titers against SARS-CoV-2 Variants
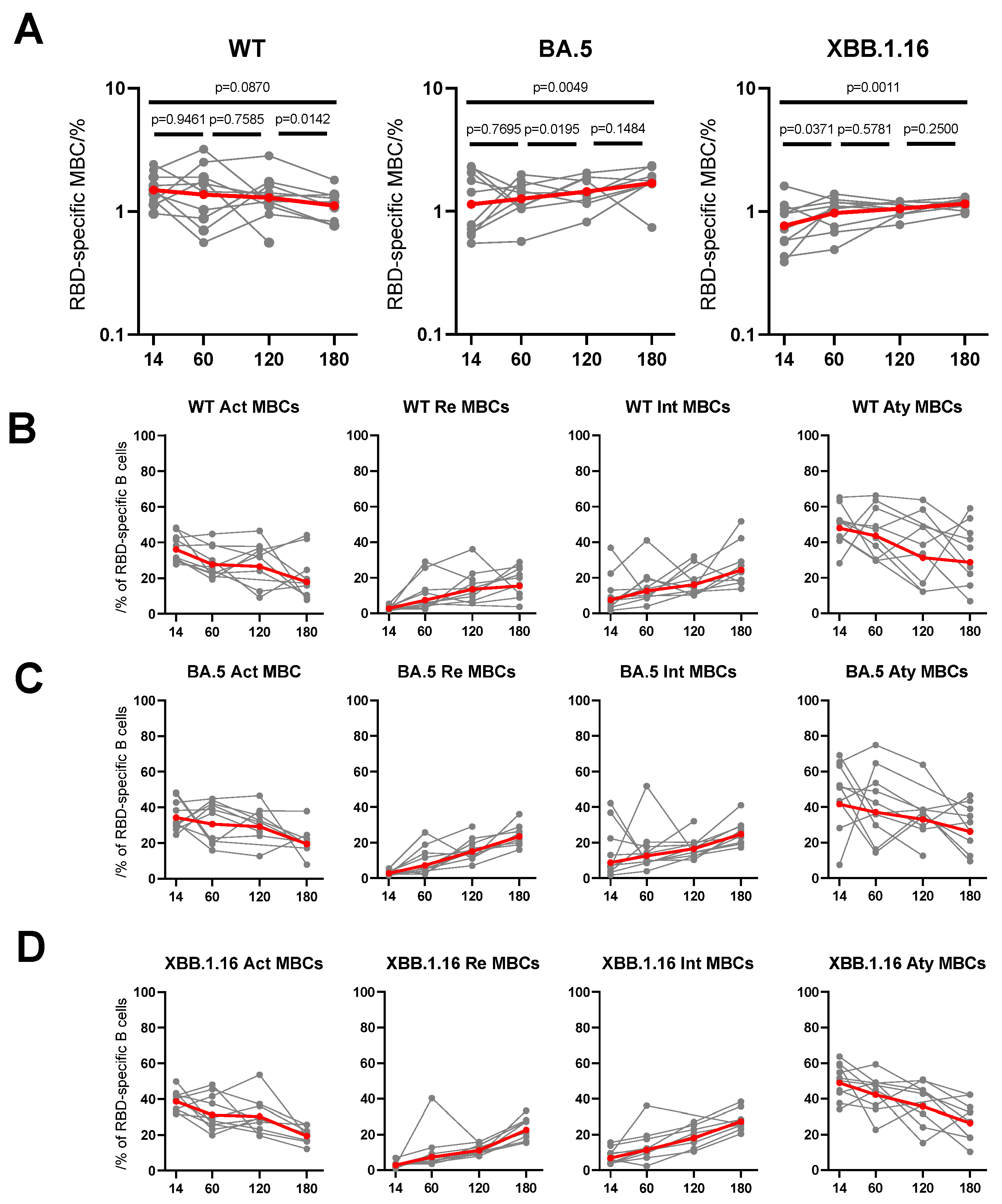
3.4. Dynamic Changes of RBD-Specific Memory B Cells to Breakthrough Infection
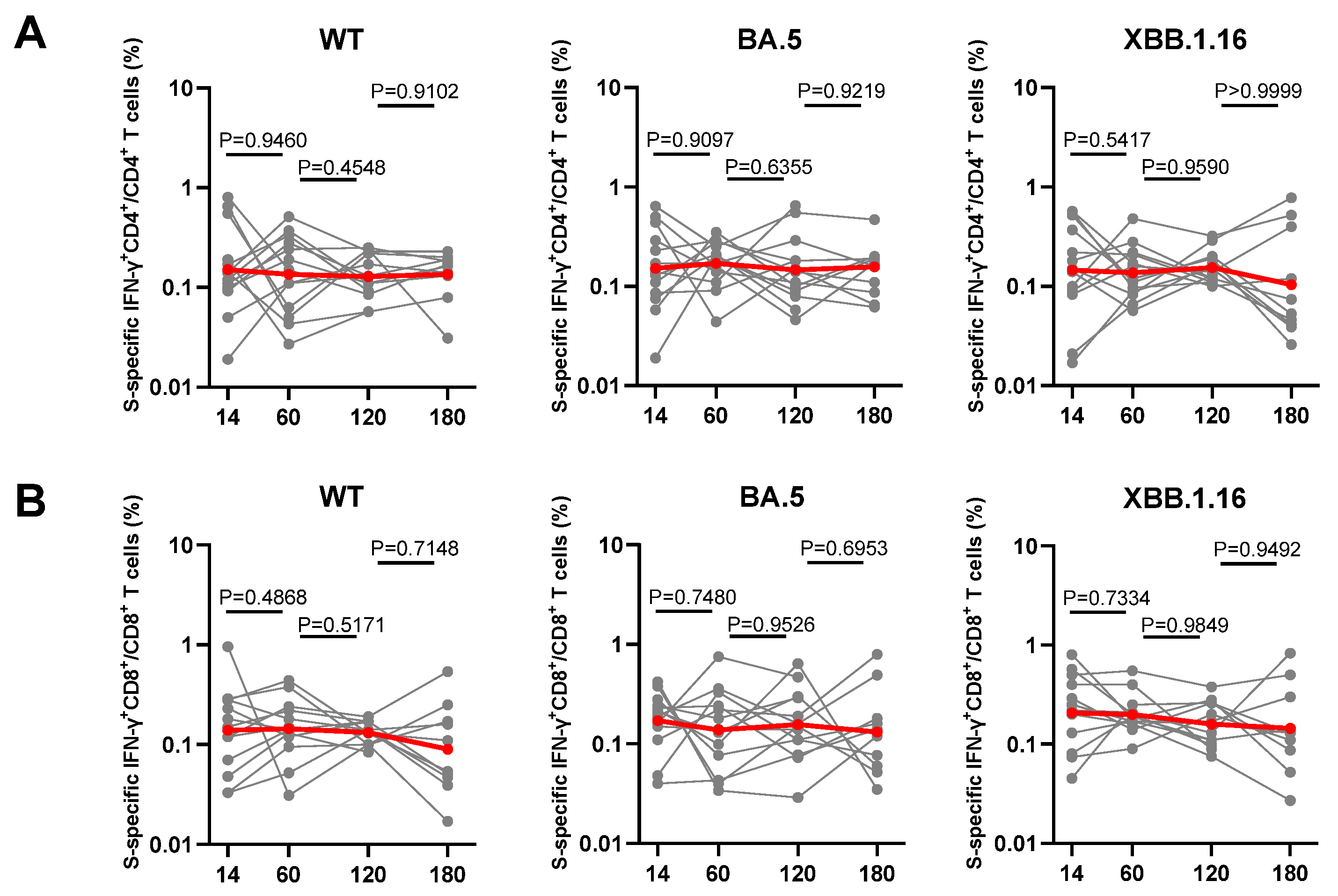
3.5. Dynamic Changes of Spike-Specific CD4+ and CD8+ T Cell Response Elicited by Breakthrough Infection
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Johns Hopkins University & Medicine Coronavirus Resource Center. COVID-19 Dashboard. Available online: https://coronavirus.jhu.edu/map.html (accessed on 29 January 2023).
- Arabi, M.; Al-Najjar, Y.; Sharma, O.; Kamal, I.; Javed, A.; Gohil, H.S.; Paul, P.; Al-Khalifa, A.M.; Laws, S.; Zakaria, D. Role of previous infection with SARS-CoV-2 in protecting against omicron reinfections and severe complications of COVID-19 compared to pre-omicron variants: A systematic review. BMC Infect. Dis. 2023, 23, 432. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Guo, Y.; Iketani, S.; Nair, M.S.; Li, Z.; Mohri, H.; Wang, M.; Yu, J.; Bowen, A.D.; Chang, J.Y.; et al. Antibody evasion by SARS-CoV-2 Omicron subvariants BA.2.12.1, BA.4 and BA.5. Nature 2022, 608, 603–608. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Iketani, S.; Li, Z.; Liu, L.; Guo, Y.; Huang, Y.; Bowen, A.D.; Liu, M.; Wang, M.; Yu, J.; et al. Alarming antibody evasion properties of rising SARS-CoV-2 BQ and XBB subvariants. Cell 2023, 186, 279–286.e8. [Google Scholar] [CrossRef] [PubMed]
- Parums, D.V. Editorial: The XBB.1.5 (‘Kraken’) Subvariant of Omicron SARS-CoV-2 and its Rapid Global Spread. Med. Sci. Monit. 2023, 29, e939580. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization (WHO), Technical Advisory Group on Virus Evolution (TAG-VE). 2023. Available online: https://www.who.int/docs/default-source/coronaviruse/11jan2023_xbb15_rapid_risk_assessment.pdf (accessed on 23 March 2023).
