The Dysregulation of the Monocyte–Dendritic Cell Interplay Is Associated with In-Hospital Mortality in COVID-19 Pneumonia
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. Frequency Distribution and Absolute Numbers of Circulating CD169-Expressing Monocytes
2.3. Frequency Distribution and Absolute Numbers of Circulating Dendritic Cell Subsets
2.4. Measurement of Serum Levels of Interleukin-6
2.5. Human Immune-Inflammatory and SARS-CoV-2 Gene Profile Analysis
2.6. Statistical Analysis
3. Results
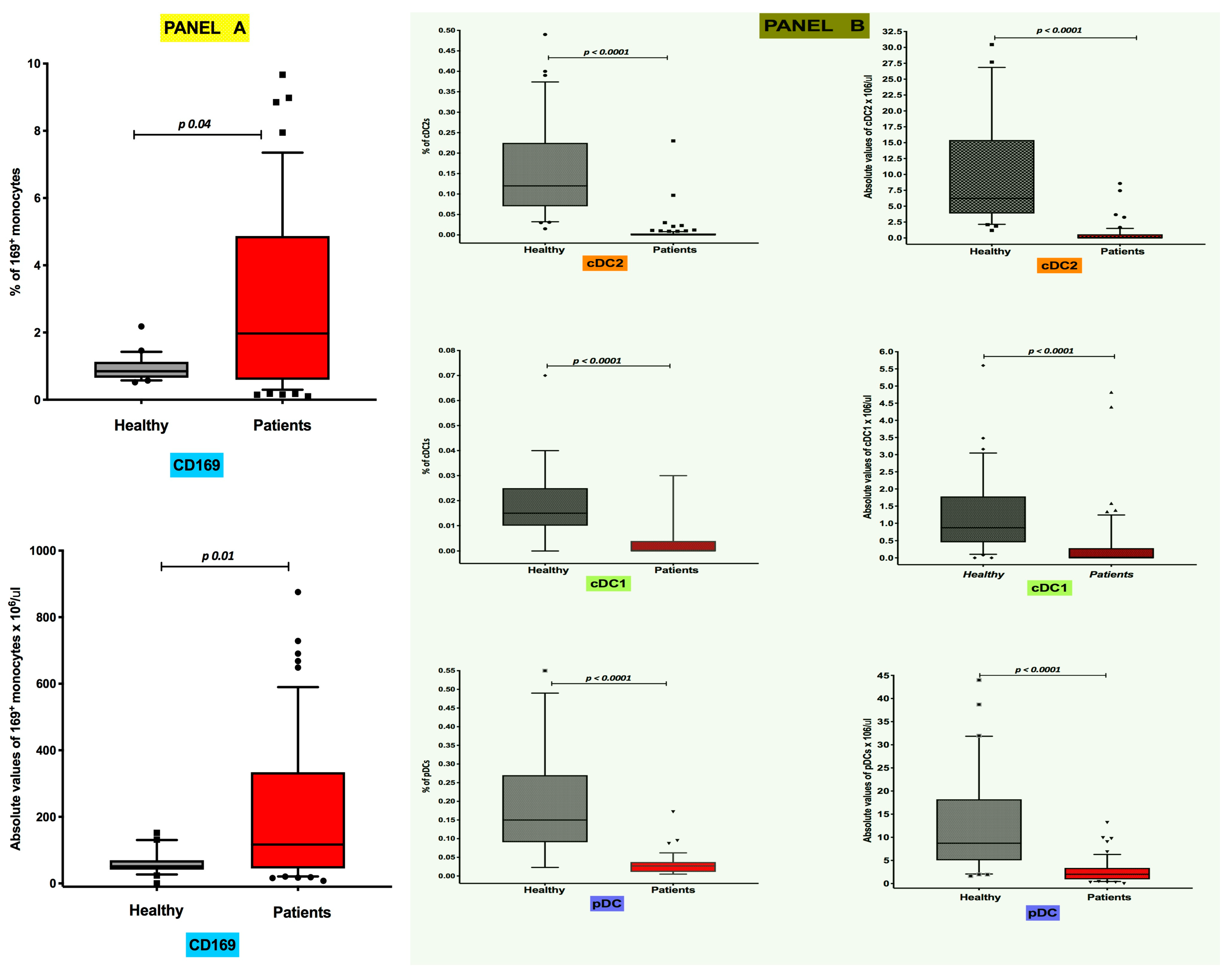
3.1. Differential Distribution of Circulating CD169+ Monocytes and Dendritic Cell Subsets in COVID-19 Pneumonia
3.2. Inverse Correlation of Absolute Levels of CD169-Positive Monocytes with CD141+ cDC1 Numbers in COVID-19 Pneumonia Patients
3.3. Short-Term Distribution of Circulating CD169+ Monocyte DC Subsets Is Not Modified by Systemic Steroids
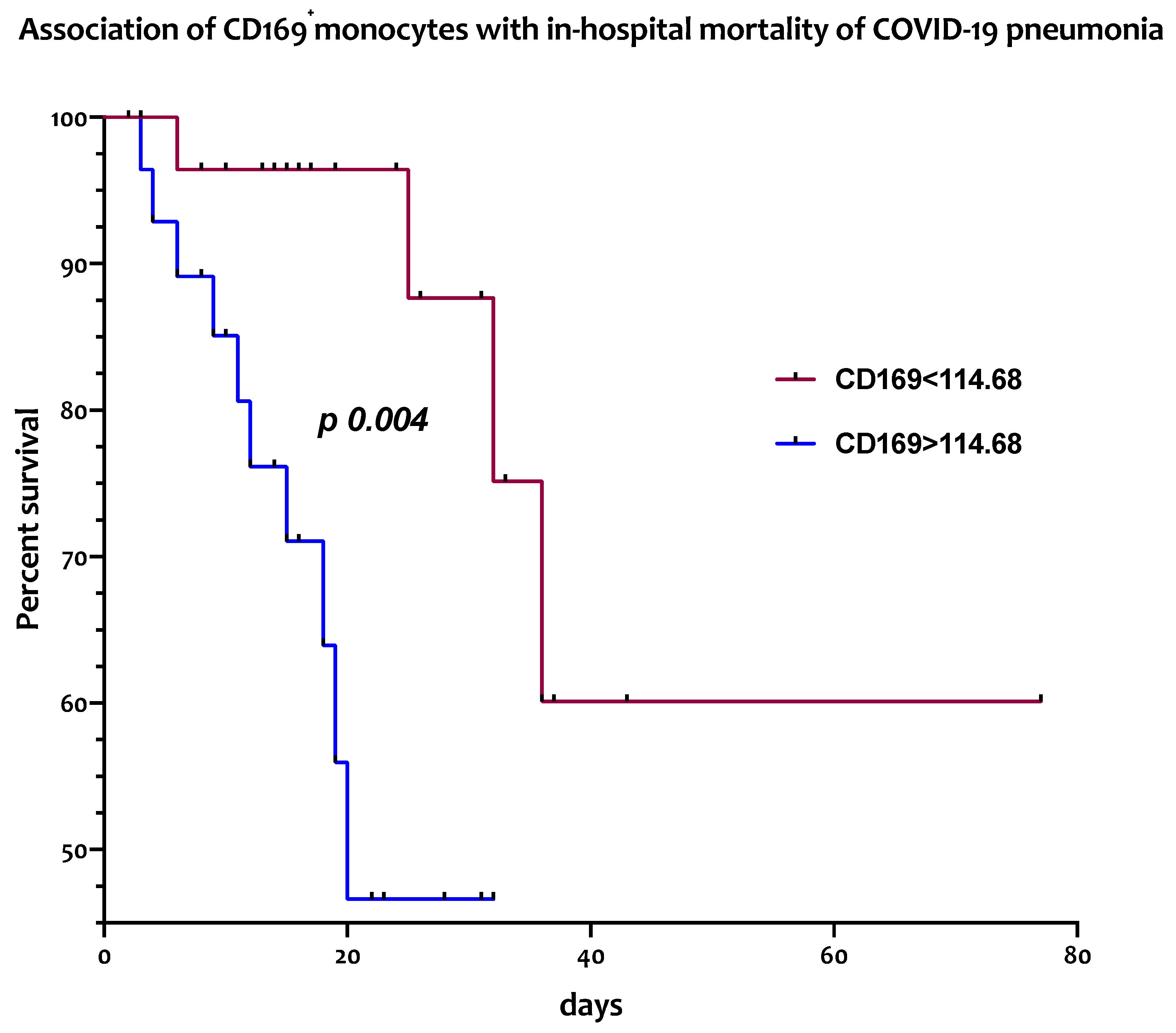
3.4. Circulating CD169+ Monocytes Are Associated with In-Hospital Mortality from COVID-19 Pneumonia
