Effect of Melittin on Metabolomic Profile and Cytokine Production in PMA-Differentiated THP-1 Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Isolation and Preparation
2.2. Cell Culture and Differentiation
2.3. Cell Viability Assay
2.4. Cytokine Production
2.5. Enzyme-Linked Immunosorbent Assay (ELISA)
2.6. Metabolite Extraction
2.7. LC-MS Conditions
2.8. Data Extraction and Statistical Analysis
3. Results
3.1. BV Fractionation and Isolation of Melittin
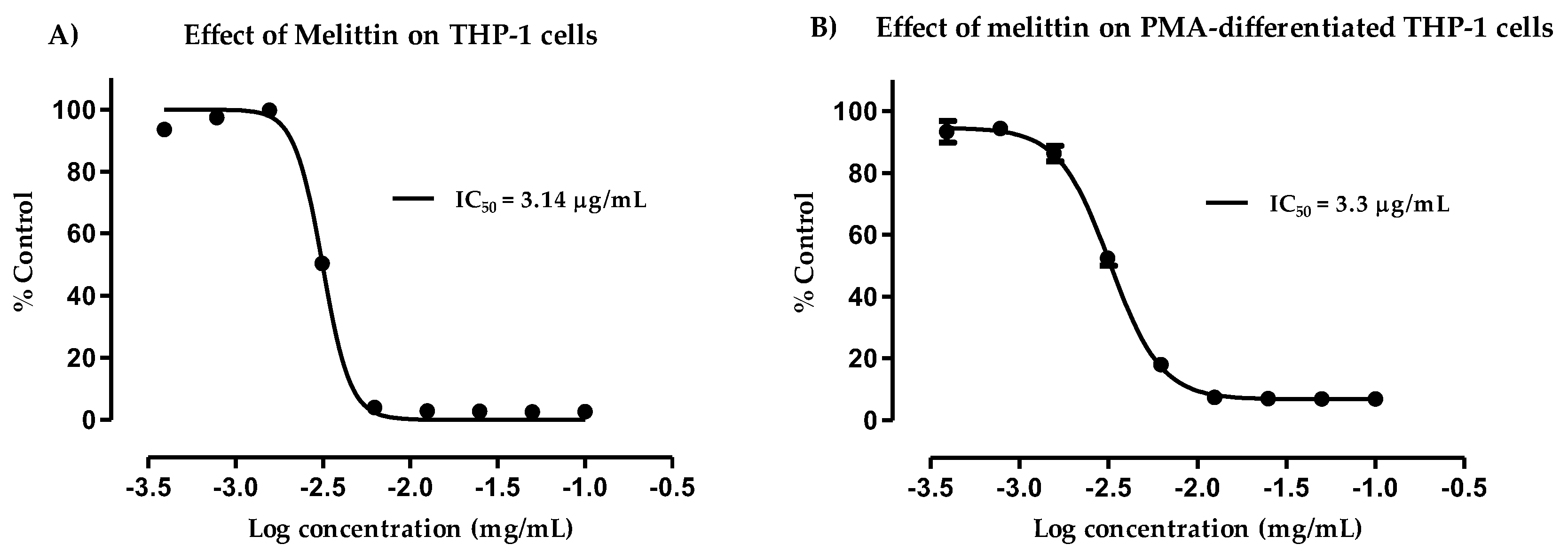
3.2. Cytotoxicity of Melittin against Normal and PMA-Differentiated THP-1 Cells
3.3. Effect of Melittin on the Production of Pro-Inflammatory TNF-α Cytokine
3.4. Effect of Melittin on the Production of Pro-Inflammatory IL-1β Cytokines
3.5. Effect of Melittin on the Production of Pro-Inflammatory Il-6 Cytokines
3.6. Effect of Melittin on the Production of Anti-Inflammatory Il-10 Cytokines
3.7. Effect of Melittin on the Cell Metabolome
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Neill, L.A.J. A critical role for citrate metabolism in LPS signalling. Biochem. J. 2011, 438, e5–e6. [Google Scholar] [CrossRef] [PubMed]
- Nathan, C. Points of control in inflammation. Nature 2002, 420, 846–852. [Google Scholar] [CrossRef] [PubMed]
- Janeway, C.A., Jr.; Medzhitov, R. Innate immune recognition. Annu. Rev. Immunol. 2002, 20, 197–216. [Google Scholar] [CrossRef] [PubMed]
- Kelly, B.; Neill, L.A.J. Metabolic reprogramming in macrophages and dendritic cells in innate immunity. Cell Res. 2015, 25, 771–784. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iwasaki, A.; Medzhitov, R. Control of adaptive immunity by the innate immune system. Nat. Immunol. 2015, 16, 343–353. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pesce, J.; Ramalingam, T.; Mentink-Kane, M.; Wilson, M.; El Kasmi, K.; Smith, A.; Thompson, R.; Cheever, A.; Murray, P.; Wynn, T.; et al. Arginase-1aExpressing Macrophages Suppress Th2 CytokineaDriven Inflammation and Fibrosis. PLoS Pathog. 2009, 5, e1000371. [Google Scholar] [CrossRef] [PubMed]
- Weinberg, S.E.; Sena, L.A.; Chandel, N.S. Mitochondria in the Regulation of Innate and Adaptive Immunity. Immunity 2015, 42, 406–417. [Google Scholar] [CrossRef] [PubMed]
- Guthrie, L.A.; McPhail, L.C.; Henson, P.M.; Johnston, R.B. Priming of neutrophils for enhanced release of oxygen metabolites by bacterial lipopolysaccharide: Evidence for increased activity of the superoxide-producing enzyme. J. Exp. Med. 1984, 160, 1656–1671. [Google Scholar] [CrossRef] [PubMed]
- Newsholme, P.; Curi, R.; Gordon, S.; Newsholme, E. Metabolism of glucose, glutamine, long-chain fatty acids and ketone bodies by murine macrophages. Biochem. J. 1986, 239, 121–125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Palsson-Mcdermott, E.M.; Curtis, A.M.; Goel, G.; Lauterbach, M.A.R.; Sheedy, F.J.; Gleeson, L.E.; Van den Bosch, M.W.M.; Quinn, S.R.; Domingo-Fernandez, R.; Johnston, D.G.W.; et al. Pyruvate Kinase M2 Regulates Hif-1α Activity and IL-1β Induction and Is a Critical Determinant of the Warburg Effect in LPS-Activated Macrophages. Cell Metab. 2015, 21, 347. [Google Scholar] [CrossRef] [PubMed]
- Haschemi, A.; Kosma, P.; Gille, L.; Evans, C.R.; Burant, C.F.; Starkl, P.; Knapp, B.; Haas, R.; Schmid, J.A.; Jandl, C.; et al. The Sedoheptulose Kinase CARKL Directs Macrophage Polarization through Control of Glucose Metabolism. Cell Metab. 2012, 15, 813–826. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tannahill, G.M.; Curtis, A.M.; Adamik, J.; Palsson-McDermott, E.M.; McGettrick, A.F.; Goel, G.; Frezza, C.; Bernard, N.J.; Kelly, B.; Foley, N.H.; et al. Succinate is an inflammatory signal that induces IL-1[beta] through HIF-1[alpha]. Nature 2013, 496, 238–242. [Google Scholar] [CrossRef] [PubMed]