- Planas, D.; Staropoli, I.; Michel, V.; Lemoine, F.; Donati, F.; Prot, M.; Porrot, F.; Guivel-Benhassine, F.; Jeyarajah, B.; Brisebarre, A.; et al. Distinct evolution of SARS-CoV-2 Omicron XBB and BA.2.86 lineages combining increased fitness and antibody evasion. bioRxiv 2023. [Google Scholar] [CrossRef]
- Yang, S.; Yu, Y.; Xu, Y.; Jian, F.; Song, W.; Yisimayi, A.; Wang, P.; Wang, J.; Liu, J.; Yu, L.; et al. Fast evolution of SARS-CoV-2 BA.2.86 to JN.1 under heavy immune pressure. Lancet Infect. Dis. 2024, 24, e70–e72. [Google Scholar] [CrossRef]
- Wang, Q.; Guo, Y.; Liu, L.; Schwanz, L.T.; Li, Z.; Nair, M.S.; Ho, J.; Zhang, R.M.; Iketani, S.; Yu, J.; et al. Antigenicity and receptor affinity of SARS-CoV-2 BA.2.86 spike. Nature 2023, 624, 639–644. [Google Scholar] [CrossRef] [PubMed]
- Lassaunière, R.; Polacek, C.; Utko, M.; Sørensen, K.M.; Baig, S.; Ellegaard, K.; Escobar-Herrera, L.A.; Fomsgaard, A.; Spiess, K.; Gunalan, V.; et al. Virus isolation and neutralisation of SARS-CoV-2 variants BA.2.86 and EG.5.1. Lancet Infect. Dis. 2023, 23, e509–e510. [Google Scholar] [CrossRef]
- Sette, A.; Crotty, S. Adaptive immunity to SARS-CoV-2 and COVID-19. Cell 2021, 184, 861–880. [Google Scholar] [CrossRef]
- Khoury, D.S.; Cromer, D.; Reynaldi, A.; Schlub, T.E.; Wheatley, A.K.; Juno, J.A.; Subbarao, K.; Kent, S.J.; Triccas, J.A.; Davenport, M.P. Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS-CoV-2 infection. Nat. Med. 2021, 27, 1205–1211. [Google Scholar] [CrossRef]
- Vanshylla, K.; Di Cristanziano, V.; Kleipass, F.; Dewald, F.; Schommers, P.; Gieselmann, L.; Gruell, H.; Schlotz, M.; Ercanoglu, M.S.; Stumpf, R.; et al. Kinetics and correlates of the neutralizing antibody response to SARS-CoV-2 infection in humans. Cell Host Microbe 2021, 29, 917–929.e4. [Google Scholar] [CrossRef]
- Sokal, A.; Chappert, P.; Barba-Spaeth, G.; Roeser, A.; Fourati, S.; Azzaoui, I.; Vandenberghe, A.; Fernandez, I.; Meola, A.; Bouvier-Alias, M.; et al. Maturation and persistence of the anti-SARS-CoV-2 memory B cell response. Cell 2021, 184, 1201–1213.e14. [Google Scholar] [CrossRef] [PubMed]
- Röltgen, K.; Boyd, S.D. Antibody and B cell responses to SARS-CoV-2 infection and vaccination. Cell Host Microbe 2021, 29, 1063–1075. [Google Scholar] [CrossRef]
- Balachandran, H.; Phetsouphanh, C.; Agapiou, D.; Adhikari, A.; Rodrigo, C.; Hammoud, M.; Shrestha, L.B.; Keoshkerian, E.; Gupta, M.; Turville, S.; et al. Maintenance of broad neutralizing antibodies and memory B cells 1 year post-infection is predicted by SARS-CoV-2-specific CD4+ T cell responses. Cell Rep. 2022, 38, 110345. [Google Scholar] [CrossRef] [PubMed]
- DiPiazza, A.T.; Graham, B.S.; Ruckwardt, T.J. T cell immunity to SARS-CoV-2 following natural infection and vaccination. Biochem. Biophys. Res. Commun. 2021, 538, 211–217. [Google Scholar] [CrossRef] [PubMed]
- Tan, A.T.; Linster, M.; Tan, C.W.; Le Bert, N.; Chia, W.N.; Kunasegaran, K.; Zhuang, Y.; Tham, C.Y.L.; Chia, A.; Smith, G.J.D.; et al. Early induction of functional SARS-CoV-2-specific T cells associates with rapid viral clearance and mild disease in COVID-19 patients. Cell Rep. 2021, 34, 108728. [Google Scholar] [CrossRef] [PubMed]
