Influenza and Memory T Cells: How to Awake the Force
Abstract
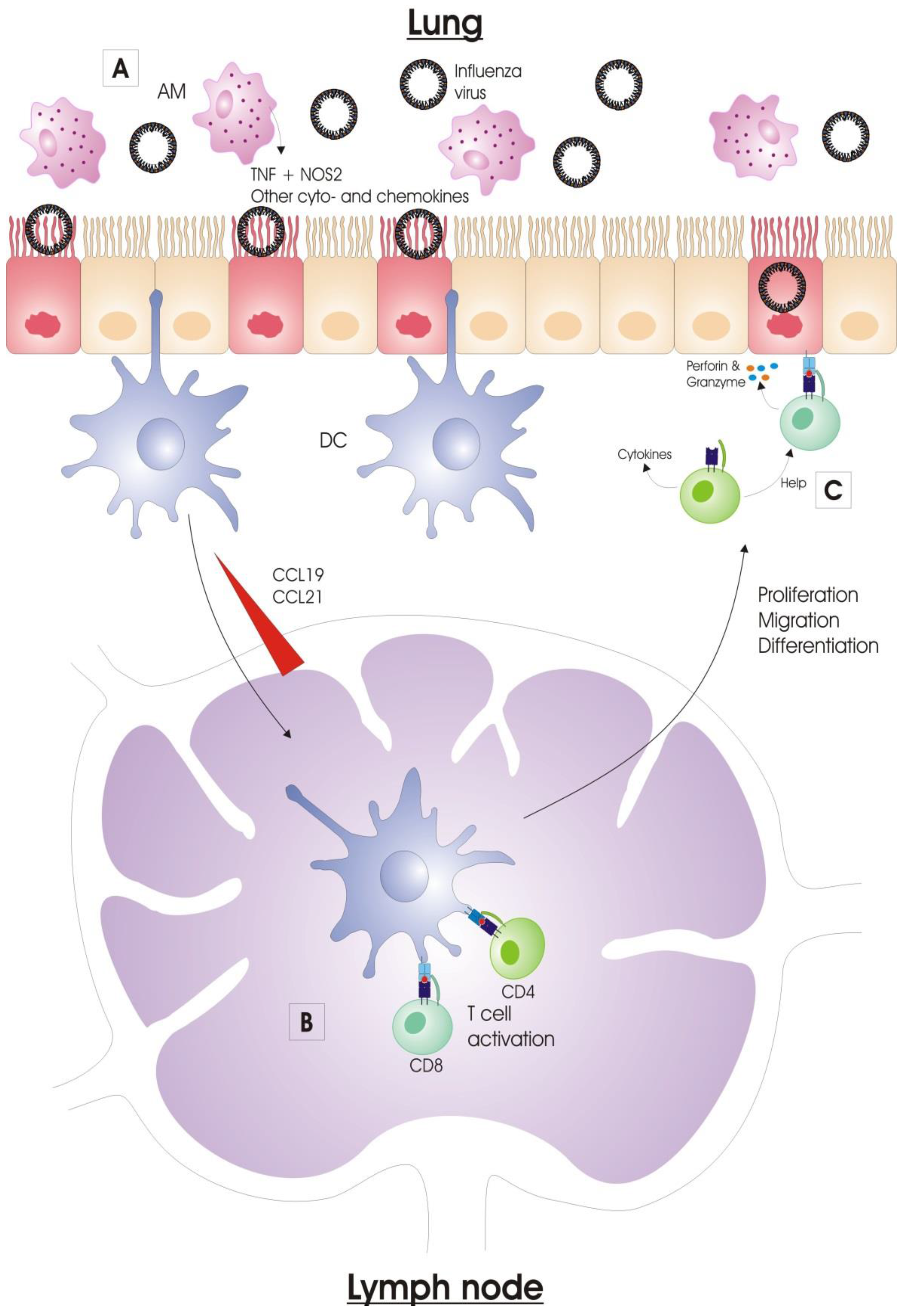
:1. Introduction
2. Immune Response to Influenza Virus Infection
2.1. Innate Immunity
2.1.1. Extracellular Barriers to Overcome
2.1.2. Sensing of Influenza Virus Infection
2.1.3. Alveolar Macrophages
2.1.4. Dendritic Cells
2.1.5. Natural Killer Cells
2.2. Adaptive Immunity
2.2.1. Activation of Antigen-Presenting Cells
2.2.2. Antigen Presentation
2.2.3. Lymphocyte Migration to the Infected Lung
2.2.4. Viral Clearance
3. T Cell Response to Influenza Virus Infection
3.1. Primary T Cell Response: Deflowering the T Cells
3.1.1. CD4+ T Cell Primary Responses
3.1.2. CD8+ T Cell Primary Responses
3.2. Memory T Cell Response: The T Cells Remember
3.2.1. CD4+ Memory T Cells
3.2.2. CD8+ Memory T Cells
4. Vaccines
4.1. Current Influenza Vaccines
4.2. T Cells in Heterologous Protection against Influenza Viruses
4.3. Vaccination-Induced T Cell Immunity: To Serve and Protect
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| ADCC | Antibody-dependent cellular cytotoxicity |
| AM | Alveolar macrophage |
| APC | Antigen-presenting cell |
| CCL | Chemokine (C-C motif) ligand |
| CCR | C-C chemokine receptor |
| CD | Cluster of differentiation |
| CD62L | L-selectin |
| Cdc | Classical dendritic cell |
| CTL | Cytotoxic T lymphocyte |
| CXCL | Chemokine (C-X-C motif) ligand |
| DC | Dendritic cell |
| DLN | Draining lymph node |
| DNA | Deoxyribonucleic acid |
| FasL | Fas ligand |
| GTP | Guanosine triphosphate |
| HA | Hemagglutinin |
| HI | Hemagglutination-inhibiting |
| IAV | Influenza A virus |
| ICAM-1 | Intercellular adhesion molecule 1 |
| IFITM3 | Interferon-induced transmembrane protein 3 |
| IFN | Interferon |
| Ig | Immunoglobulin |
| IL | Interleukin |
| ISG | Interferon-stimulated gene |
| iTreg | Induced regulatory T cell |
| LAIV | Live-attenuated influenza vaccine |
| LFA-1 | Lymphocyte function-associated antigen 1 |
| M | Matrix protein |
| MCP | Monocyte chemoattractant protein |
| MHC | Major histocompatibility complex |
| MIP | Macrophage inflammatory protein |
| mRNA | Messenger ribonucleic acid |
| MVA | Modified Vaccinia Ankara |
| Mx | Myxovirus resistant |
| NA | Neuraminidase |
| NK | Natural killer |
| NLRP3 | NOD-like receptor family pyrin domain containing 3 protein |
| NOS2 | Nitric oxide synthase 2 |
| NP | Nucleoprotein |
| nTreg | Natural regulatory T cell |
| PB | polybasic |
| PBMC | Peripheral blood mononuclear cell |
| PRR | Pattern recognition receptor |
| QIV | Quadrivalent inactivated influenza vaccine |
| RIG-I | Retinoic acid-inducible gene I |
| RNA | Ribonucleic acid |
| SP | Surfactant protein |
| Tcm | Central memory T cell |
| TCR | T cell receptor |
| Tem | Effector memory T cell |
| Tfh | Follicular helper T cell |
| Th | T helper cell |
| TIV | Trivalent inactivated influenza vaccine |
| TNF | Tumor necrosis factor |
| TRAIL | TNF-related apoptosis-inducing ligand |
| Treg | regulatory T cell |
| Trm | Resident memory T cell |
| WHO | World Health Organization |
References
- WHO. Influenza (Seasonal). Available online: http://www.who.int/mediacentre/factsheets/fs211/en/ (accessed on 24 February 2016).
- Palese, P. Influenza: Old and new threats. Nat. Med. 2004, 10, S82–S87. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization (WHO). Pandemic (H1N1) 2009–Update 112. Available online: http://www.who.int/csr/don/2010_08_06/en/ (accessed on 24 February 2016).