- El Kasmi, K.C.; Stenmark, K.R. Contribution of metabolic reprogramming to macrophage plasticity and function. Semin. Immunol. 2015, 27, 267–275. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meera, E.; Ingrid, E.; Pascale, E.; Ellen, I.C.; Markus, E. Metabolism via arginase or nitric oxide synthase: Two competing arginine pathways in macrophages. Front. Immunol. 2014, 5, 532. [Google Scholar] [CrossRef]
- Clementi, E.; Brown, G.; Feelisch, M.; Moncada, S. Persistent inhibition of cell respiration by nitric oxide: Crucial role of S-nitrosylation of mitochondrial complex I and protective action of glutathione. Proc. Natl. Acad. Sci. USA 1998, 95, 7631–7636. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kihira, Y.; Yamano, N.; Izawa-Ishizawa, Y.; Ishizawa, K.; Ikeda, Y.; Tsuchiya, K.; Tamaki, T.; Tomita, S. Basic fibroblast growth factor regulates glucose metabolism through glucose transporter 1 induced by hypoxia-inducible factor-1α in adipocytes. Int. J. Biochem. Cell Biol. 2011, 43, 1602–1611. [Google Scholar] [CrossRef] [PubMed]
- Semenza, G.; Jiang, B.-H.; Leung, S.; Passantino, R.; Concordet, J.P.; Maire, P.; Giallongo, A. Hypoxia response elements in the aldolase A, enolase 1, and lactate dehydrogenase A gene promoters contain essential binding sites for hypoxia-inducible factor 1. J. Biol. Chem. 1996, 271, 32529–32537. [Google Scholar] [CrossRef] [PubMed]
- Huo, Y.; Iadevaia, V.; Yao, Z.; Kelly, I.; Cosulich, S.; Guichard, S.; Foster, L.J.; Proud, C.G. Stable isotope-labelling analysis of the impact of inhibition of the mammalian target of rapamycin on protein synthesis. Biochem. J. 2012, 444, 141–151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodríguez-Prados, J.C.; Través, P.G.; Cuenca, J.; Rico, D.; Aragone, J.; Martín-Sanz, P.; Cascante, M.; Boscá, L. Substrate fate in activated macrophages: A comparison between innate, classic, and alternative activation. J. Immunol. 2010, 185, 605–614. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sag, D.; Carling, D.; Stout, R.D.; Suttles, J. Adenosine 5′-monophosphate-activated protein kinase promotes macrophage polarization to an anti-inflammatory functional phenotype. J. Immunol. 2008, 181, 8633–8641. [Google Scholar] [CrossRef] [PubMed]
- Stuhlmeier, K.M. Apis mellifera venom and melittin block neither NF-kappa B-p50-DNA interactions nor the activation of NF-kappa B, instead they activate the transcription of proinflammatory genes and the release of reactive oxygen intermediates. J. Immunol. 2007, 179, 655–664. [Google Scholar] [CrossRef] [PubMed]
- Terwilliger, T.C.; Eisenberg, D. The structure of melittin. I. Structure determination and partial refinement. J. Biol. Chem. 1982, 257, 6010–6015. [Google Scholar] [PubMed]
- Terwilliger, T.C.; Eisenberg, D. The structure of melittin. II. Interpretation of the structure. J. Biol. Chem. 1982, 257, 6016–6022. [Google Scholar] [PubMed]
- Alonezi, S.; Tusiimire, J.; Wallace, J.; Dufton, M.; Parkinson, J.; Young, L.; Clements, C.; Park, J.; Jeon, J.; Ferro, V.; et al. Metabolomic Profiling of the Effects of Melittin on Cisplatin Resistant and Cisplatin Sensitive Ovarian Cancer Cells Using Mass Spectrometry and Biolog Microarray Technology. Metabolites 2016, 6, 35. [Google Scholar] [CrossRef] [PubMed]
- Rady, I.; Siddiqui, I.; Rady, M.; Mukhtar, H. Melittin, a major peptide component of bee venom, and its conjugates in cancer therapy. Cancer Lett. 2017, 402, 16–31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Skalickova, S.; Heger, Z.; Krejcova, L.; Pekarik, V.; Bastl, K.; Janda, J.; Kostolansky, F.; Vareckova, E.; Zitka, O.; Adam, V.; et al. Perspective of Use of Antiviral Peptides against Influenza Virus. Viruses 2015, 7, 5428–5442. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adade, C.M.; Oliveira, I.R.S.; Pais, J.A.R.; Souto-Padrón, T. Melittin peptide kills Trypanosoma cruzi parasites by inducing different cell death pathways. Toxicon 2013, 69, 227–239. [Google Scholar] [CrossRef] [PubMed]
- Shi, W.; Li, C.; Li, M.; Zong, X.; Han, D.; Chen, Y. Antimicrobial peptide melittin against Xanthomonas oryzae pv. oryzae, the bacterial leaf blight pathogen in rice. Appl. Microbiol. Biotechnol. 2016, 100, 5059–5067. [Google Scholar] [CrossRef] [PubMed]