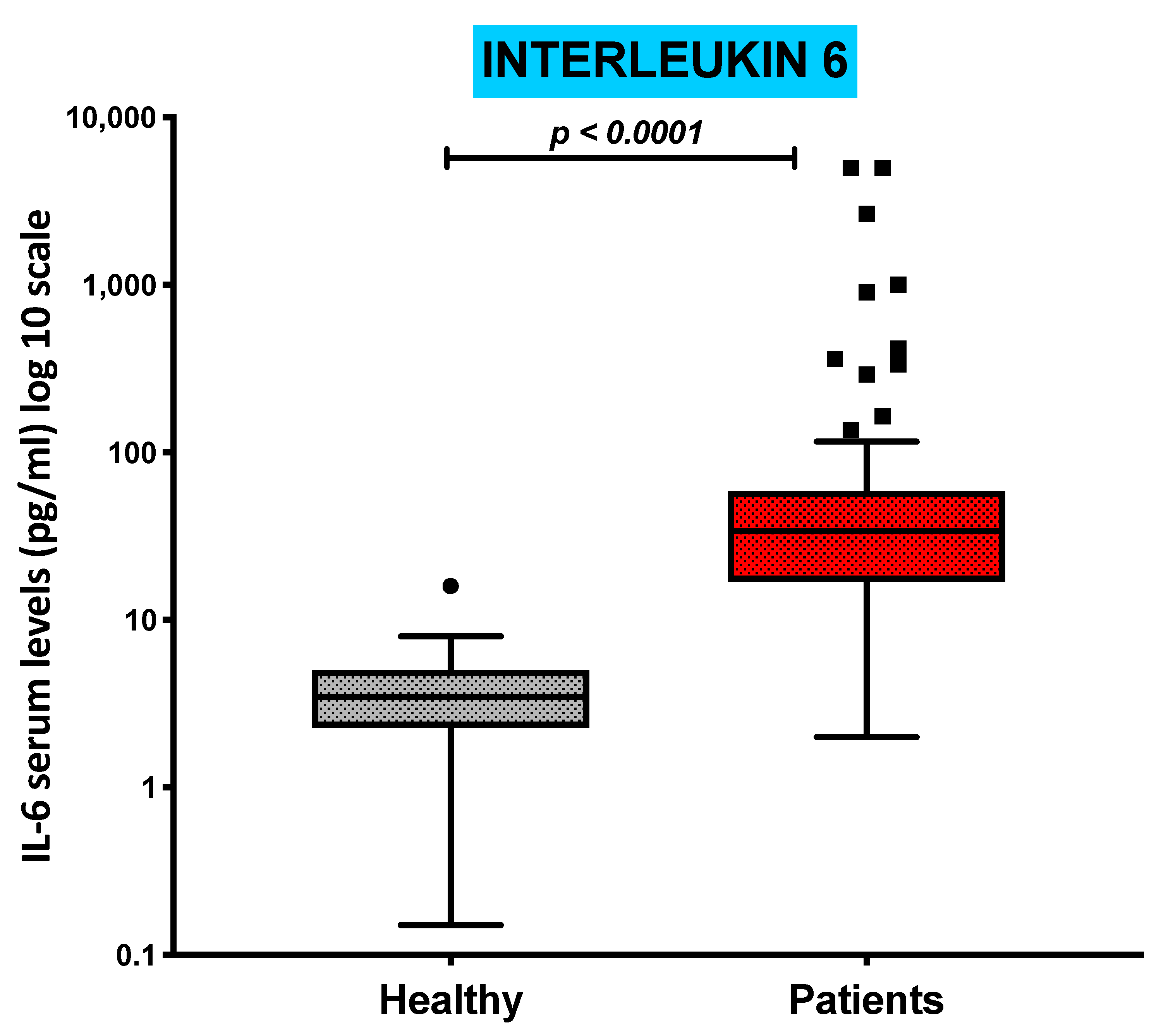
3.5. Serum Levels of Pro-Inflammatory Cytokine IL-6 Are Increased in COVID-19 Patients and Inversely Correlated with the Absolute Numbers of cDC1s and pDCs
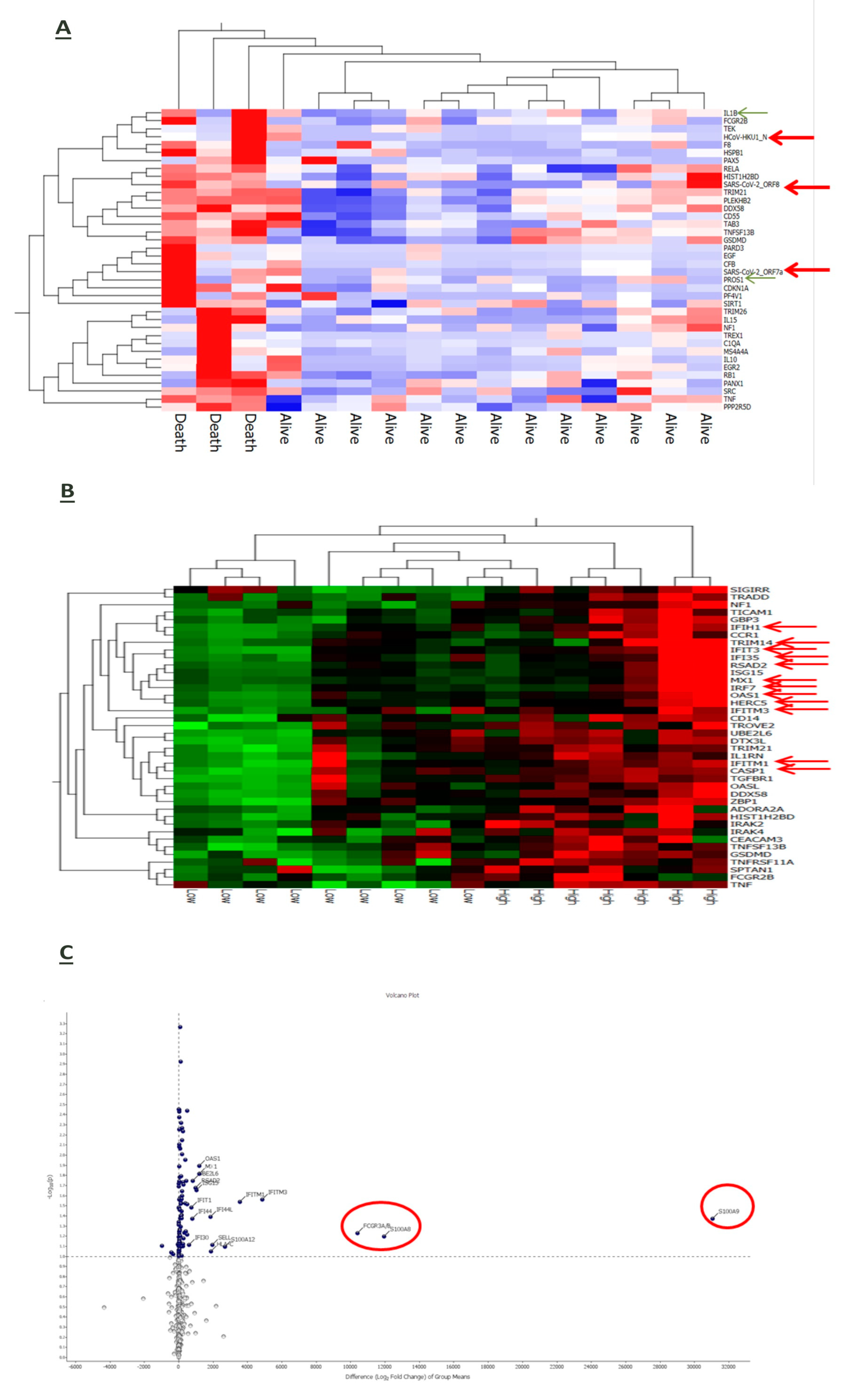
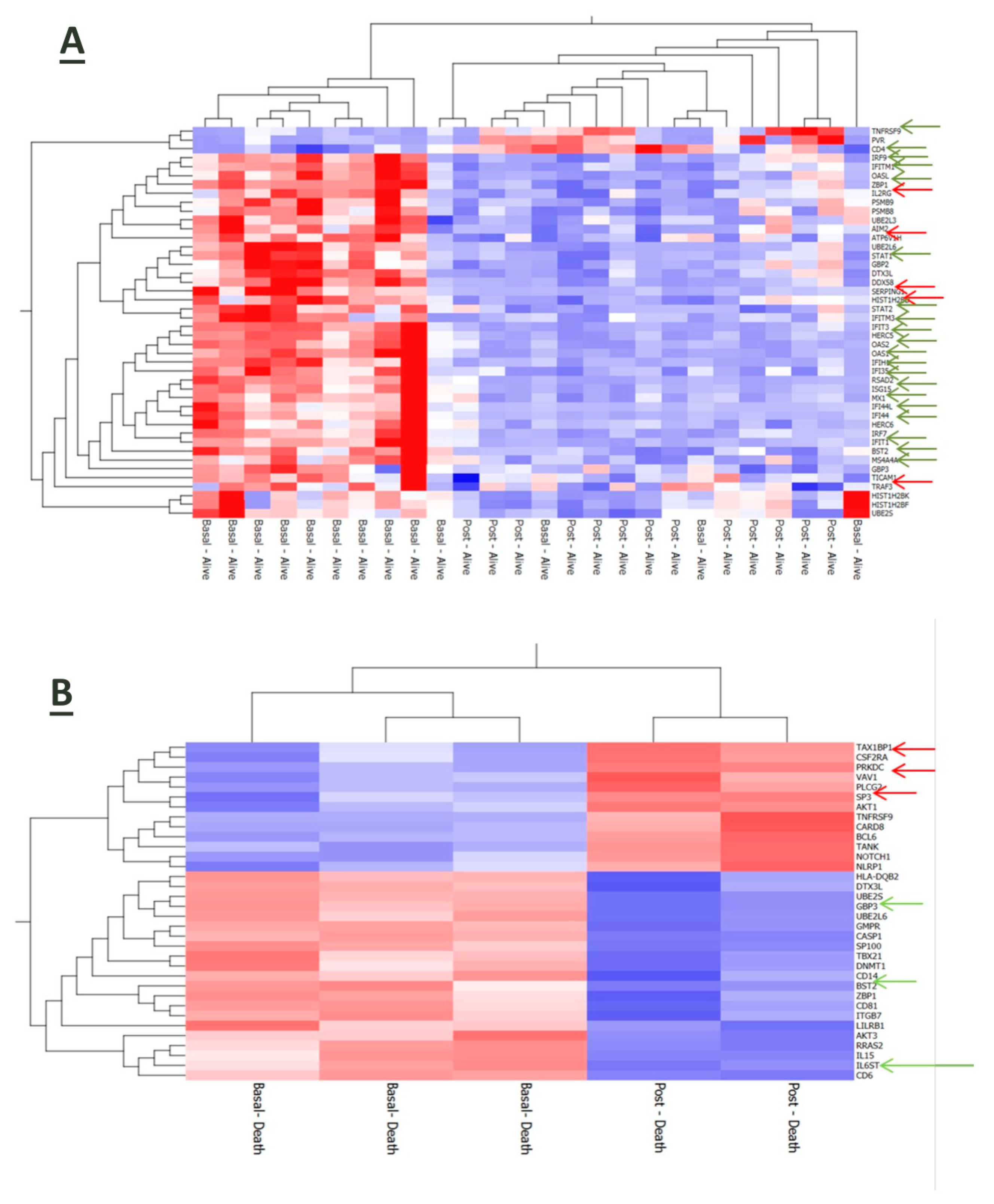
3.6. Interplay of Host Immune-Inflammatory Genes and SARS-CoV-2 Gene Signature in COVID-19 Pneumonia
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Osuchowski, M.F.; Winkler, M.S.; Skirecki, T.; Cajander, S.; Shankar-Hari, M.; Lachmann, G.; Monneret, G.; Venet, F.; Bauer, M.; Brunkhorst, F.M.; et al. The COVID-19 puzzle: Deciphering pathophysiology and phenotypes of a new disease entity. Lancet Respir. Med. 2021, 9, 622–642. [Google Scholar] [CrossRef]
- Shi, Y.; Wang, Y.; Shao, C.; Huang, J.; Gan, J.; Huang, X.; Bucci, E.; Piacentini, M.; Ippolito, G.; Melino, G. COVID-19 infection: The perspectives on immune responses. Cell Death Differ. 2020, 27, 1451–1454. [Google Scholar] [CrossRef]