- Dawood, F.S.; Iuliano, A.D.; Reed, C.; Meltzer, M.I.; Shay, D.K.; Cheng, P.Y.; Bandaranayake, D.; Breiman, R.F.; Brooks, W.A.; Buchy, P.; et al. Estimated global mortality associated with the first 12 months of 2009 pandemic influenza A H1N1 virus circulation: A modelling study. Lancet Infect. Dis. 2012, 12, 687–695. [Google Scholar] [CrossRef]
- Waffarn, E.E.; Baumgarth, N. Protective B cell responses to flu—No fluke! J. Immunol. 2011, 186, 3823–3829. [Google Scholar] [CrossRef] [PubMed]
- Monto, A.S.; Petrie, J.G.; Cross, R.T.; Johnson, E.; Liu, M.; Zhong, W.; Levine, M.; Katz, J.M.; Ohmit, S.E. Antibody to Influenza Virus Neuraminidase: An Independent Correlate of Protection. J. Infect. Dis. 2015, 212, 1191–1199. [Google Scholar] [CrossRef] [PubMed]
- Nobusawa, E.; Sato, K. Comparison of the mutation rates of human influenza A and B viruses. J. Virol. 2006, 80, 3675–3678. [Google Scholar] [CrossRef] [PubMed]
- Gotch, F.; McMichael, A.; Smith, G.; Moss, B. Identification of viral molecules recognized by influenza-specific human cytotoxic T lymphocytes. J. Exp. Med. 1987, 165, 408–416. [Google Scholar] [CrossRef] [PubMed]
- Yewdell, J.W.; Bennink, J.R.; Smith, G.L.; Moss, B. Influenza A virus nucleoprotein is a major target antigen for cross-reactive anti-influenza A virus cytotoxic T lymphocytes. Proc. Natl. Acad. Sci. USA 1985, 82, 1785–1789. [Google Scholar] [CrossRef] [PubMed]
- Rimmelzwaan, G.F.; Boon, A.C.; Voeten, J.T.; Berkhoff, E.G.; Fouchier, R.A.; Osterhaus, A.D. Sequence variation in the influenza A virus nucleoprotein associated with escape from cytotoxic T lymphocytes. Virus Res. 2004, 103, 97–100. [Google Scholar] [CrossRef] [PubMed]
- Valkenburg, S.A.; Quiñones-Parra, S.; Gras, S.; Komadina, N.; McVernon, J.; Wang, Z.; Halim, H.; Iannello, P.; Cole, C.; Laurie, K.; et al. Acute emergence and reversion of influenza A virus quasispecies within CD8+ T cell antigenic peptides. Nat. Commun. 2013. [Google Scholar] [CrossRef] [PubMed]
- Cox, N.J.; Subbarao, K. Influenza. Lancet 1999, 354, 1277–1282. [Google Scholar] [CrossRef]
- Grohskopf, L.A.; Sokolow, L.Z.; Olsen, S.J.; Bresee, J.S.; Broder, K.R.; Karron, R.A. Prevention and Control of Influenza with Vaccines: Recommendations of the Advisory Committee on Immunization Practices, United States, 2015–16 Influenza Season. Morb. Mortal. Wkly. Rep. 2015, 64, 818–825. [Google Scholar] [CrossRef]
- CDC (Centers for Disease Control and Prevention). Cell-Based Flu Vaccines. 2016. Available online: http://www.cdc.gov/flu/protect/vaccine/cell-based.htm (accessed on 12 September 2016). [Google Scholar]
- Belshe, R.; Lee, M.S.; Walker, R.E.; Stoddard, J.; Mendelman, P.M. Safety, immunogenicity and efficacy of intranasal, live attenuated influenza vaccine. Expert Rev. Vaccin. 2004, 3, 643–654. [Google Scholar] [CrossRef] [PubMed]
- Chung, J.R.; Flannery, B.; Thompson, M.G.; Gaglani, M.; Jackson, M.L.; Monto, A.S.; Nowalk, M.P.; Talbot, H.K.; Treanor, J.J.; Belongia, E.A.; et al. Seasonal Effectiveness of Live Attenuated and Inactivated Influenza Vaccine. Pediatrics 2016, 137, e20153279. [Google Scholar] [CrossRef] [PubMed]
- Flannery, B.; Chung, J. Influenza Vaccine Effectiveness, Including LAIV vs IIV in Children and Adolescents, US Flu VE Network, 2015–2016. Available online: https://www.cdc.gov/vaccines/acip/meetings/downloads/slides-2016-06/influenza-05-flannery.pdf (accessed on 10 September 2016).
- CDC (Centers for Disease Control and Prevention). ACIP Votes down Use of LAIV for 2016–2017 Flu Season. 2016. Available online: http://www.cdc.gov/media/releases/2016/s0622-laiv-flu.html (accessed on 12 September 2016). [Google Scholar]
- Tricco, A.C.; Chit, A.; Soobiah, C.; Hallett, D.; Meier, G.; Chen, M.H.; Tashkandi, M.; Bauch, C.T.; Loeb, M. Comparing influenza vaccine efficacy against mismatched and matched strains: A systematic review and meta-analysis. BMC Med. 2013. [Google Scholar] [CrossRef] [PubMed]
- Subbramanian, R.A.; Basha, S.; Shata, M.T.; Brady, R.C.; Bernstein, D.I. Pandemic and seasonal H1N1 influenza hemagglutinin-specific T cell responses elicited by seasonal influenza vaccination. Vaccine 2010, 28, 8258–8267. [Google Scholar] [CrossRef] [PubMed]
- He, X.S.; Holmes, T.H.; Zhang, C.; Mahmood, K.; Kemble, G.W.; Lewis, D.B.; Dekker, C.L.; Greenberg, H.B.; Arvin, A.M. Cellular immune responses in children and adults receiving inactivated or live attenuated influenza vaccines. J. Virol. 2006, 80, 11756–11766. [Google Scholar] [CrossRef] [PubMed]
- Bodewes, R.; Fraaij, P.L.; Geelhoed-Mieras, M.M.; van Baalen, C.A.; Tiddens, H.A.; van Rossum, A.M.; van der Klis, F.R.; Fouchier, R.A.; Osterhaus, A.D.; Rimmelzwaan, G.F. Annual vaccination against influenza virus hampers development of virus-specific CD8(+) T cell immunity in children. J. Virol. 2011, 85, 11995–12000. [Google Scholar] [CrossRef] [PubMed]
- Ehre, C.; Worthington, E.N.; Liesman, R.M.; Grubb, B.R.; Barbier, D.; O’Neal, W.K.; Sallenave, J.M.; Pickles, R.J.; Boucher, R.C. Overexpressing mouse model demonstrates the protective role of Muc5ac in the lungs. Proc. Natl. Acad. Sci. USA 2012, 109, 16528–16533. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, S.; Horie, M.; Daidoji, T.; Honda, T.; Yasugi, M.; Kuno, A.; Komori, T.; Okuzaki, D.; Narimatsu, H.; Nakaya, T.; et al. Influenza A Virus-Induced Expression of a GalNAc Transferase, GALNT3, via MicroRNAs Is Required for Enhanced Viral Replication. J. Virol. 2015, 90, 1788–1801. [Google Scholar] [CrossRef] [PubMed]
- Matrosovich, M.N.; Matrosovich, T.Y.; Gray, T.; Roberts, N.A.; Klenk, H.D. Neuraminidase is important for the initiation of influenza virus infection in human airway epithelium. J. Virol. 2004, 78, 12665–12667. [Google Scholar] [CrossRef] [PubMed]
- Benne, C.A.; Kraaijeveld, C.A.; Van Strijp, J.A.; Brouwer, E.; Harmsen, M.; Verhoef, J.; Van Golde, L.M.; Van Iwaarden, J.F. Interactions of surfactant protein A with influenza A viruses: Binding and neutralization. J. Infect. Dis. 1995, 171, 335–341. [Google Scholar] [CrossRef] [PubMed]