- Park, H.J.; Lee, S.H.; Son, D.J.; Oh, K.W.; Kim, K.H.; Song, H.S.; Kim, G.J.; Oh, G.T.; Yoon, D.Y.; Hong, J.T. Antiarthritic effect of bee venom: Inhibition of inflammation mediator generation by suppression of NF-κB through interaction with the p50 subunit. Arthritis Rheum. 2004, 50, 3504–3515. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.-R.; Kim, K.-H.; An, H.-J.; Kim, J.-Y.; Han, S.-M.; Lee, K.-G.; Park, K.-K. Protective effect of melittin against inflammation and apoptosis on Propionibacterium acnes-induced human THP-1 monocytic cell. Eur. J. Pharmacol. 2014, 740, 218–226. [Google Scholar] [CrossRef] [PubMed]
- Phelps, C.B.; Sengchanthalangsy, L.L.; Malek, S.; Ghosh, G. Mechanism of κB DNA binding by Rel/ NF-κb dimers. J. Biol. Chem. 2000, 275, 24392–24399. [Google Scholar] [CrossRef] [PubMed]
- Baeuerle, P.A. Pro-inflammatory signaling: Last pieces in the NF-κB puzzle? Curr. Biol. 1998, 8, R19–R22. [Google Scholar] [CrossRef]
- Srivastava, R.M.; Srivastava, S.; Singh, M.; Bajpai, V.K.; Ghosh, J.K. Consequences of alteration in leucine zipper sequence of melittin in its neutralization of lipopolysaccharide-induced proinflammatory response in macrophage cells and interaction with lipopolysaccharide. J. Biol. Chem. 2012, 287, 1980–1995. [Google Scholar] [CrossRef] [PubMed]
- Tusiimire, J.; Wallace, J.; Woods, N.; Dufton, M.J.; Parkinson, J.A.; Abbott, G.; Clements, C.J.; Young, L.; Park, J.K.; Jeon, J.W.; et al. Effect of Bee Venom and Its Fractions on the Release of Pro-Inflammatory Cytokines in PMA-Differentiated U937 Cells Co-Stimulated with LPS. Vaccines 2016, 4, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, P.; Davis, P.; Liu, H.; Krishnan, T.R. Evaluation of cytotoxicity and absorption enhancing effects of melittin—A novel absorption enhancer. Eur. J. Pharm. Biopharm. 1999, 48, 85–87. [Google Scholar] [CrossRef]
- Alpar, H.O.; Eyles, J.E.; Williamson, E.D.; Somavarapu, S. Intranasal vaccination against plague, tetanus and diphtheria. Adv. Drug Deliv. Rev. 2001, 51, 173–201. [Google Scholar] [CrossRef]
- Bramwell, V.; Somavarapu, S.; Outschoorn, I.; Alpar, H. Adjuvant action of melittin following intranasal immunisation with tetanus and diphtheria toxoids. J. Drug Target. 2003, 11, 525–530. [Google Scholar] [CrossRef] [PubMed]
- Beger, R.D.; Dunn, W.; Schmidt, M.A.; Gross, S.S.; Kirwan, J.A.; Cascante, M.; Brennan, L.; Wishart, D.S.; Oresic, M.; Hankemeier, T.; et al. Metabolomics enables precision medicine: “A White Paper, Community Perspective”. Metabolomics 2016, 12, 149. [Google Scholar] [CrossRef] [PubMed]
- Alonezi, S.; Tusiimire, J.; Wallace, J.; Dufton, M.J.; Parkinson, J.A.; Young, L.C.; Clements, C.J.; Park, J.-K.; Jeon, J.-W.; Ferro, V.A.; et al. Metabolomic profiling of the synergistic effects of melittin in combination with cisplatin on ovarian cancer cells. Metabolites 2017, 7, 14. [Google Scholar] [CrossRef] [PubMed]
- Howe, C.C.F.; Alshehri, A.; Muggeridge, D.; Mullen, A.B.; Boyd, M.; Spendiff, O.; Moir, H.J.; Watson, D.G. Untargeted metabolomics profiling of an 80.5 km simulated treadmill ultramarathon. Metabolites 2018, 8, 14. [Google Scholar] [CrossRef] [PubMed]
- Sugimoto, M.; Sakagami, H.; Yokote, Y.; Onuma, H.; Kaneko, M.; Mori, M.; Sakaguchi, Y.; Soga, T.; Tomita, M. Non-targeted metabolite profiling in activated macrophage secretion. Metabolomics 2012, 8, 624–633. [Google Scholar] [CrossRef]
- Liu, K.; Pi, F.; Zhang, H.; Ji, J.; Xia, S.; Cui, F.; Sun, J.; Sun, X. Metabolomics Analysis to Evaluate the Anti-Inflammatory Effects of Polyphenols: Glabridin Reversed Metabolism Change Caused by LPS in RAW 264.7 Cells. J. Agric. Food Chem. 2017, 65, 6070–6079. [Google Scholar] [CrossRef] [PubMed]