- Azevedo, F.; Pavlović, T.; Rêgo, G.G.; Ay, F.C.; Gjoneska, B.; Etienne, T.W.; Ross, R.M.; Schönegger, P.; Riaño-Moreno, J.C.; Cichocka, A.; et al. Social and moral psychology of COVID-19 across 69 countries. Sci. Data 2023, 10, 272. [Google Scholar] [CrossRef]
- Kihlström, L.; Siemes, L.; Huhtakangas, M.; Keskimäki, I.; Tynkkynen, L.K. Power and politics in a pandemic: Insights from Finnish health system leaders during COVID-19. Soc. Sci. Med. 2023, 321, 115783. [Google Scholar] [CrossRef] [PubMed]
- Yisimayi, A.; Song, W.; Wang, J.; Jian, F.; Yu, Y.; Chen, X.; Xu, Y.; Yang, S.; Niu, X.; Xiao, T.; et al. Repeated Omicron exposures override ancestral SARS-CoV-2 immune imprinting. Nature 2023, 625, 148–156. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Zhang, L.; Liang, Z.; Wang, N.; Liu, S.; Li, T.; Yu, Y.; Cui, Q.; Wu, X.; Nie, J.; et al. Cross-reactivity of eight SARS-CoV-2 variants rationally predicts immunogenicity clustering in sarbecoviruses. Signal Transduct. Target. Ther. 2022, 7, 256. [Google Scholar] [CrossRef]
- Wang, X.; Jiang, S.; Ma, W.; Li, X.; Wei, K.; Xie, F.; Zhao, C.; Zhao, X.; Wang, S.; Li, C.; et al. Enhanced neutralization of SARS-CoV-2 variant BA.2.86 and XBB sub-lineages by a tetravalent COVID-19 vaccine booster. Cell Host Microbe 2024, 32, 25–34.e5. [Google Scholar] [CrossRef] [PubMed]
- Lyke, K.E.; Atmar, R.L.; Islas, C.D.; Posavad, C.M.; Szydlo, D.; Paul Chourdhury, R.; Deming, M.E.; Eaton, A.; Jackson, L.A.; Branche, A.R.; et al. Rapid decline in vaccine-boosted neutralizing antibodies against SARS-CoV-2 Omicron variant. Cell Rep. Med. 2022, 3, 100679. [Google Scholar] [CrossRef]
- Sherina, N.; Piralla, A.; Du, L.; Wan, H.; Kumagai-Braesch, M.; Andréll, J.; Braesch-Andersen, S.; Cassaniti, I.; Percivalle, E.; Sarasini, A.; et al. Persistence of SARS-CoV-2-specific B and T cell responses in convalescent COVID-19 patients 6-8 months after the infection. Med 2021, 2, 281–295.e4. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Jiang, S.; Jiang, S.; Li, X.; Ai, J.; Lin, K.; Lv, S.; Zhang, S.; Li, M.; Li, J.; et al. Neutralization of SARS-CoV-2 BQ.1.1, CH.1.1, and XBB.1.5 by breakthrough infection sera from previous and recent waves in China. Cell Discov. 2023, 9, 64. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Ai, J.; Li, X.; Zhao, X.; Wu, J.; Zhang, H.; He, X.; Zhao, C.; Qiao, R.; Li, M.; et al. Neutralization of Omicron BA.4/BA.5 and BA.2.75 by booster vaccination or BA.2 breakthrough infection sera. Cell Discov. 2022, 8, 110. [Google Scholar] [CrossRef] [PubMed]
- Tarke, A.; Coelho, C.H.; Zhang, Z.; Dan, J.M.; Yu, E.D.; Methot, N.; Bloom, N.I.; Goodwin, B.; Phillips, E.; Mallal, S.; et al. SARS-CoV-2 vaccination induces immunological T cell memory able to cross-recognize variants from Alpha to Omicron. Cell 2022, 185, 847–859.e11. [Google Scholar] [CrossRef] [PubMed]