- Hartshorn, K.L.; White, M.R.; Shepherd, V.; Reid, K.; Jensenius, J.C.; Crouch, E.C. Mechanisms of anti-influenza activity of surfactant proteins A and D: Comparison with serum collectins. Am. J. Physiol. 1997, 273, L1156–L1166. [Google Scholar] [PubMed]
- Stauffer, S.; Feng, Y.; Nebioglu, F.; Heilig, R.; Picotti, P.; Helenius, A. Stepwise priming by acidic pH and a high K+ concentration is required for efficient uncoating of influenza A virus cores after penetration. J. Virol. 2014, 88, 13029–13046. [Google Scholar] [CrossRef] [PubMed]
- Luo, M. Influenza virus entry. Adv. Exp. Med. Biol. 2012, 726, 201–221. [Google Scholar] [PubMed]
- Pang, I.K.; Iwasaki, A. Inflammasomes as mediators of immunity against influenza virus. Trends Immunol. 2011, 32, 34–41. [Google Scholar] [CrossRef] [PubMed]
- Patel, J.R.; Garcia-Sastre, A. Activation and regulation of pathogen sensor RIG-I. Cytokine Growth Factor Rev. 2014, 25, 513–523. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Hamming, O.J.; Ank, N.; Paludan, S.R.; Nielsen, A.L.; Hartmann, R. Type III interferon (IFN) induces a type I IFN-like response in a restricted subset of cells through signaling pathways involving both the Jak-STAT pathway and the mitogen-activated protein kinases. J. Virol. 2007, 81, 7749–7758. [Google Scholar] [CrossRef] [PubMed]
- Verhelst, J.; Hulpiau, P.; Saelens, X. Mx proteins: Antiviral gatekeepers that restrain the uninvited. Microbiol. Mol. Biol. Rev. 2013, 77, 551–566. [Google Scholar] [CrossRef] [PubMed]
- Longhi, M.P.; Trumpfheller, C.; Idoyaga, J.; Caskey, M.; Matos, I.; Kluger, C.; Salazar, A.M.; Colonna, M.; Steinman, R.M. Dendritic cells require a systemic type I interferon response to mature and induce CD4+ Th1 immunity with poly IC as adjuvant. J. Exp. Med. 2009, 206, 1589–1602. [Google Scholar] [CrossRef] [PubMed]
- Ichinohe, T.; Pang, I.K.; Iwasaki, A. Influenza virus activates inflammasomes via its intracellular M2 ion channel. Nat. Immunol. 2010, 11, 404–410. [Google Scholar] [CrossRef] [PubMed]
- Acosta-Rodriguez, E.V.; Napolitani, G.; Lanzavecchia, A.; Sallusto, F. Interleukins 1beta and 6 but not transforming growth factor-beta are essential for the differentiation of interleukin 17-producing human T helper cells. Nat. Immunol. 2007, 8, 942–949. [Google Scholar] [CrossRef] [PubMed]
- Ben-Sasson, S.Z.; Hu-Li, J.; Quiel, J.; Cauchetaux, S.; Ratner, M.; Shapira, I.; Dinarello, C.A.; Paul, W.E. IL-1 acts directly on CD4 T cells to enhance their antigen-driven expansion and differentiation. Proc. Natl. Acad. Sci. USA 2009, 106, 7119–7124. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.M.; Lee, Y.W.; Lee, K.J.; Kim, H.S.; Cho, S.W.; van Rooijen, N.; Guan, Y.; Seo, S.H. Alveolar macrophages are indispensable for controlling influenza viruses in lungs of pigs. J. Virol. 2008, 82, 4265–4274. [Google Scholar] [CrossRef] [PubMed]
- Tumpey, T.M.; Garcia-Sastre, A.; Taubenberger, J.K.; Palese, P.; Swayne, D.E.; Pantin-Jackwood, M.J.; Schultz-Cherry, S.; Solorzano, A.; Van Rooijen, N.; Katz, J.M.; et al. Pathogenicity of influenza viruses with genes from the 1918 pandemic virus: Functional roles of alveolar macrophages and neutrophils in limiting virus replication and mortality in mice. J. Virol. 2005, 79, 14933–14944. [Google Scholar] [CrossRef] [PubMed]
- Jayasekera, J.P.; Vinuesa, C.G.; Karupiah, G.; King, N.J. Enhanced antiviral antibody secretion and attenuated immunopathology during influenza virus infection in nitric oxide synthase-2-deficient mice. J. Gen. Virol. 2006, 87, 3361–3371. [Google Scholar] [CrossRef] [PubMed]
- Peper, R.L.; Van Campen, H. Tumor necrosis factor as a mediator of inflammation in influenza A viral pneumonia. Microb. Pathog. 1995, 19, 175–183. [Google Scholar] [CrossRef] [PubMed]
- Wijburg, O.L.; DiNatale, S.; Vadolas, J.; van Rooijen, N.; Strugnell, R.A. Alveolar macrophages regulate the induction of primary cytotoxic T-lymphocyte responses during influenza virus infection. J. Virol. 1997, 71, 9450–9457. [Google Scholar] [PubMed]
- El Bakkouri, K.; Descamps, F.; De Filette, M.; Smet, A.; Festjens, E.; Birkett, A.; Van Rooijen, N.; Verbeek, S.; Fiers, W.; Saelens, X. Universal vaccine based on ectodomain of matrix protein 2 of influenza A: Fc receptors and alveolar macrophages mediate protection. J. Immunol. 2011, 186, 1022–1031. [Google Scholar] [CrossRef] [PubMed]
- Lambrecht, B.N.; Hammad, H. Lung dendritic cells in respiratory viral infection and asthma: From protection to immunopathology. Annu. Rev. Immunol. 2012, 30, 243–270. [Google Scholar] [CrossRef] [PubMed]
- GeurtsvanKessel, C.H.; Lambrecht, B.N. Division of labor between dendritic cell subsets of the lung. Mucosal. Immunol. 2008, 1, 442–450. [Google Scholar] [CrossRef] [PubMed]
- Banchereau, J.; Steinman, R.M. Dendritic cells and the control of immunity. Nature 1998, 392, 245–252. [Google Scholar] [CrossRef] [PubMed]
- Guermonprez, P.; Valladeau, J.; Zitvogel, L.; Thery, C.; Amigorena, S. Antigen presentation and T cell stimulation by dendritic cells. Annu. Rev. Immunol. 2002, 20, 621–667. [Google Scholar] [CrossRef] [PubMed]
- Mandelboim, O.; Lieberman, N.; Lev, M.; Paul, L.; Arnon, T.I.; Bushkin, Y.; Davis, D.M.; Strominger, J.L.; Yewdell, J.W.; Porgador, A. Recognition of haemagglutinins on virus-infected cells by NKp46 activates lysis by human NK cells. Nature 2001, 409, 1055–1060. [Google Scholar] [CrossRef] [PubMed]
- Arnon, T.I.; Lev, M.; Katz, G.; Chernobrov, Y.; Porgador, A.; Mandelboim, O. Recognition of viral hemagglutinins by NKp44 but not by NKp30. Eur. J. Immunol. 2001, 31, 2680–2689. [Google Scholar] [CrossRef]
- Hashimoto, G.; Wright, P.F.; Karzon, D.T. Antibody-dependent cell-mediated cytotoxicity against influenza virus-infected cells. J. Infect. Dis. 1983, 148, 785–794. [Google Scholar] [CrossRef] [PubMed]
- Del Rio, M.L.; Rodriguez-Barbosa, J.I.; Kremmer, E.; Forster, R. CD103− and CD103+ bronchial lymph node dendritic cells are specialized in presenting and cross-presenting innocuous antigen to CD4+ and CD8+ T cells. J. Immunol. 2007, 178, 6861–6866. [Google Scholar] [CrossRef] [PubMed]