- Traves, P.; de Atauri, P.; Marin, S.; Pimentel-Santillana, M.; Rodriguez-Prados, J.-C.; Marin de Mas, I.; Selivanov, V.; Martin-Sanz, P.; Bosca, L.; Cascante, M. Relevance of the MEK/ ERK Signaling Pathway in the Metabolism of Activated Macrophages: A Metabolomic Approach. J. Immunol. 2012, 188, 1402–1410. [Google Scholar] [CrossRef] [PubMed]
- Berg, H.A.; Scherer, E.P. Adipose Tissue, Inflammation, and Cardiovascular Disease. Circ. Res. 2005, 96, 939–949. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fitzpatrick, M.; Young, S.P. Metabolomics—A novel window into inflammatory disease. Swiss Med. Wkly. 2013, 143, w13743. [Google Scholar] [CrossRef] [PubMed]
- McClenathan, B.M.; Stewart, D.A.; Spooner, C.E.; Pathmasiri, W.W.; Burgess, J.P.; McRitchie, S.L.; Choi, Y.S.; Sumner, S.C.J. Metabolites as biomarkers of adverse reactions following vaccination: A pilot study using nuclear magnetic resonance metabolomics. Vaccine 2017, 35, 1238–1245. [Google Scholar] [CrossRef] [PubMed]
- Engler, R.J.M.; Nelson, M.R.; Collins, L.C., Jr.; Spooner, C.; Hemann, B.A.; Gibbs, B.T.; Atwood, J.E.; Howard, R.S.; Chang, A.S.; Cruser, D.L.; et al. A Prospective Study of the Incidence of Myocarditis/Pericarditis and New Onset Cardiac Symptoms following Smallpox and Influenza Vaccination. PLoS ONE 2015, 10, e0118283. [Google Scholar] [CrossRef] [PubMed]
- Gray, D.W.; Welsh, M.D.; Doherty, S.; Mansoor, F.; Chevallier, O.P.; Elliott, C.T.; Mooney, M.H. Identification of systemic immune response markers through metabolomic profiling of plasma from calves given an intra-nasally delivered respiratory vaccine. Vet. Res. 2015, 46, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tusiimire, J.; Wallace, J.; Dufton, M.; Parkinson, J.; Clements, C.; Young, L.; Park, J.; Jeon, J.; Watson, D. An LCMS method for the assay of melittin in cosmetic formulations containing bee venom. Anal. Bioanal. Chem. 2015, 407, 3627–3635. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Watson, D.G.; Wang, L.; Westrop, G.D.; Coombs, G.H.; Zhang, T. Evaluation of mobile phase characteristics on three zwitterionic columns in hydrophilic interaction liquid chromatography mode for liquid chromatography-high resolution mass spectrometry based untargeted metabolite profiling of Leishmania parasites. J. Chromatogr. A 2014, 1362, 168–179. [Google Scholar] [CrossRef] [PubMed]
- Daigneault, M.; Preston, J.; Marriott, H.; Whyte, M.; Dockrell, D. The Identification of Markers of Macrophage Differentiation in PMA-Stimulated THP-1 Cells and Monocyte-Derived Macrophages. PLoS ONE 2010, 5, e8668. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.; Bae, S.-J.S.; Joo, H.; Bae, H. Melittin suppresses tumor progression by regulating tumor-associated macrophages in a Lewis lung carcinoma mouse model. Oncotarget 2017, 8, 54951–54965. [Google Scholar] [CrossRef] [PubMed]
- Alberta Di, P.; Scott, P.; Fernanda Tavares Da, S.; Nathalie, G. Vaccine Adjuvants: From 1920 to 2015 and Beyond. Vaccines 2015, 3, 320–343. [Google Scholar] [CrossRef]
- Reed, S.G.; Pihl, D.L.; Conlon, P.J.; Grabstein, K.H. IL-1 as adjuvant. Role of T cells in augmentation of specific antibody production by recombinant human IL-1α. J. Immunol. 1989, 142, 3129–3133. [Google Scholar] [PubMed]
- Pape, K.; Khoruts, A.; Mondino, A.; Jenkins, M. Inflammatory cytokines enhance the in vivo clonal expansion and differentiation of antigen-activated CD4 super(+) T cells. J. Immunol. 1997, 159, 591–598. [Google Scholar] [PubMed]
- Li, H.; Nookala, S.; Re, F. Aluminum Hydroxide Adjuvants Activate Caspase-1 and Induce IL-1 beta and IL-18 Release. J. Immunol. 2007, 178, 5271–5276. [Google Scholar] [CrossRef] [PubMed]
- Bomalaski, J.; Ford, T.; Hudson, M.A.; Clark, J. Phospholipase A sub(2)-activating protein induces the synthesis of IL-1 and TNF in human monocytes. J. Immunol. 1995, 154, 4027–4031. [Google Scholar] [PubMed]
- Bomalaski, J.S.; Baker, D.G.; Brophy, L.; Resurreccion, N.V.; Spilberg, I.; Muniain, M.; Clark, M.A. A phospholipase A 2—Activating protein (PLAP) stimulates human neutrophil aggregation and release of lysosomal enzymes, superoxide, and eicosanoids. J. Immunol. 1989, 142, 3957–3962. [Google Scholar] [PubMed]