- Wu, Z.; McGoogan, J.M. Characteristics of and Important Lessons from the Coronavirus Disease 2019 (COVID-19) Outbreak in China: Summary of a Report of 72314 Cases from the Chinese Center for Disease Control and Prevention. JAMA 2020, 323, 1239–1242. [Google Scholar] [CrossRef]
- Zhang, Q.; Wang, Z.; Lv, Y.; Zhao, J.; Dang, Q.; Xu, D.; Zhao, D.; Liu, H.; Wang, Z.; Zhao, X.; et al. Clinical features and prognostic factors of patients with COVID-19 in Henan Province, China. Hum. Cell 2021, 34, 419–435. [Google Scholar] [CrossRef]
- Casalino, L.; Gaieb, Z.; Goldsmith, J.A.; Hjorth, C.K.; Dommer, A.C.; Harbison, A.M.; Fogarty, C.A.; Barros, E.P.; Taylor, B.C.; McLellan, J.S.; et al. Beyond Shielding: The Roles of Glycans in the SARS-CoV-2 Spike Protein. ACS Cent. Sci. 2020, 6, 1722–1734. [Google Scholar] [CrossRef]
- Gadanec, L.K.; McSweeney, K.R.; Qaradakhi, T.; Ali, B.; Zulli, A.; Apostolopoulos, V. Can SARS-CoV-2 Virus Use Multiple Receptors to Enter Host Cells? Int. J. Mol. Sci. 2021, 22, 992. [Google Scholar] [CrossRef]