- Hao, X.; Kim, T.S.; Braciale, T.J. Differential response of respiratory dendritic cell subsets to influenza virus infection. J. Virol. 2008, 82, 4908–4919. [Google Scholar] [CrossRef] [PubMed]
- Ioannidis, L.J.; Verity, E.E.; Crawford, S.; Rockman, S.P.; Brown, L.E. Abortive replication of influenza virus in mouse dendritic cells. J. Virol. 2012, 86, 5922–5925. [Google Scholar] [CrossRef] [PubMed]
- Herrera, O.B.; Golshayan, D.; Tibbott, R.; Salcido Ochoa, F.; James, M.J.; Marelli-Berg, F.M.; Lechler, R.I. A novel pathway of alloantigen presentation by dendritic cells. J. Immunol. 2004, 173, 4828–4837. [Google Scholar] [CrossRef] [PubMed]
- Joly, E.; Hudrisier, D. What is trogocytosis and what is its purpose? Nat. Immunol. 2003. [Google Scholar] [CrossRef] [PubMed]
- Yewdell, J.W.; Dolan, B.P. Immunology: Cross-dressers turn on T cells. Nature 2011, 471, 581–582. [Google Scholar] [CrossRef] [PubMed]
- Hintzen, G.; Ohl, L.; del Rio, M.L.; Rodriguez-Barbosa, J.I.; Pabst, O.; Kocks, J.R.; Krege, J.; Hardtke, S.; Forster, R. Induction of tolerance to innocuous inhaled antigen relies on a CCR7-dependent dendritic cell-mediated antigen transport to the bronchial lymph node. J. Immunol. 2006, 177, 7346–7354. [Google Scholar] [CrossRef] [PubMed]
- Hufford, M.M.; Kim, T.S.; Sun, J.; Braciale, T.J. The effector T cell response to influenza infection. Curr. Top. Microbiol. Immunol. 2015, 386, 423–455. [Google Scholar] [PubMed]
- Helft, J.; Manicassamy, B.; Guermonprez, P.; Hashimoto, D.; Silvin, A.; Agudo, J.; Brown, B.D.; Schmolke, M.; Miller, J.C.; Leboeuf, M.; et al. Cross-presenting CD103+ dendritic cells are protected from influenza virus infection. J. Clin. Investig. 2012, 122, 4037–4047. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.S.; Braciale, T.J. Respiratory dendritic cell subsets differ in their capacity to support the induction of virus-specific cytotoxic CD8+ T cell responses. PLoS ONE 2009, 4, e4204. [Google Scholar] [CrossRef] [PubMed]
- Moltedo, B.; Li, W.; Yount, J.S.; Moran, T.M. Unique type I interferon responses determine the functional fate of migratory lung dendritic cells during influenza virus infection. PLoS Pathog. 2011, 7, e1002345. [Google Scholar] [CrossRef] [PubMed]
- Nakano, H.; Lin, K.L.; Yanagita, M.; Charbonneau, C.; Cook, D.N.; Kakiuchi, T.; Gunn, M.D. Blood-derived inflammatory dendritic cells in lymph nodes stimulate acute T helper type 1 immune responses. Nat. Immunol. 2009, 10, 394–402. [Google Scholar] [CrossRef] [PubMed]
- Von Andrian, U.H.; Mackay, C.R. T-cell function and migration. Two sides of the same coin. N. Engl. J. Med. 2000, 343, 1020–1034. [Google Scholar] [PubMed]
- O’Shea, J.J.; Paul, W.E. Mechanisms underlying lineage commitment and plasticity of helper CD4+ T cells. Science 2010, 327, 1098–1102. [Google Scholar] [CrossRef] [PubMed]
- Sallusto, F.; Kremmer, E.; Palermo, B.; Hoy, A.; Ponath, P.; Qin, S.; Förster, R.; Lipp, M.; Lanzavecchia, A. Switch in chemokine receptor expression upon TCR stimulation reveals novel homing potential for recently activated T cells. Eur J. Immunol. 1999, 29, 2037–2045. [Google Scholar] [CrossRef]
- Warnock, R.A.; Askari, S.; Butcher, E.C.; Von Andrian, U.H. Molecular mechanisms of lymphocyte homing to peripheral lymph nodes. J. Exp. Med. 1998, 187, 205–216. [Google Scholar] [CrossRef] [PubMed]
- Griffith, J.W.; Sokol, C.L.; Luster, A.D. Chemokines and chemokine receptors: Positioning cells for host defense and immunity. Annu. Rev. Immunol. 2014, 32, 659–702. [Google Scholar] [CrossRef] [PubMed]
- Thatte, J.; Dabak, V.; Williams, M.B.; Braciale, T.J.; Ley, K. LFA-1 is required for retention of effector CD8 T cells in mouse lungs. Blood 2003, 101, 4916–4922. [Google Scholar] [CrossRef] [PubMed]
- Mikhak, Z.; Strassner, J.P.; Luster, A.D. Lung dendritic cells imprint T cell lung homing and promote lung immunity through the chemokine receptor CCR4. J. Exp. Med. 2013, 210, 1855–1869. [Google Scholar] [CrossRef] [PubMed]
- Galkina, E.; Thatte, J.; Dabak, V.; Williams, M.B.; Ley, K.; Braciale, T.J. Preferential migration of effector CD8+ T cells into the interstitium of the normal lung. J. Clin. Investig. 2005, 115, 3473–3483. [Google Scholar] [CrossRef] [PubMed]
- Slütter, B.; Pewe, L.L.; Kaech, S.M.; Harty, J.T. Lung airway-surveilling CXCR3(hi) memory CD8(+) T cells are critical for protection against influenza A virus. Immunity 2013, 39, 939–948. [Google Scholar] [CrossRef] [PubMed]
- Lim, K.; Hyun, Y.M.; Lambert-Emo, K.; Capece, T.; Bae, S.; Miller, R.; Topham, D.J.; Kim, M. Neutrophil trails guide influenza-specific CD8(+) T cells in the airways. Science 2015. [Google Scholar] [CrossRef] [PubMed]
- Russell, J.H.; Ley, T.J. Lymphocyte-mediated cytotoxicity. Annu. Rev. Immunol. 2002, 20, 323–370. [Google Scholar] [CrossRef] [PubMed]
- Topham, D.J.; Tripp, R.A.; Sarawar, S.R.; Sangster, M.Y.; Doherty, P.C. Immune CD4+ T cells promote the clearance of influenza virus from major histocompatibility complex class II−/− respiratory epithelium. J. Virol. 1996, 70, 1288–1291. [Google Scholar] [PubMed]
- Topham, D.J.; Tripp, R.A.; Hamilton-Easton, A.M.; Sarawar, S.R.; Doherty, P.C. Quantitative analysis of the influenza virus-specific CD4+ T cell memory in the absence of B cells and Ig. J. Immunol. 1996, 157, 2947–2952. [Google Scholar] [PubMed]
- Topham, D.J.; Doherty, P.C. Clearance of an influenza A virus by CD4+ T cells is inefficient in the absence of B cells. J. Virol. 1998, 72, 882–885. [Google Scholar] [PubMed]
- Hua, L.; Yao, S.; Pham, D.; Jiang, L.; Wright, J.; Sawant, D.; Dent, A.L.; Braciale, T.J.; Kaplan, M.H.; Sun, J. Cytokine-dependent induction of CD4+ T cells with cytotoxic potential during influenza virus infection. J. Virol. 2013, 87, 11884–11893. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Yamane, H.; Paul, W.E. Differentiation of effector CD4 T cell populations (*). Annu. Rev. Immunol. 2010, 28, 445–489. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Paul, W.E. Heterogeneity and plasticity of T helper cells. Cell Res. 2010, 20, 4–12. [Google Scholar] [CrossRef] [PubMed]