- Jha, A.K.; Huang, S.C.-C.; Sergushichev, A.; Lampropoulou, V.; Ivanova, Y.; Loginicheva, E.; Chmielewski, K.; Stewart, K.M.; Ashall, J.; Everts, B.; et al. Network Integration of Parallel Metabolic and Transcriptional Data Reveals Metabolic Modules that Regulate Macrophage Polarization. Immunity 2015, 42, 419–430. [Google Scholar] [CrossRef] [PubMed]
- Qualls, J.E.; Subramanian, C.; Rafi, W.; Smith, A.M.; Balouzian, L.; Defreitas, A.A.; Shirey, K.A.; Reutterer, B.; Kernbauer, E.; Stockinger, S.; et al. Sustained Generation of Nitric Oxide and Control of Mycobacterial Infection Requires Argininosuccinate Synthase 1. Cell Host Microbe 2012, 12, 313–323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brown, G.C. Nitric oxide and mitochondrial respiration. BBA Bioenerg. 1999, 1411, 351–369. [Google Scholar] [CrossRef]
- Mateo, R.B.; Reichner, J.S.; Mastrofrancesco, B.; Kraft-Stolar, D.; Albina, J.E. Impact of nitric oxide on macrophage glucose metabolism and glyceraldehyde-3-phosphate dehydrogenase activity. Am. J. Physiol. Cell Physiol. 1995, 268, C669–C675. [Google Scholar] [CrossRef] [PubMed]
- Albina, J.; Mastrofrancesco, B. Modulation of glucose metabolism in macrophages by products of nitric oxide synthase. Am. J. Physiol. 1993, 33, C1594–C1599. [Google Scholar] [CrossRef] [PubMed]
- Everts, B.; Amiel, E.; van der Windt, G.J.; Freitas, T.C.; Chott, R.; Yarasheski, K.E.; Pearce, E.L.; Pearce, E.J. Commitment to glycolysis sustains survival of nitric oxide-producing inflammatory dendritic cells. Blood 2012, 120, 1422–1431. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Chen, T.; Zhang, N.; Yang, M.; Li, B.; Lü, X.; Cao, X.; Ling, C. Melittin, a major component of bee venom, sensitizes human hepatocellular carcinoma cells to tumor necrosis factor-related apoptosis-inducing ligand (TRAIL)-induced apoptosis by activating CaMKII-TAK1-JNK/p38 and inhibiting IkappaBalpha kinase-NFkappaB. J. Biol. Chem. 2009, 284, 3804–3813. [Google Scholar] [CrossRef] [PubMed]
- Freemerman, A.J.; Johnson, A.R.; Sacks, G.N.; Milner, J.J.; Makowski, L.; Kirk, E.L.; Troester, M.A.; Macintyre, A.N.; Goraksha-Hicks, P.; Rathmell, J.C. Metabolic reprogramming of macrophages: Glucose transporter 1 (GLUT1)-mediated glucose metabolism drives a proinflammatory phenotype. J. Biol. Chem. 2014, 289, 7884–7896. [Google Scholar] [CrossRef] [PubMed]
- Blagih, J.; Jones, R.G. Polarizing Macrophages through Reprogramming of Glucose Metabolism. Cell Metab. 2012, 15, 793–795. [Google Scholar] [CrossRef] [PubMed]
- Barnes, V.M.; Teles, R.; Trivedi, H.M.; Devizio, W.; Xu, T.; Mitchell, M.W.; Milburn, M.V.; Guo, L. Acceleration of Purine Degradation by Periodontal Diseases. J. Dent. Res. 2009, 88, 851–855. [Google Scholar] [CrossRef] [PubMed]
- Gudbjornsson, B.; Hiillgren, R.; Zak, A.; Niklasson, F. Hypoxanthine, xanthine, and urate in synovial fluid from patients with inflammatory arthritides. Ann. Rheum. Dis. 1991, 50, 669–672. [Google Scholar] [CrossRef] [PubMed]
- Madsen, R.K.; Lundstedt, T.; Gabrielsson, J.; Sennbro, C.J.; Alenius, G.M.; Moritz, T.; Rantapää-Dahlqvist, S.; Trygg, J. Diagnostic properties of metabolic perturbations in rheumatoid arthritis. Arthritis Res. Ther. 2011, 13, R19. [Google Scholar] [CrossRef] [PubMed]
- Infantino, V.; Convertini, P.; Di Noia, M.A.; Calvello, R.; Palmieri, F.; Iacobazzi, V.; Cucci, L.; Panaro, M.A. The mitochondrial citrate carrier: A new player in inflammation. Biochem. J. 2011, 438, 433–436. [Google Scholar] [CrossRef] [PubMed]
- Wellen, K.E.; Hatzivassiliou, G.; Sachdeva, U.M.; Bui, T.V.; Cross, J.R.; Thompson, C.B. ATP-citrate lyase links cellular metabolism to histone acetylation. Science 2009, 324, 1076–1080. [Google Scholar] [CrossRef] [PubMed]