- Walls, A.C.; Park, Y.J.; Tortorici, M.A.; Wall, A.; McGuire, A.T.; Veesler, D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell 2020, 183, 1735. [Google Scholar] [CrossRef]
- Guan, W.J.; Ni, Z.Y.; Hu, Y.; Liang, W.H.; Ou, C.Q.; He, J.X.; Liu, L.; Shan, H.; Lei, C.L.; Hui, D.S.C.; et al. Clinical Characteristics of Coronavirus Disease 2019 in China. N. Engl. J. Med. 2020, 382, 1708–1720. [Google Scholar] [CrossRef]
- Pedersen, S.F.; Ho, Y.C. SARS-CoV-2: A storm is raging. J. Clin. Investig. 2020, 130, 2202–2205. [Google Scholar] [CrossRef]
- Vellingiri, B.; Jayaramayya, K.; Iyer, M.; Narayanasamy, A.; Govindasamy, V.; Giridharan, B.; Ganesan, S.; Venugopal, A.; Venkatesan, D.; Ganesan, H.; et al. COVID-19: A promising cure for the global panic. Sci. Total Environ. 2020, 725, 138277. [Google Scholar] [CrossRef]
- Zhou, Z.; Ren, L.; Zhang, L.; Zhong, J.; Xiao, Y.; Jia, Z.; Guo, L.; Yang, J.; Wang, C.; Jiang, S.; et al. Heightened Innate Immune Responses in the Respiratory Tract of COVID-19 Patients. Cell Host Microbe 2020, 27, 883–890.e882. [Google Scholar] [CrossRef]
- Merad, M.; Martin, J.C. Pathological inflammation in patients with COVID-19: A key role for monocytes and macrophages. Nat. Rev. Immunol. 2020, 20, 355–362. [Google Scholar] [CrossRef]
- Chevrier, S.; Zurbuchen, Y.; Cervia, C.; Adamo, S.; Raeber, M.E.; de Souza, N.; Sivapatham, S.; Jacobs, A.; Bachli, E.; Rudiger, A.; et al. A distinct innate immune signature marks progression from mild to severe COVID-19. Cell Rep. Med. 2021, 2, 100166. [Google Scholar] [CrossRef]
- Martinez, F.O.; Combes, T.W.; Orsenigo, F.; Gordon, S. Monocyte activation in systemic COVID-19 infection: Assay and rationale. EBioMedicine 2020, 59, 102964. [Google Scholar] [CrossRef]
- Ait Belkacem, I.; Mossadegh-keller, N.; Bourgoin, P.; Arnoux, I.; Loosveld, M.; Morange, P.-e.; Markarian, T.; Michelet, P.; Busnel, J.M.; Roulland, S.; et al. Cell Analysis from Dried Blood Spots: New Opportunities in Immunology, Hematology, and Infectious Diseases. Adv. Sci. 2021, 8, 2100323. [Google Scholar] [CrossRef]