- Mosmann, T.R.; Cherwinski, H.; Bond, M.W.; Giedlin, M.A.; Coffman, R.L. Two types of murine helper T cell clone. I. Definition according to profiles of lymphokine activities and secreted proteins. J. Immunol. 1986, 136, 2348–2357. [Google Scholar] [PubMed]
- De Vries, J.E.; Punnonen, J.; Cocks, B.G.; de Waal Malefyt, R.; Aversa, G. Regulation of the human IgE response by IL4 and IL13. Res. Immunol. 1993, 144, 597–601. [Google Scholar] [CrossRef]
- Roman, E.; Miller, E.; Harmsen, A.; Wiley, J.; Von Andrian, U.H.; Huston, G.; Swain, S.L. CD4 effector T cell subsets in the response to influenza: Heterogeneity, migration, and function. J. Exp. Med. 2002, 196, 957–968. [Google Scholar] [CrossRef] [PubMed]
- Gerhard, W.; Mozdzanowska, K.; Furchner, M.; Washko, G.; Maiese, K. Role of the B-cell response in recovery of mice from primary influenza virus infection. Immunol. Rev. 1997, 159, 95–103. [Google Scholar] [CrossRef] [PubMed]
- Divekar, A.A.; Zaiss, D.M.; Lee, F.E.; Liu, D.; Topham, D.J.; Sijts, A.J.; Mosmann, T.R. Protein vaccines induce uncommitted IL-2-secreting human and mouse CD4 T cells, whereas infections induce more IFN-gamma-secreting cells. J. Immunol. 2006, 176, 1465–1473. [Google Scholar] [CrossRef] [PubMed]
- Park, H.; Li, Z.; Yang, X.O.; Chang, S.H.; Nurieva, R.; Wang, Y.H.; Wang, Y.; Hood, L.; Zhu, Z.; Tian, Q.; et al. A distinct lineage of CD4 T cells regulates tissue inflammation by producing interleukin 17. Nat. Immunol. 2005, 6, 1133–1141. [Google Scholar] [CrossRef] [PubMed]
- Liang, S.C.; Tan, X.Y.; Luxenberg, D.P.; Karim, R.; Dunussi-Joannopoulos, K.; Collins, M.; Fouser, L.A. Interleukin (IL)-22 and IL-17 are coexpressed by Th17 cells and cooperatively enhance expression of antimicrobial peptides. J. Exp. Med. 2006, 203, 2271–2279. [Google Scholar] [CrossRef] [PubMed]
- Kudva, A.; Scheller, E.V.; Robinson, K.M.; Crowe, C.R.; Choi, S.M.; Slight, S.R.; Khader, S.A.; Dubin, P.J.; Enelow, R.I.; Kolls, J.K.; et al. Influenza A inhibits Th17-mediated host defense against bacterial pneumonia in mice. J. Immunol. 2011, 186, 1666–1674. [Google Scholar] [CrossRef] [PubMed]
- Hamada, H.; Garcia-Hernandez Mde, L.; Reome, J.B.; Misra, S.K.; Strutt, T.M.; McKinstry, K.K.; Cooper, A.M.; Swain, S.L.; Dutton, R.W. Tc17, a unique subset of CD8 T cells that can protect against lethal influenza challenge. J. Immunol. 2009, 182, 3469–3481. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Chan, C.C.; Yang, M.; Deng, J.; Poon, V.K.; Leung, V.H.; Ko, K.H.; Zhou, J.; Yuen, K.Y.; Zheng, B.J.; et al. A critical role of IL-17 in modulating the B-cell response during H5N1 influenza virus infection. Cell. Mol. Immunol. 2011, 8, 462–468. [Google Scholar] [CrossRef] [PubMed]
- Crowe, C.R.; Chen, K.; Pociask, D.A.; Alcorn, J.F.; Krivich, C.; Enelow, R.I.; Ross, T.M.; Witztum, J.L.; Kolls, J.K. Critical role of IL-17RA in immunopathology of influenza infection. J. Immunol. 2009, 183, 5301–5310. [Google Scholar] [CrossRef] [PubMed]
- Gopal, R.; Rangel-Moreno, J.; Fallert Junecko, B.A.; Mallon, D.J.; Chen, K.; Pociask, D.A.; Connell, T.D.; Reinhart, T.A.; Alcorn, J.F.; Ross, T.M.; et al. Mucosal pre-exposure to Th17-inducing adjuvants exacerbates pathology after influenza infection. Am. J. Pathol. 2014, 184, 55–63. [Google Scholar] [CrossRef] [PubMed]
- Larkin, J., 3rd; Rankin, A.L.; Picca, C.C.; Riley, M.P.; Jenks, S.A.; Sant, A.J.; Caton, A.J. CD4+CD25+ regulatory T cell repertoire formation shaped by differential presentation of peptides from a self-antigen. J. Immunol. 2008, 180, 2149–2157. [Google Scholar] [CrossRef] [PubMed]
- Curotto de Lafaille, M.A.; Lino, A.C.; Kutchukhidze, N.; Lafaille, J.J. CD25- T cells generate CD25+Foxp3+ regulatory T cells by peripheral expansion. J. Immunol. 2004, 173, 7259–7268. [Google Scholar] [CrossRef] [PubMed]
- Betts, R.J.; Prabhu, N.; Ho, A.W.; Lew, F.C.; Hutchinson, P.E.; Rotzschke, O.; Macary, P.A.; Kemeny, D.M. Influenza A virus infection results in a robust, antigen-responsive, and widely disseminated Foxp3+ regulatory T cell response. J. Virol. 2012, 86, 2817–2825. [Google Scholar] [CrossRef] [PubMed]
- Crotty, S. T follicular helper cell differentiation, function, and roles in disease. Immunity 2014, 41, 529–542. [Google Scholar] [CrossRef] [PubMed]
- Good-Jacobson, K.L.; Szumilas, C.G.; Chen, L.; Sharpe, A.H.; Tomayko, M.M.; Shlomchik, M.J. PD-1 regulates germinal center B cell survival and the formation and affinity of long-lived plasma cells. Nat. Immunol. 2010, 11, 535–542. [Google Scholar] [CrossRef] [PubMed]
- Schwickert, T.A.; Victora, G.D.; Fooksman, D.R.; Kamphorst, A.O.; Mugnier, M.R.; Gitlin, A.D.; Dustin, M.L.; Nussenzweig, M.C. A dynamic T cell-limited checkpoint regulates affinity-dependent B cell entry into the germinal center. J. Exp. Med. 2011, 208, 1243–1252. [Google Scholar] [CrossRef] [PubMed]
- Bentebibel, S.E.; Lopez, S.; Obermoser, G.; Schmitt, N.; Mueller, C.; Harrod, C.; Flano, E.; Mejias, A.; Albrecht, R.A.; Blankenship, D.; et al. Induction of ICOS+CXCR3+CXCR5+ TH cells correlates with antibody responses to influenza vaccination. Sci. Transl. Med. 2013. [Google Scholar] [CrossRef] [PubMed]
- Bousso, P.; Robey, E. Dynamics of CD8+ T cell priming by dendritic cells in intact lymph nodes. Nat. Immunol. 2003, 4, 579–585. [Google Scholar] [CrossRef] [PubMed]
- Kagi, D.; Vignaux, F.; Ledermann, B.; Burki, K.; Depraetere, V.; Nagata, S.; Hengartner, H.; Golstein, P. Fas and perforin pathways as major mechanisms of T cell-mediated cytotoxicity. Science 1994, 265, 528–530. [Google Scholar] [CrossRef] [PubMed]
- Brincks, E.L.; Katewa, A.; Kucaba, T.A.; Griffith, T.S.; Legge, K.L. CD8 T cells utilize TRAIL to control influenza virus infection. J. Immunol. 2008, 181, 4918–4925. [Google Scholar] [CrossRef] [PubMed]
- Welsh, R.M.; McNally, J.M. Immune deficiency, immune silencing, and clonal exhaustion of T cell responses during viral infections. Curr. Opin. Microbiol. 1999, 2, 382–327. [Google Scholar] [CrossRef]