- Mallah, S.I.; Ghorab, O.K.; Al-Salmi, S.; Abdellatif, O.S.; Tharmaratnam, T.; Iskandar, M.A.; Sefen, J.A.N.; Sidhu, P.; Atallah, B.; El-Lababidi, R.; et al. COVID-19: Breaking down a global health crisis. Ann. Clin. Microbiol. Antimicrob. 2021, 20, 35. [Google Scholar] [CrossRef]
- Ma, K.C.; Shirk, P.; Lambrou, A.S.; Hassell, N.; Zheng, X.Y.; Payne, A.B.; Ali, A.R.; Batra, D.; Caravas, J.; Chau, R.; et al. Genomic Surveillance for SARS-CoV-2 Variants: Circulation of Omicron Lineages—United States, January 2022–May 2023. MMWR Morb. Mortal. Wkly. Rep. 2023, 72, 651–656. [Google Scholar] [CrossRef]
- Viana, R.; Moyo, S.; Amoako, D.G.; Tegally, H.; Scheepers, C.; Althaus, C.L.; Anyaneji, U.J.; Bester, P.A.; Boni, M.F.; Chand, M.; et al. Rapid epidemic expansion of the SARS-CoV-2 Omicron variant in southern Africa. Nature 2022, 603, 679–686. [Google Scholar] [CrossRef]
- Shrestha, L.B.; Foster, C.; Rawlinson, W.; Tedla, N.; Bull, R.A. Evolution of the SARS-CoV-2 omicron variants BA.1 to BA.5: Implications for immune escape and transmission. Rev. Med. Virol. 2022, 32, e2381. [Google Scholar] [CrossRef]
- Kaku, Y.; Okumura, K.; Padilla-Blanco, M.; Kosugi, Y.; Uriu, K.; Hinay, A.A., Jr.; Chen, L.; Plianchaisuk, A.; Kobiyama, K.; Ishii, K.J.; et al. Virological characteristics of the SARS-CoV-2 JN.1 variant. Lancet Infect. Dis. 2024, 24, e82. [Google Scholar] [CrossRef] [PubMed]
- Agergaard, J.; Gunst, J.D.; Schiøttz-Christensen, B.; Østergaard, L.; Wejse, C. Long-term Prognosis at 1.5 years after Infection with Wild-type strain of SARS-CoV-2 and Alpha, Delta, as well as Omicron Variants. Int. J. Infect. Dis. 2023, 137, 126–133. [Google Scholar] [CrossRef] [PubMed]
- Cao, C.; Guo, Q.; Dai, Y.; Jiang, J.; Liu, M.; Ji, T.; Zhou, P.; Gong, F. Neutralizing activity of a third dose of CoronaVac against Omicron subvariants within a 20-month follow-up study. Hum. Vaccines Immunother. 2023, 19, 2242217. [Google Scholar] [CrossRef] [PubMed]
- Feng, C.; Shi, J.; Fan, Q.; Wang, Y.; Huang, H.; Chen, F.; Tang, G.; Li, Y.; Li, P.; Li, J.; et al. Protective humoral and cellular immune responses to SARS-CoV-2 persist up to 1 year after recovery. Nat. Commun. 2021, 12, 4984. [Google Scholar] [CrossRef] [PubMed]
- Gao, B.; He, L.; Bao, Y.; Chen, Y.; Lu, G.; Zhang, Y.; Xu, Y.; Su, B.; Xu, J.; Wang, Y.; et al. Repeated vaccination of inactivated SARS-CoV-2 vaccine dampens neutralizing antibodies against Omicron variants in breakthrough infection. Cell Res. 2023, 33, 258–261. [Google Scholar] [CrossRef]
- Wang, H.; Xue, Q.; Zhang, H.; Yuan, G.; Wang, X.; Sheng, K.; Li, C.; Cai, J.; Sun, Y.; Zhao, J.; et al. Neutralization against Omicron subvariants after BA.5/BF.7 breakthrough infection weakened as virus evolution and aging despite repeated prototype-based vaccination(1). Emerg. Microbes Infect. 2023, 12, 2249121. [Google Scholar] [CrossRef] [PubMed]
- Tan, H.X.; Juno, J.A. Interplay of infection and vaccination in long-term protection from COVID-19. Lancet Infect. Dis. 2022, 22, 744–745. [Google Scholar] [CrossRef] [PubMed]
- Tenforde, M.W.; Link-Gelles, R.; Patel, M.M. Long-term Protection Associated With COVID-19 Vaccination and Prior Infection. JAMA 2022, 328, 1402–1404. [Google Scholar] [CrossRef]