- Do, N.; Weindl, G.; Grohmann, L.; Salwiczek, M.; Koksch, B.; Korting, H.C.; Schäfer-Korting, M. Cationic membrane-active peptides—Anticancer and antifungal activity as well as penetration into human skin. Exp. Dermatol. 2014, 23, 326–331. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.Y.; Park, H.S.; Lee, S.J.; Choi, M.-U. Melittin Exerts Multiple Effects on the Release of Free Fatty Acids from L1210 Cells: Lack of Selective Activation of Phospholipase A2by Melittin. Arch. Biochem. Biophys. 2001, 389, 57–67. [Google Scholar] [CrossRef] [PubMed]
- Palomba, L.; Bianchi, M.; Persichini, T.; Magnani, M.; Colasanti, M.; Cantoni, O. Downregulation of nitric oxide formation by cytosolic phospholipase A 2-released arachidonic acid. Free Radic. Biol. Med. 2004, 36, 319–329. [Google Scholar] [CrossRef] [PubMed]
- Geddis, M.S.; Tornieri, K.; Giesecke, A.; Rehder, V. PLA 2 and secondary metabolites of arachidonic acid control filopodial behavior in neuronal growth cones. Cell Motil. Cytoskel. 2004, 57, 53–67. [Google Scholar] [CrossRef] [PubMed]
- Norris, P.C.; Reichart, D.; Dumlao, D.S.; Glass, C.K.; Dennis, E.A. Specificity of eicosanoid production depends on the TLR-4-stimulated macrophage phenotype. J. Leukocyte Biol. 2011, 90, 563–574. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hanna, V.S.; Hafez, E.A.A. Synopsis of arachidonic acid metabolism: A review. J. Adv. Res. 2018, 11, 23–32. [Google Scholar] [CrossRef] [PubMed]







| Mass | Rt | Putative Metabolite | LPS | Mel | Mel + LPS | |||
|---|---|---|---|---|---|---|---|---|
| Ratio | p Value | Ratio | p Value | Ratio | p Value | |||
| Arginine and proline metabolism | ||||||||
| 175.096 | 15.25 | L-Citrulline * | 1.825 | 0.048 | 7.482 | 0.001 | 8.657 | 0.001 |
| 174.112 | 26.50 | L-Arginine * | 0.255 | <0.001 | 1.111 | ns | 0.916 | ns |
| 240.122 | 15.55 | Homocarnosine | 0.640 | 0.005 | 1.570 | 0.002 | 1.481 | 0.001 |
| 145.085 | 14.52 | 4-Guanidinobutanoate | 1.122 | 0.014 | 0.792 | <0.001 | 1.611 | <0.001 |
| 246.133 | 13.55 | N2-(D-1-Carboxyethyl)-l-arginine | 0.295 | <0.001 | 0.951 | ns | 1.544 | <0.001 |
| 113.059 | 9.30 | Creatinine | 0.288 | <0.001 | 1.045 | ns | 1.234 | ns |
| 129.043 | 13.62 | Oxoproline | 0.135 | <0.001 | 0.192 | <0.001 | 0.182 | <0.001 |
| 290.123 | 15.83 | N-(L-Arginino) succinate | 0.562 | <0.001 | 1.367 | 0.006 | 0.832 | 0.040 |
| 132.090 | 23.04 | L-Ornithine * | 0.387 | <0.001 | 1.051 | ns | 1.177 | 0.004 |
| 147.053 | 13.45 | L-Glutamate * | 0.661 | <0.001 | 0.917 | ns | 0.855 | 0.026 |
| 398.137 | 16.01 | S-Adenosyl-l-methionine | 0.768 | ns | 5.717 | <0.001 | 4.267 | <0.001 |
| 115.063 | 12.14 | L-Proline * | 0.439 | <0.001 | 0.909 | ns | 1.103 | ns |
| Glycolysis/TCA cycle | ||||||||
| 339.996 | 17.18 | D-Fructose 1,6-bisphosphate * | 1.397 | ns | 0.544 | <0.001 | 0.671 | 0.009 |
| 260.030 | 15.02 | D-Fructose 6-phosphate * | 1.210 | ns | 1.620 | <0.001 | 1.590 | <0.001 |
| 260.020 | 15.94 | D-Glucose 6-sulfate | 2.193 | 0.000 | 1.359 | ns | 1.190 | 0.018 |
| 260.030 | 15.83 | D-Glucose 1-phosphate * | 1.366 | 0.004 | 2.264 | <0.001 | 1.909 | <0.001 |
| 809.125 | 12.25 | Acetyl-CoA | 0.713 | 0.027 | 0.946 | ns | 0.792 | 0.028 |
| 185.993 | 15.94 | 3-Phospho-D-glycerate | 0.753 | 0.001 | 0.610 | <0.001 | 0.501 | <0.001 |
| 169.998 | 15.10 | Glycerone phosphate | 0.381 | <0.001 | 0.315 | <0.001 | 0.249 | <0.001 |
| 192.027 | 17.28 | Citrate * | 0.318 | <0.001 | 1.089 | ns | 1.297 | 0.001 |
| 192.027 | 18.47 | Isocitrate * | 0.489 | <0.001 | 0.580 | <0.001 | 0.466 | <0.001 |
| 116.011 | 14.82 | Fumarate * | 0.751 | 0.013 | 0.891 | ns | 0.677 | 0.001 |
| 665.125 | 13.23 | NADH * | 0.324 | <0.001 | 0.492 | <0.001 | 0.584 | <0.001 |
| 663.109 | 14.17 | NAD+ * | 0.482 | <0.001 | 0.923 | ns | 0.671 | <0.001 |
| OXPHOS/Pentose phosphate pathway | ||||||||
| 506.996 | 15.76 | ATP * | 0.751 | 0.006 | 0.987 | ns | 0.585 | <0.001 |
| 427.030 | 14.31 | ADP * | 0.534 | <0.001 | 0.812 | 0.012 | 0.640 | <0.001 |
| 258.014 | 16.69 | D-Glucono-1,5-lactone 6-phosphate | 1.214 | ns | 1.207 | ns | 1.407 | 0.026 |