- Razvi, E.S.; Jiang, Z.; Woda, B.A.; Welsh, R.M. Lymphocyte apoptosis during the silencing of the immune response to acute viral infections in normal, LPR, and BCL-2-transgenic mice. Am. J. Pathol. 1995, 147, 79–91. [Google Scholar] [PubMed]
- Buchholz, V.R.; Schumacher, T.N.; Busch, D.H. T Cell Fate at the Single-Cell Level. Annu. Rev. Immunol. 2016, 34, 65–92. [Google Scholar] [CrossRef] [PubMed]
- Kondrack, R.M.; Harbertson, J.; Tan, J.T.; McBreen, M.E.; Surh, C.D.; Bradley, L.M. Interleukin 7 regulates the survival and generation of memory CD4 cells. J. Exp. Med. 2003, 198, 1797–1806. [Google Scholar] [CrossRef] [PubMed]
- Ku, C.C.; Murakami, M.; Sakamoto, A.; Kappler, J.; Marrack, P. Control of homeostasis of CD8+ memory T cells by opposing cytokines. Science 2000, 288, 675–678. [Google Scholar] [CrossRef] [PubMed]
- Purton, J.F.; Tan, J.T.; Rubinstein, M.P.; Kim, D.M.; Sprent, J.; Surh, C.D. Antiviral CD4+ memory T cells are IL-15 dependent. J. Exp. Med. 2007, 204, 951–961. [Google Scholar] [CrossRef] [PubMed]
- Tan, J.T.; Dudl, E.; LeRoy, E.; Murray, R.; Sprent, J.; Weinberg, K.I.; Surh, C.D. IL-7 is critical for homeostatic proliferation and survival of naive T cells. Proc. Natl. Acad. Sci. USA 2001, 98, 8732–8737. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Sun, S.; Hwang, I.; Tough, D.F.; Sprent, J. Potent and selective stimulation of memory-phenotype CD8+ T cells in vivo by IL-15. Immunity 1998, 8, 591–599. [Google Scholar] [CrossRef]
- Kaech, S.M.; Wherry, E.J. Heterogeneity and cell-fate decisions in effector and memory CD8+ T cell differentiation during viral infection. Immunity 2007, 27, 393–405. [Google Scholar] [CrossRef] [PubMed]
- Sallusto, F.; Lenig, D.; Forster, R.; Lipp, M.; Lanzavecchia, A. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature 1999, 401, 708–712. [Google Scholar] [CrossRef] [PubMed]
- Gebhardt, T.; Wakim, L.M.; Eidsmo, L.; Reading, P.C.; Heath, W.R.; Carbone, F.R. Memory T cells in nonlymphoid tissue that provide enhanced local immunity during infection with herpes simplex virus. Nat. Immunol. 2009, 10, 524–530. [Google Scholar] [CrossRef] [PubMed]
- Teijaro, J.R.; Turner, D.; Pham, Q.; Wherry, E.J.; Lefrancois, L.; Farber, D.L. Cutting edge: Tissue-retentive lung memory CD4 T cells mediate optimal protection to respiratory virus infection. J. Immunol. 2011, 187, 5510–5514. [Google Scholar] [CrossRef] [PubMed]
- Laidlaw, B.J.; Zhang, N.; Marshall, H.D.; Staron, M.M.; Guan, T.; Hu, Y.; Cauley, L.S.; Craft, J.; Kaech, S.M. CD4+ T cell help guides formation of CD103+ lung-resident memory CD8+ T cells during influenza viral infection. Immunity 2014, 41, 633–645. [Google Scholar] [CrossRef] [PubMed]
- Hogan, R.J.; Usherwood, E.J.; Zhong, W.; Roberts, A.A.; Dutton, R.W.; Harmsen, A.G.; Woodland, D.L. Activated antigen-specific CD8+ T cells persist in the lungs following recovery from respiratory virus infections. J. Immunol. 2001, 166, 1813–1822. [Google Scholar] [CrossRef] [PubMed]
- Piet, B.; de Bree, G.J.; Smids-Dierdorp, B.S.; van der Loos, C.M.; Remmerswaal, E.B.; von der Thusen, J.H.; van Haarst, J.M.; Eerenberg, J.P.; ten Brinke, A.; van der Bij, W.; et al. CD8(+) T cells with an intraepithelial phenotype upregulate cytotoxic function upon influenza infection in human lung. J. Clin. Investig. 2011, 121, 2254–2263. [Google Scholar] [CrossRef] [PubMed]
- Turner, D.L.; Bickham, K.L.; Thome, J.J.; Kim, C.Y.; D’ovidio, F.; Wherry, E.J.; Farber, D.L. Lung niches for the generation and maintenance of tissue-resident memory T cells. Mucosal. Immunol. 2014, 7, 501–510. [Google Scholar] [CrossRef] [PubMed]
- Foulds, K.E.; Zenewicz, L.A.; Shedlock, D.J.; Jiang, J.; Troy, A.E.; Shen, H. Cutting edge: CD4 and CD8 T cells are intrinsically different in their proliferative responses. J. Immunol. 2002, 168, 1528–1532. [Google Scholar] [CrossRef] [PubMed]
- Hogan, R.J.; Zhong, W.; Usherwood, E.J.; Cookenham, T.; Roberts, A.D.; Woodland, D.L. Protection from respiratory virus infections can be mediated by antigen-specific CD4(+) T cells that persist in the lungs. J. Exp. Med. 2001, 193, 981–986. [Google Scholar] [CrossRef] [PubMed]
- Lee, L.Y.; Ha do, L.A.; Simmons, C.; de Jong, M.D.; Chau, N.V.; Schumacher, R.; Peng, Y.C.; McMichael, A.J.; Farrar, J.J.; Smith, G.L.; et al. Memory T cells established by seasonal human influenza A infection cross-react with avian influenza A (H5N1) in healthy individuals. J. Clin. Investig. 2008, 118, 3478–3490. [Google Scholar] [CrossRef] [PubMed]
- Alexander, J.; Bilsel, P.; del Guercio, M.F.; Stewart, S.; Marinkovic-Petrovic, A.; Southwood, S.; Crimi, C.; Vang, L.; Walker, L.; Ishioka, G.; et al. Universal influenza DNA vaccine encoding conserved CD4+ T cell epitopes protects against lethal viral challenge in HLA-DR transgenic mice. Vaccine 2010, 28, 664–672. [Google Scholar] [CrossRef] [PubMed]
- Teijaro, J.R.; Verhoeven, D.; Page, C.A.; Turner, D.; Farber, D.L. Memory CD4 T cells direct protective responses to influenza virus in the lungs through helper-independent mechanisms. J. Virol. 2010, 84, 9217–9226. [Google Scholar] [CrossRef] [PubMed]
- Belz, G.T.; Wodarz, D.; Diaz, G.; Nowak, M.A.; Doherty, P.C. Compromised influenza virus-specific CD8(+)-T-cell memory in CD4(+)-T-cell-deficient mice. J. Virol. 2002, 76, 12388–12393. [Google Scholar] [CrossRef] [PubMed]
- MacLeod, M.K.; David, A.; McKee, A.S.; Crawford, F.; Kappler, J.W.; Marrack, P. Memory CD4 T cells that express CXCR5 provide accelerated help to B cells. J. Immunol. 2011, 186, 2889–2896. [Google Scholar] [CrossRef] [PubMed]
- McKinstry, K.K.; Strutt, T.M.; Kuang, Y.; Brown, D.M.; Sell, S.; Dutton, R.W.; Swain, S.L. Memory CD4+ T cells protect against influenza through multiple synergizing mechanisms. J. Clin. Investig. 2012, 122, 2847–2856. [Google Scholar] [CrossRef] [PubMed]
- Nolz, J.C.; Harty, J.T. Protective capacity of memory CD8+ T cells is dictated by antigen exposure history and nature of the infection. Immunity 2011, 34, 781–793. [Google Scholar] [CrossRef] [PubMed]