- Lin, D.Y.; Gu, Y.; Xu, Y.; Wheeler, B.; Young, H.; Sunny, S.K.; Moore, Z.; Zeng, D. Association of Primary and Booster Vaccination and Prior Infection With SARS-CoV-2 Infection and Severe COVID-19 Outcomes. JAMA 2022, 328, 1415–1426. [Google Scholar] [CrossRef]
- Sokal, A.; Barba-Spaeth, G.; Hunault, L.; Fernández, I.; Broketa, M.; Meola, A.; Fourati, S.; Azzaoui, I.; Vandenberghe, A.; Lagouge-Roussey, P.; et al. SARS-CoV-2 Omicron BA.1 breakthrough infection drives late remodeling of the memory B cell repertoire in vaccinated individuals. Immunity 2023, 56, 2137–2151.e7. [Google Scholar] [CrossRef]
- Korenkov, M.; Zehner, M.; Cohen-Dvashi, H.; Borenstein-Katz, A.; Kottege, L.; Janicki, H.; Vanshylla, K.; Weber, T.; Gruell, H.; Koch, M.; et al. Somatic hypermutation introduces bystander mutations that prepare SARS-CoV-2 antibodies for emerging variants. Immunity 2023, 56, 2803–2815.e6. [Google Scholar] [CrossRef] [PubMed]
- Lam, J.H.; Smith, F.L.; Baumgarth, N. B Cell Activation and Response Regulation During Viral Infections. Viral Immunol. 2020, 33, 294–306. [Google Scholar] [CrossRef] [PubMed]
- Chemaitelly, H.; Nagelkerke, N.; Ayoub, H.H.; Coyle, P.; Tang, P.; Yassine, H.M.; Al-Khatib, H.A.; Smatti, M.K.; Hasan, M.R.; Al-Kanaani, Z.; et al. Duration of immune protection of SARS-CoV-2 natural infection against reinfection. J. Travel. Med. 2022, 29, taac109. [Google Scholar] [CrossRef]
- Zhang, Z.; Mateus, J.; Coelho, C.H.; Dan, J.M.; Moderbacher, C.R.; Gálvez, R.I.; Cortes, F.H.; Grifoni, A.; Tarke, A.; Chang, J.; et al. Humoral and cellular immune memory to four COVID-19 vaccines. Cell 2022, 185, 2434–2451.e17. [Google Scholar] [CrossRef]
- Cui, Z.; Luo, W.; Chen, R.; Li, Y.; Wang, Z.; Liu, Y.; Liu, S.; Feng, L.; Jia, Z.; Cheng, R.; et al. Comparing T- and B-cell responses to COVID-19 vaccines across varied immune backgrounds. Signal Transduct. Target. Ther. 2023, 8, 179. [Google Scholar] [CrossRef]
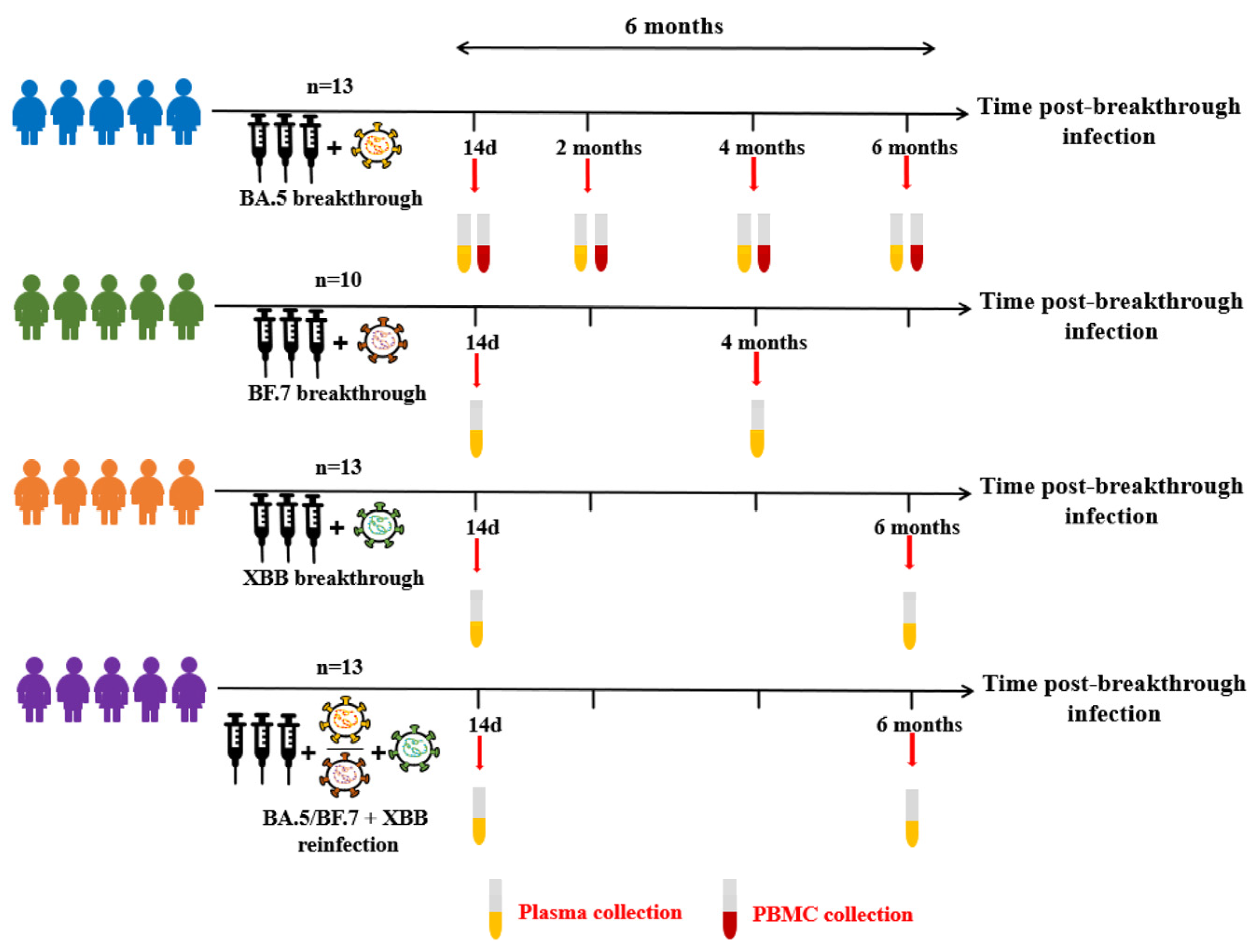

| BA.5 Breakthrough Infection Individuals (n = 13) | BF.7 Breakthrough Infection Individuals (n = 10) | XBB Breakthrough Infection Individuals (n = 13) | BA.5/BF.7-XBB Reinfection Individuals (n = 13) | |
|---|---|---|---|---|
| Age (year), median (range) | 31.6 (22–47) | 40.1 (34–50) | 32.33 (19–46) | 34.23 (18–69) |
| Male, n (%) | 13 (59.09%) | 4 (28.6%) | 7 (53.84%) | 6 (46.15%) |
| BMI (kg/m2), mean (SD) | 23.0 (3.9) | 23.9 (3.3) | 23.3 (3.0) | 21.4 (2.5) |
| Breakthrough infections months after the last COVID-19 vaccines, median (range) | 10.8 (2–19) | 10.6 (12–14) | 16.6 (13–25) | 10.6 (5–23) |
| Vaccinated days after the last COVID-19 vaccines, median (range) | 427.2 (242–607) | 407.4 (363–432) | 535.8 (419–790) | 337.7 (172–735) |
| Comorbidities (%) | N/A | N/A | N/A | N/A |
| Any, n (%) | 0 (0%) | 0 (0%) | 1 (7.69%) | 0 (0%) |
| HTN, n (%) | 0 (0%) | 0 (0%) | 1 (7.69%) | 2 (15.38%) |
| CAD, n (%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) |