| 290.041 | 15.23 | D-Sedoheptulose 7-phosphate | 1.831 | <0.001 | 1.181 | 0.002 | 1.489 | <0.001 |
| 230.019 | 14.60 | D-Ribulose 5-phosphate | 0.932 | ns | 0.579 | 0.000 | 0.713 | 0.005 |
| 370.007 | 17.42 | D-Sedoheptulose 1,7-bisphosphate | 1.725 | 0.002 | 0.758 | ns | 1.065 | ns |
| 745.091 | 16.74 | NADPH * | 1.845 | 0.049 | 1.521 | ns | 0.508 | 0.048 |
| 743.075 | 16.51 | NADP+ | 0.693 | 0.030 | 0.764 | 0.031 | 0.820 | 0.047 |
| Purine metabolism | ||||||||
| 151.049 | 11.82 | Guanine * | 1.300 | ns | 0.632 | 0.010 | 1.707 | 0.001 |
| 283.092 | 12.93 | Guanosine | 1.310 | ns | 0.619 | 0.015 | 2.185 | 0.001 |
| 348.047 | 14.40 | IMP * | 2.093 | ns | 2.305 | 0.021 | 1.727 | ns |
| 268.081 | 10.32 | Inosine * | 4.079 | <0.001 | 1.852 | 0.001 | 6.051 | <0.001 |
| 168.028 | 11.38 | Urate * | 0.168 | <0.001 | 1.372 | 0.001 | 1.838 | <0.001 |
| 136.039 | 9.74 | Hypoxanthine * | 7.345 | <0.001 | 4.852 | <0.001 | 33.584 | <0.001 |
| 152.034 | 10.46 | Xanthine | 0.283 | <0.001 | 1.109 | 0.043 | 1.264 | 0.003 |
| 284.076 | 9.72 | Xanthosine | 0.270 | <0.001 | 1.019 | ns | 1.259 | 0.015 |
| 135.054 | 9.22 | Adenine | 1.246 | ns | 2.237 | 0.009 | 1.649 | ns |
| 347.063 | 12.62 | AMP * | 0.538 | 0.002 | 0.873 | ns | 0.339 | <0.001 |
| Pyrimidine metabolism | ||||||||
| 403.018 | 16.15 | CDP * | 0.584 | <0.001 | 0.690 | 0.001 | 0.650 | 0.002 |
| 111.043 | 11.38 | Cytosine | 0.684 | ns | 2.434 | ns | 3.660 | 0.021 |
| 482.985 | 17.60 | CTP * | 0.659 | 0.001 | 0.719 | 0.001 | 0.463 | <0.001 |
| 243.086 | 11.36 | Cytidine * | 0.528 | <0.001 | 2.375 | <0.001 | 3.628 | <0.001 |
| 483.969 | 17.02 | UTP * | 0.617 | <0.001 | 0.685 | <0.001 | 0.437 | <0.001 |
| 324.036 | 15.17 | UMP * | 0.721 | 0.011 | 0.630 | <0.001 | 0.589 | <0.001 |
| 404.003 | 15.58 | UDP * | 0.422 | <0.001 | 0.666 | 0.001 | 0.534 | <0.001 |
| 580.035 | 17.95 | UDP-glucuronate | 0.604 | <0.001 | 0.560 | <0.001 | 0.495 | <0.001 |
| 566.055 | 16.00 | UDP-glucose | 0.637 | 0.000 | 0.555 | 0.000 | 0.457 | <0.001 |
| 605.077 | 17.17 | GDP-mannose | 1.812 | <0.001 | 1.996 | <0.001 | 1.532 | <0.001 |
| Fatty acids and metabolites C4 | ||||||||
| 306.256 | 12.45 | Eicosatrienoic acid * | 1.650 | 0.004 | 1.983 | ns | 1.977 | <0.001 |
| 328.240 | 18.27 | Docosahexaenoic acid * | 0.346 | ns | 6.658 | 0.002 | 5.846 | 0.002 |
| 318.219 | 4.10 | Leukotriene A4 or isomer | 1.115 | ns | 1.469 | ns | 2.183 | 0.002 |
| 310.287 | 17.73 | Eicosenoic acid * | 1.328 | ns | 0.477 | ns | 0.313 | 0.009 |
| 356.256 | 14.78 | Prostaglandin F1alpha or isomer | 0.276 | 0.007 | 0.479 | 0.024 | 0.482 | 0.048 |
| 336.230 | 4.55 | Prostaglandin B1 or isomer | 1.004 | ns | 1.432 | ns | 2.125 | <0.001 |
| 402.225 | 4.00 | 5S-HETE di-endoperoxide or isomer | 1.709 | ns | 0.850 | ns | 0.650 | 0.007 |
| 282.256 | 20.06 | Oleic acid * | 1.239 | 0.036 | 9.007 | <0.001 | 8.089 | <0.001 |
| 256.240 | 7.84 | Hexadecanoic acid isomer | 1.397 | 0.005 | 7.235 | <0.001 | 13.602 | 0.004 |
| 338.319 | 24.61 | Docosenoic acid * | 1.246 | 0.005 | 2.740 | <0.001 | 2.941 | <0.001 |
| 268.240 | 11.26 | Heptadecenoic acid | 1.362 | 0.013 | 1.782 | <0.001 | 1.211 | ns |
| 366.350 | 26.59 | Tetracosenoic acid * | 1.354 | 0.038 | 2.665 | <0.001 | 2.800 | <0.001 |
| 304.240 | 18.36 | Eeicosatetraenoic acid * | 1.049 | ns | 46.370 | <0.001 | 42.444 | <0.001 |
| 332.272 | 20.30 | Docosatetraenoic acid | 0.929 | ns | 25.404 | <0.001 | 21.474 | <0.001 |
| 242.225 | 17.97 | Pentadecanoic acid * | 0.967 | ns | 1.787 | <0.001 | 1.758 | <0.001 |
| 334.287 | 21.82 | Docosatrienoic acid * | 0.776 | ns | 8.679 | 0.001 | 7.684 | 0.001 |
| 226.193 | 15.25 | Tetradecenoic acid | 0.890 | ns | 3.570 | 0.002 | 3.367 | 0.003 |
| 270.256 | 12.86 | Heptadecanoic acid | 0.874 | ns | 4.801 | 0.010 | 1.854 | 0.009 |
| 278.225 | 17.08 | Linolenic acid * | 0.789 | 0.029 | 3.716 | <0.001 | 2.861 | <0.001 |
| 280.240 | 18.38 | Linoleate * | 1.105 | ns | 9.760 | <0.001 | 8.943 | <0.001 |