- Sridhar, S.; Begom, S.; Bermingham, A.; Hoschler, K.; Adamson, W.; Carman, W.; Bean, T.; Barclay, W.; Deeks, J.J.; Lalvani, A. Cellular immune correlates of protection against symptomatic pandemic influenza. Nat. Med. 2013, 19, 1305–1312. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, T.M.; Li, C.K.; Chui, C.S.; Huang, A.K.; Perkins, M.; Liebner, J.C.; Lambkin-Williams, R.; Gilbert, A.; Oxford, J.; Nicholas, B.; et al. Preexisting influenza-specific CD4+ T cells correlate with disease protection against influenza challenge in humans. Nat. Med. 2012, 18, 274–280. [Google Scholar] [CrossRef] [PubMed]
- Wakim, L.M.; Gupta, N.; Mintern, J.D.; Villadangos, J.A. Enhanced survival of lung tissue-resident memory CD8(+) T cells during infection with influenza virus due to selective expression of IFITM3. Nat. Immunol. 2013, 14, 238–245. [Google Scholar] [CrossRef] [PubMed]
- Wakim, L.M.; Woodward-Davis, A.; Liu, R.; Hu, Y.; Villadangos, J.; Smyth, G.; Bevan, M.J. The molecular signature of tissue resident memory CD8 T cells isolated from the brain. J. Immunol. 2012, 189, 3462–3471. [Google Scholar] [CrossRef] [PubMed]
- Desai, T.M.; Marin, M.; Chin, C.R.; Savidis, G.; Brass, A.L.; Melikyan, G.B. IFITM3 restricts influenza A virus entry by blocking the formation of fusion pores following virus-endosome hemifusion. PLoS Pathog. 2014, 10, e1004048. [Google Scholar] [CrossRef] [PubMed]
- Treanor, J.J.; Kotloff, K.; Betts, R.F.; Belshe, R.; Newman, F.; Iacuzio, D.; Wittes, J.; Bryant, M. Evaluation of trivalent, live, cold-adapted (CAIV-T) and inactivated (TIV) influenza vaccines in prevention of virus infection and illness following challenge of adults with wild-type influenza A (H1N1), A (H3N2), and B viruses. Vaccine 1999, 18, 899–906. [Google Scholar] [CrossRef]
- Beyer, W.E.; Palache, A.M.; de Jong, J.C.; Osterhaus, A.D. Cold-adapted live influenza vaccine versus inactivated vaccine: Systemic vaccine reactions, local and systemic antibody response, and vaccine efficacy. A meta-analysis. Vaccine 2002, 20, 1340–1353. [Google Scholar] [CrossRef]
- Abramson, J.S. Intranasal, cold-adapted, live, attenuated influenza vaccine. Pediatr. Infect. Dis. J. 1999, 18, 1103–1104. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.; Zengel, J.R.; Suguitan, A.L., Jr.; Xu, Q.; Wang, W.; Lin, J.; Jin, H. Evaluation of the humoral and cellular immune responses elicited by the live attenuated and inactivated influenza vaccines and their roles in heterologous protection in ferrets. J. Infect. Dis. 2013, 208, 594–602. [Google Scholar] [CrossRef] [PubMed]
- Belshe, R.B.; Edwards, K.M.; Vesikari, T.; Black, S.V.; Walker, R.E.; Hultquist, M.; Kemble, G.; Connor, E.M. Live attenuated versus inactivated influenza vaccine in infants and young children. N. Engl. J. Med. 2007, 356, 685–696. [Google Scholar] [CrossRef] [PubMed]
- Ambrose, C.S.; Wu, X.; Knuf, M.; Wutzler, P. The efficacy of intranasal live attenuated influenza vaccine in children 2 through 17 years of age: A meta-analysis of 8 randomized controlled studies. Vaccine 2012, 30, 886–892. [Google Scholar] [CrossRef] [PubMed]
- Jin, H.; Subbarao, K. Live attenuated influenza vaccine. Curr. Top. Microbiol. Immunol. 2015, 386, 181–204. [Google Scholar] [PubMed]
- Monto, A.S.; Ohmit, S.E.; Petrie, J.G.; Johnson, E.; Truscon, R.; Teich, E.; Rotthoff, J.; Boulton, M.; Victor, J.C. Comparative efficacy of inactivated and live attenuated influenza vaccines. N. Engl. J. Med. 2009, 361, 1260–1267. [Google Scholar] [CrossRef] [PubMed]
- Nichol, K.L.; Mendelman, P.M.; Mallon, K.P.; Jackson, L.A.; Gorse, G.J.; Belshe, R.B.; Glezen, W.P.; Wittes, J. Effectiveness of live, attenuated intranasal influenza virus vaccine in healthy, working adults: A randomized controlled trial. JAMA 1999, 282, 137–144. [Google Scholar] [CrossRef] [PubMed]
- De Villiers, P.J.; Steele, A.D.; Hiemstra, L.A.; Rappaport, R.; Dunning, A.J.; Gruber, W.C.; Forrest, B.D. Efficacy and safety of a live attenuated influenza vaccine in adults 60 years of age and older. Vaccine 2009, 28, 228–234. [Google Scholar] [CrossRef] [PubMed]
- Hobson, D.; Curry, R.L.; Beare, A.S.; Ward-Gardner, A. The role of serum haemagglutination-inhibiting antibody in protection against challenge infection with influenza A2 and B viruses. J. Hyg. 1972, 70, 767–777. [Google Scholar] [CrossRef] [PubMed]
- Black, S.; Nicolay, U.; Vesikari, T.; Knuf, M.; Del Giudice, G.; Della Cioppa, G.; Tsai, T.; Clemens, R.; Rappuoli, R. Hemagglutination inhibition antibody titers as a correlate of protection for inactivated influenza vaccines in children. Pediatr. Infect. Dis. J. 2011, 30, 1081–1085. [Google Scholar] [CrossRef] [PubMed]
- Gravenstein, S.; Drinka, P.; Duthie, E.H.; Miller, B.A.; Brown, C.S.; Hensley, M.; Circo, R.; Langer, E.; Ershler, W.B. Efficacy of an influenza hemagglutinin-diphtheria toxoid conjugate vaccine in elderly nursing home subjects during an influenza outbreak. J. Am. Geriatr. Soc. 1994, 42, 245–251. [Google Scholar] [CrossRef] [PubMed]
- Glezen, W.P.; Keitel, W.A.; Taber, L.H.; Piedra, P.A.; Clover, R.D.; Couch, R.B. Age distribution of patients with medically-attended illnesses caused by sequential variants of influenza A/H1N1: Comparison to age-specific infection rates, 1978–1989. Am. J. Epidemiol. 1991, 133, 296–304. [Google Scholar] [PubMed]
- Jordan, W.S., Jr.; Badger, G.F.; Dingle, J.H. A study of illness in a group of Cleveland families. XVI. The epidemiology of influenza, 1948–1953. Am. J. Hyg. 1958, 68, 169–189. [Google Scholar] [PubMed]
- Epstein, S.L. Prior H1N1 influenza infection and susceptibility of Cleveland Family Study participants during the H2N2 pandemic of 1957: An experiment of nature. J. Infect. Dis. 2006, 193, 49–53. [Google Scholar] [CrossRef] [PubMed]
- Hayward, A.C.; Wang, L.; Goonetilleke, N.; Fragaszy, E.B.; Bermingham, A.; Copas, A.; Dukes, O.; Millett, E.R.; Nazareth, I.; Nguyen-Van-Tam, J.S.; et al. Natural T Cell-mediated Protection against Seasonal and Pandemic Influenza. Results of the Flu Watch Cohort Study. Am. J. Respir. Crit. Care Med. 2015, 191, 1422–1431. [Google Scholar] [CrossRef] [PubMed]