| DM, n (%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) |
| NAFLD, n (%) | 0 (0%) | 0 (0%) | 1 (7.69%) | 0 (0%) |
| Hyperlipidemia, n (%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) |
| Obesity, n (%) | 2 (9.09%) | 0 (0%) | 0 (0%) | 0 (0%) |
| Arrhy, n (%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) |
| Asthma, n (%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) |
| Rhinitis, n (%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) |
| Urticaria, n (%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) |
| Variant | Protein Sequence |
|---|---|
| WT | MFVFLVLLPLVSSQCVNLTTRTQLPPAYTNSFTRGVYYPDKVFRSSVLHSTQDLFLPFFSNVTWFHAIHVSGTNGTKRFDNPVLPFNDGVYFASTEKSNIIRGWIFGTTLDSKTQSLLIVNNATNVVIKVCEFQFCNDPFLGVYYHKNNKSWMESEFRVYSSANNCTFEYVSQPFLMDLEGKQGNFKNLREFVFKNIDGYFKIYSKHTPINLVRDLPQGFSALEPLVDLPIGINITRFQTLLALHRSYLTPGDSSSGWTAGAAAYYVGYLQPRTFLLKYNENGTITDAVDCALDPLSETKCTLKSFTVEKGIYQTSNF“RVQPTESIVRFPNITNLCPFGEVFNATRFASVYAWNRKRISNCVADYSVLYNSASFSTFKCYGVSPTKLNDLCFTNVYADSFVIRGDEVRQIAPGQTGKIADYNYKLPDDFTGCVIAWNSNNLDSKVGGNYNYLYRLFRKSNLKPFERDISTEIYQAGSTPCNGVEGFNCYFPLQSYGFQPTNGVGYQPYRVVVLSFELLHAPATVCGPKKSTNLVKNKCVNF”NFNGLTGTGVLTESNKKFLPFQQFGRDIADTTDAVRDPQTLEILDITPCSFGGVSVITPGTNTSNQVAVLYQGVNCTEVPVAIHADQLTPTWRVYSTGSNVFQTRAGCLIGAEHVNNSYECDIPIGAGICASYQTQTNSPRRARSVASQSIIAYTMSLGAENSVAYSNNSIAIPTNFTISVTTEILPVSMTKTSVDCTMYICGDSTECSNLLLQYGSFCTQLNRALTGIAVEQDKNTQEVFAQVKQIYKTPPIKDFGGFNFSQILPDPSKPSKRSFIEDLLFNKVTLADAGFIKQYGDCLGDIAARDLICAQKFNGLTVLPPLLTDEMIAQYTSALLAGTITSGWTFGAGAALQIPFAMQMAYRFNGIGVTQNVLYENQKLIANQFNSAIGKIQDSLSSTASALGKLQDVVNQNAQALNTLVKQLSSNFGAISSVLNDILSRLDKVEAEVQIDRLITGRLQSLQTYVTQQLIRAAEIRASANLAATKMSECVLGQSKRVDFCGKGYHLMSFPQSAPHGVVFLHVTYVPAQEKNFTTAPAICHDGKAHFPREGVFVSNGTHWFVTQRNFYEPQIITTDNTFVSGNCDVVIGIVNNTVYDPLQPELDSFKEELDKYFKNHTSPDVDLGDISGINASVVNIQKEIDRLNEVAKNLNESLIDLQELGKYEQYIKWPWYIWLGFIAGLIAIVMVTIMLCCMTSCCSCLKGCCSCGSCCK_ |
| BA.2 | WT+T19I, Del24-26, A27S, G142D, V213G, G339D, S371F, S373P, S375F, T376A, D405N, R408S, K417N, N440K, S477N, T478K, E484A, Q493R, Q498R, N501Y, Y505H, H655Y, N679K, P681H, N764K, D796Y, Q954H, N969K |
| BA.5 | WT+T19I, Del24-26, A27S, Del69-70, G142D, V213G, G339D, S371F, S373P, S375F, T376A, D405N, R408S, K417N, N440K, L452R, S477N, T478K, E484A, F486V, Q498R, N501Y, Y505H, H655Y, N679K, P681H, N764K, D796Y, Q954H, N969K |
| BF.7 | WT+T19I, Del24-26, A27S, Del69-70, G142D, V213G, G339D, R346T, S371F, S373P, S375F, T376A, D405N, R408S, K417N, N440K, L452R, S477N, T478K, E484A, F486V, Q498R, N501Y, Y505H, H655Y, N679K, P681H, N764K, D796Y, Q954H, N969K |
| BQ.1.1 | WT+T19I, Del24-26, A27S, Del69-70, G142D, V213G, G339D, R346T, S371F, S373P, S375F, T376A, D405N, R408S, K417N, N440K, K444T, L452R, N460K, S477N, T478K, E484A, F486V, Q498R, N501Y, Y505H, H655Y, N679K, P681H, N764K, D796Y, Q954H, N969K |
| CH.1.1 | WT+T19I, Del24-26, A27S, G142D, K147E, W152R, F157L, I210V, V213G, G257S, G339H, S371F, S373P, S375F, T376A, D405N, R408S, K417N, N440K, K444T, G446S, L452R, N460K, S477N, T478K, E484A, F486S, Q498R, N501Y, Y505H, H655Y, N679K, P681H, N764K, D796Y, Q954H, N969K |