| Glycerophospholipids C4 | ||||||||
| 721.467 | 12.69 | PC 32:6 | 0.672 | ns | 0.214 | 0.016 | 0.217 | 0.023 |
| 832.654 | 30.65 | PG 41:0 ether | 0.665 | 0.028 | 0.623 | 0.019 | 0.402 | 0.003 |
| 738.448 | 12.65 | PG34:6 | 0.949 | ns | 0.357 | 0.020 | 0.308 | 0.015 |
| 484.280 | 13.33 | Lyso PG 16:0 | 1.589 | ns | 0.449 | 0.034 | 0.283 | 0.013 |
| 800.449 | 12.56 | PI 32:5 | 0.885 | ns | 0.365 | 0.009 | 0.329 | 0.005 |
| 584.333 | 13.28 | Lyso PI 18:0 Ether | 0.941 | ns | 0.558 | 0.020 | 0.499 | 0.004 |
| 858.526 | 13.47 | PI 36:4 | 1.841 | ns | 0.441 | 0.025 | 0.316 | 0.027 |
| 814.501 | 13.55 | PI 34:4 ether | 2.088 | ns | 0.486 | 0.025 | 0.317 | 0.019 |
| 785.521 | 32.15 | PS 36:3 | 0.789 | 0.008 | 0.782 | 0.005 | 0.779 | 0.007 |
| 517.244 | 12.56 | Lyso PS 18:4 | 1.184 | ns | 2.645 | <0.001 | 1.659 | 0.022 |
| 833.520 | 22.16 | PS 40:7 | 1.118 | ns | 0.439 | 0.004 | 0.470 | 0.009 |
| 777.457 | 13.50 | PS 36:7 | 0.970 | ns | 0.432 | 0.023 | 0.371 | 0.010 |
| Miscellaneous | ||||||||
| 131.058 | 13.88 | N-Acetyl-beta-alanine | 2.017 | 0.004 | 4.879 | 0.001 | 5.950 | 0.001 |
| 217.143 | 15.88 | beta-Alanyl-L-lysine | 1.543 | 0.041 | 3.816 | <0.001 | 1.705 | 0.021 |
| 182.079 | 13.10 | D-Sorbitol * | 0.325 | <0.001 | 2.226 | <0.001 | 4.443 | <0.001 |
| 180.064 | 12.66 | D-Mannose * | 0.712 | <0.001 | 1.538 | <0.001 | 1.744 | <0.001 |
| 180.064 | 14.19 | D-Galactose | 1.326 | ns | 3.785 | 0.001 | 4.184 | 0.001 |
| 221.090 | 11.16 | N-Acetyl-D-glucosamine * | 0.464 | <0.001 | 0.953 | ns | 0.806 | 0.008 |
| 146.069 | 14.34 | L-Glutamine * | 0.370 | <0.001 | 1.085 | ns | 1.322 | 0.002 |
| 129.043 | 10.45 | 5-Oxoproline * | 2.424 | 0.018 | 0.897 | ns | 1.192 | ns |
| 220.085 | 8.96 | 5-Hydroxy-L-tryptophan * | 7.211 | <0.001 | 1.131 | 0.010 | 4.305 | <0.001 |
| 204.090 | 11.14 | L-Tryptophan * | 0.190 | <0.001 | 1.189 | ns | 1.311 | 0.046 |
| 181.074 | 12.34 | L-Tyrosine * | 0.309 | <0.001 | 1.050 | ns | 1.265 | 0.042 |
| 131.095 | 10.77 | L-Leucine * | 0.217 | 0.008 | 1.038 | ns | 1.226 | 0.048 |
| 117.079 | 11.94 | L-Valine * | 1.615 | 0.001 | 6.492 | 0.000 | 2.822 | 0.002 |
| 612.152 | 16.40 | Glutathione disulfide | 0.652 | <0.001 | 1.186 | 0.047 | 1.109 | ns |
| 142.074 | 13.04 | Ectoine * | 1.164 | ns | 0.553 | <0.001 | 0.196 | <0.001 |
| 131.069 | 13.99 | Creatine * | 0.547 | <0.001 | 0.615 | <0.001 | 0.439 | ns |
| 103.100 | 20.48 | Choline * | 0.444 | <0.001 | 3.571 | <0.001 | 2.827 | <0.001 |
| 105.043 | 15.44 | L-Serine * | 1.518 | <0.001 | 1.737 | <0.001 | 1.707 | <0.001 |
| 121.020 | 15.41 | L-Cysteine * | 0.581 | <0.001 | 2.182 | <0.001 | 4.862 | <0.001 |
| 155.070 | 15.87 | L-Histidine * | 1.088 | ns | 2.089 | <0.001 | 3.122 | <0.001 |
| 161.105 | 12.59 | L-Carnitine * | 0.608 | <0.001 | 0.822 | 0.008 | 0.744 | 0.002 |
| 238.230 | 12.66 | 2-trans-Hexadecenal C4 | 0.795 | ns | 16.865 | 0.000 | 12.016 | 0.000 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alqarni, A.M.; Ferro, V.A.; Parkinson, J.A.; Dufton, M.J.; Watson, D.G. Effect of Melittin on Metabolomic Profile and Cytokine Production in PMA-Differentiated THP-1 Cells. Vaccines 2018, 6, 72. https://doi.org/10.3390/vaccines6040072
Alqarni AM, Ferro VA, Parkinson JA, Dufton MJ, Watson DG. Effect of Melittin on Metabolomic Profile and Cytokine Production in PMA-Differentiated THP-1 Cells. Vaccines. 2018; 6(4):72. https://doi.org/10.3390/vaccines6040072
Chicago/Turabian StyleAlqarni, Abdulmalik M., Valerie A. Ferro, John A. Parkinson, Mark J. Dufton, and David G. Watson. 2018. "Effect of Melittin on Metabolomic Profile and Cytokine Production in PMA-Differentiated THP-1 Cells" Vaccines 6, no. 4: 72. https://doi.org/10.3390/vaccines6040072
APA StyleAlqarni, A. M., Ferro, V. A., Parkinson, J. A., Dufton, M. J., & Watson, D. G. (2018). Effect of Melittin on Metabolomic Profile and Cytokine Production in PMA-Differentiated THP-1 Cells. Vaccines, 6(4), 72. https://doi.org/10.3390/vaccines6040072