- McMichael, A.J.; Gotch, F.M.; Noble, G.R.; Beare, P.A. Cytotoxic T-cell immunity to influenza. N. Engl. J. Med. 1983, 309, 13–17. [Google Scholar] [CrossRef] [PubMed]
- Tetsutani, K.; Ishii, K.J. Adjuvants in influenza vaccines. Vaccine 2012, 30, 7658–7661. [Google Scholar] [CrossRef] [PubMed]
- Wan, H.; Gao, J.; Xu, K.; Chen, H.; Couzens, L.K.; Rivers, K.H.; Easterbrook, J.D.; Yang, K.; Zhong, L.; Rajabi, M.; et al. Molecular basis for broad neuraminidase immunity: Conserved epitopes in seasonal and pandemic H1N1 as well as H5N1 influenza viruses. J. Virol. 2013, 87, 9290–9300. [Google Scholar] [CrossRef] [PubMed]
- Berthoud, T.K.; Hamill, M.; Lillie, P.J.; Hwenda, L.; Collins, K.A.; Ewer, K.J.; Milicic, A.; Poyntz, H.C.; Lambe, T.; Fletcher, H.A.; et al. Potent CD8+ T-cell immunogenicity in humans of a novel heterosubtypic influenza A vaccine, MVA-NP+M1. Clin. Infect. Dis. 2011, 52, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Antrobus, R.D.; Lillie, P.J.; Berthoud, T.K.; Spencer, A.J.; McLaren, J.E.; Ladell, K.; Lambe, T.; Milicic, A.; Price, D.A.; Hill, A.V.; et al. A T cell-inducing influenza vaccine for the elderly: Safety and immunogenicity of MVA-NP+M1 in adults aged over 50 years. PLoS ONE 2012, 7, e48322. [Google Scholar] [CrossRef] [PubMed]
- Powell, T.J.; Peng, Y.; Berthoud, T.K.; Blais, M.E.; Lillie, P.J.; Hill, A.V.; Rowland-Jones, S.L.; McMichael, A.J.; Gilbert, S.C.; Dong, T. Examination of influenza specific T cell responses after influenza virus challenge in individuals vaccinated with MVA-NP+M1 vaccine. PLoS ONE 2013, 8, e62778. [Google Scholar] [CrossRef] [PubMed]
- Antrobus, R.D.; Berthoud, T.K.; Mullarkey, C.E.; Hoschler, K.; Coughlan, L.; Zambon, M.; Hill, A.V.; Gilbert, S.C. Coadministration of seasonal influenza vaccine and MVA-NP+M1 simultaneously achieves potent humoral and cell-mediated responses. Mol. Ther. 2014, 22, 233–238. [Google Scholar] [CrossRef] [PubMed]
- Stoloff, G.A.; Caparros-Wanderley, W. Synthetic multi-epitope peptides identified in silico induce protective immunity against multiple influenza serotypes. Eur. J. Immunol. 2007, 37, 2441–2449. [Google Scholar] [CrossRef] [PubMed]
- Pleguezuelos, O.; Robinson, S.; Stoloff, G.A.; Caparros-Wanderley, W. Synthetic Influenza vaccine (FLU-v) stimulates cell mediated immunity in a double-blind, randomised, placebo-controlled Phase I trial. Vaccine 2012, 30, 4655–4660. [Google Scholar] [CrossRef] [PubMed]
- Pleguezuelos, O.; Robinson, S.; Fernandez, A.; Stoloff, G.A.; Mann, A.; Gilbert, A.; Balaratnam, G.; Wilkinson, T.; Lambkin-Williams, R.; Oxford, J.; et al. A Synthetic Influenza Virus Vaccine Induces a Cellular Immune Response That Correlates with Reduction in Symptomatology and Virus Shedding in a Randomized Phase Ib Live-Virus Challenge in Humans. Clin. Vaccine Immunol. 2015, 22, 828–835. [Google Scholar] [CrossRef] [PubMed]
- Francis, J.N.; Bunce, C.J.; Horlock, C.; Watson, J.M.; Warrington, S.J.; Georges, B.; Brown, C.B. A novel peptide-based pan-influenza A vaccine: A double blind, randomised clinical trial of immunogenicity and safety. Vaccine 2015, 33, 396–402. [Google Scholar] [CrossRef] [PubMed]
- Atsmon, J.; Kate-Ilovitz, E.; Shaikevich, D.; Singer, Y.; Volokhov, I.; Haim, K.Y.; Ben-Yedidia, T. Safety and immunogenicity of multimeric-001—A novel universal influenza vaccine. J. Clin. Immunol. 2012, 32, 595–603. [Google Scholar] [CrossRef] [PubMed]
- Waithman, J.; Zanker, D.; Xiao, K.; Oveissi, S.; Wylie, B.; Ng, R.; Togel, L.; Chen, W. Resident CD8(+) and migratory CD103(+) dendritic cells control CD8 T cell immunity during acute influenza infection. PLoS ONE 2013, 8, e66136. [Google Scholar] [CrossRef] [PubMed]
- Cohn, L.; Delamarre, L. Dendritic cell-targeted vaccines. Front. Immunol. 2014. [Google Scholar] [CrossRef] [PubMed]
- Wakim, L.M.; Smith, J.; Caminschi, I.; Lahoud, M.H.; Villadangos, J.A. Antibody-targeted vaccination to lung dendritic cells generates tissue-resident memory CD8 T cells that are highly protective against influenza virus infection. Mucosal. Immunol. 2015, 8, 1060–1071. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.; Hu, Y.; Lee, Y.T.; Bouchard, K.R.; Benechet, A.; Khanna, K.; Cauley, L.S. Lung-resident memory CD8 T cells (TRM) are indispensable for optimal cross-protection against pulmonary virus infection. J. Leukoc. Biol. 2014, 95, 215–224. [Google Scholar] [CrossRef] [PubMed]
- Slutter, B.; Pewe, L.L.; Lauer, P.; Harty, J.T. Cutting edge: Rapid boosting of cross-reactive memory CD8 T cells broadens the protective capacity of the Flumist vaccine. J. Immunol. 2013, 190, 3854–3858. [Google Scholar] [CrossRef] [PubMed]
- Goenka, R.; Barnett, L.G.; Silver, J.S.; O’Neill, P.J.; Hunter, C.A.; Cancro, M.P.; Laufer, T.M. Cutting edge: Dendritic cell-restricted antigen presentation initiates the follicular helper T cell program but cannot complete ultimate effector differentiation. J. Immunol. 2011, 187, 1091–1095. [Google Scholar] [CrossRef] [PubMed]
- McKinstry, K.K.; Strutt, T.M.; Bautista, B.; Zhang, W.; Kuang, Y.; Cooper, A.M.; Swain, S.L. Effector CD4 T-cell transition to memory requires late cognate interactions that induce autocrine IL-2. Nat. Commun. 2014. [Google Scholar] [CrossRef] [PubMed]

© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Spitaels, J.; Roose, K.; Saelens, X. Influenza and Memory T Cells: How to Awake the Force. Vaccines 2016, 4, 33. https://doi.org/10.3390/vaccines4040033
Spitaels J, Roose K, Saelens X. Influenza and Memory T Cells: How to Awake the Force. Vaccines. 2016; 4(4):33. https://doi.org/10.3390/vaccines4040033
Chicago/Turabian StyleSpitaels, Jan, Kenny Roose, and Xavier Saelens. 2016. "Influenza and Memory T Cells: How to Awake the Force" Vaccines 4, no. 4: 33. https://doi.org/10.3390/vaccines4040033
APA StyleSpitaels, J., Roose, K., & Saelens, X. (2016). Influenza and Memory T Cells: How to Awake the Force. Vaccines, 4(4), 33. https://doi.org/10.3390/vaccines4040033