| XBB.1.5 | WT+T19I, Del24-26, A27S, V83A, G142D, Del144, H146Q, Q183E, V213E, G252V, G339H, R346T, L368I, S371F, S373P, S375F, T376A, D405N, R408S, K417N, N440K, V445P, G446S, N460K, S477N, T478K, E484A, F486P, F490S, Q498R, N501Y, Y505H, H655Y, N679K, P681H, N764K, D796Y, Q954H, N969K |
| XBB.1.16 | WT+T19I, Del24-26, A27S, V83A, G142D, Del144, H146Q, E180V, Q183E, V213E, G252V, G339H, R346T, L368I, S371F, S373P, S375F, T376A, D405N, R408S, K417N, N440K, V445P, G446S, N460K, S477N, T478R, E484A, F486P, F490S, Q498R, N501Y, Y505H, H655Y, N679K, P681H, N764K, D796Y, Q954H, N969K |
| EG.5.1 | WT+T19I, Del24-26, A27S, Q52H, V83A, G142D, Del144, H146Q, Q183E, V213E, G252V, G339H, R346T, L368I, S371F, S373P, S375F, T376A, D405N, R408S, K417N, N440K, V445P, G446S, F456L, N460K, S477N, T478K, E484A, F486P, F490S, Q498R, N501Y, Y505H, H655Y, N679K, P681H, N764K, D796Y, Q954H, N969K |
| HK.3 | WT+T19I, Del24-26, A27S, Q52H, V83A, G142D, Del144, H146Q, Q183E, V213E, G252V, G339H, R346T, L368I, S371F, S373P, S375F, T376A, D405N, R408S, K417N, N440K, V445P, G446S, L455F, F456L, N460K, S477N, T478K, E484A, F486P, F490S, Q498R, N501Y, Y505H, H655Y, N679K, P681H, N764K, D796Y, Q954H, N969K |
| BA.2.86 | WT+ Ins16MPLF, T19I, R21T, Del24-26, A27S, S50L, Del69-70, V127F, G142D, Del144, F157S, R158G, Del211, L212I, V213G, L216F, H245N, A264D, I332V, G339H, K356T, S371F, S373P, S375F, T376A, R403K, D405N, R408S, K417N, N440K, V445H, G446S, N450D, L452W, N460K, S477N, T478K, N481K, Del483, E484K, F486P, Q498R, N501Y, Y505H, E554K, A570V, H655Y, N679K, P681R, N764K, D796Y, S939F, Q954H, N969K, P1143L |
| JN.1 | WT+Ins16MPLF, T19I, R21T, Del24-26, A27S, S50L, Del69-70, V127F, G142D, Del144, F157S, R158G, Del211, L212I, V213G, L216F, H245N, A264D, I332V, G339H, K356T, S371F, S373P, S375F, T376A, R403K, D405N, R408S, K417N, N440K, V445H, G446S, N450D, L452W, L455S, N460K, S477N, T478K, N481K, Del483, E484K, F486P, Q498R, N501Y, Y505H, E554K, A570V, H655Y, N679K, P681R, N764K, D796Y, S939F, Q954H, N969K, P1143L |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, X.; Zhang, M.; Wei, K.; Li, C.; Yang, J.; Jiang, S.; Zhao, C.; Zhao, X.; Qiao, R.; Cui, Y.; et al. Longitudinal Analysis of Humoral and Cellular Immune Response up to 6 Months after SARS-CoV-2 BA.5/BF.7/XBB Breakthrough Infection and BA.5/BF.7-XBB Reinfection. Vaccines 2024, 12, 464. https://doi.org/10.3390/vaccines12050464
Wang X, Zhang M, Wei K, Li C, Yang J, Jiang S, Zhao C, Zhao X, Qiao R, Cui Y, et al. Longitudinal Analysis of Humoral and Cellular Immune Response up to 6 Months after SARS-CoV-2 BA.5/BF.7/XBB Breakthrough Infection and BA.5/BF.7-XBB Reinfection. Vaccines. 2024; 12(5):464. https://doi.org/10.3390/vaccines12050464
Chicago/Turabian StyleWang, Xun, Meng Zhang, Kaifeng Wei, Chen Li, Jinghui Yang, Shujun Jiang, Chaoyue Zhao, Xiaoyu Zhao, Rui Qiao, Yuchen Cui, and et al. 2024. "Longitudinal Analysis of Humoral and Cellular Immune Response up to 6 Months after SARS-CoV-2 BA.5/BF.7/XBB Breakthrough Infection and BA.5/BF.7-XBB Reinfection" Vaccines 12, no. 5: 464. https://doi.org/10.3390/vaccines12050464