- Fanelli, M.; Petrone, V.; Maracchioni, C.; Chirico, R.; Cipriani, C.; Coppola, L.; Malagnino, V.; Teti, E.; Sorace, C.; Zordan, M.; et al. Persistence of circulating CD169+monocytes and HLA-DR downregulation underline the immune response impairment in PASC individuals: The potential contribution of different COVID-19 pandemic waves. Curr. Res. Microb. Sci. 2024, 6, 100215. [Google Scholar] [CrossRef]
- Affandi, A.J.; Olesek, K.; Grabowska, J.; Nijen Twilhaar, M.K.; Rodriguez, E.; Saris, A.; Zwart, E.S.; Nossent, E.J.; Kalay, H.; de Kok, M.; et al. CD169 Defines Activated CD14(+) Monocytes With Enhanced CD8(+) T Cell Activation Capacity. Front. Immunol. 2021, 12, 697840. [Google Scholar] [CrossRef]
- Lucas, C.; Wong, P.; Klein, J.; Castro, T.B.R.; Silva, J.; Sundaram, M.; Ellingson, M.K.; Mao, T.; Oh, J.E.; Israelow, B.; et al. Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature 2020, 584, 463–469. [Google Scholar] [CrossRef]
- Chu, H.; Chan, J.F.; Wang, Y.; Yuen, T.T.; Chai, Y.; Hou, Y.; Shuai, H.; Yang, D.; Hu, B.; Huang, X.; et al. Comparative replication and immune activation profiles of SARS-CoV-2 and SARS-CoV in human lungs: An ex vivo study with implications for the pathogenesis of COVID-19. Clin. Infect. Dis. 2020, 71, 1400–1409. [Google Scholar] [CrossRef]
- Parackova, Z.; Zentsova, I.; Bloomfield, M.; Vrabcova, P.; Smetanova, J.; Klocperk, A.; Meseznikov, G.; Casas Mendez, L.F.; Vymazal, T.; Sediva, A. Disharmonic Inflammatory Signatures in COVID-19: Augmented Neutrophils’ but Impaired Monocytes’ and Dendritic Cells’ Responsiveness. Cells 2020, 9, 2206. [Google Scholar] [CrossRef]
- Ragab, D.; Salah Eldin, H.; Taeimah, M.; Khattab, R.; Salem, R. The COVID-19 Cytokine Storm; What We Know So Far. Front. Immunol. 2020, 11, 1446. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Wu, D.; Guo, W.; Cao, Y.; Huang, D.; Wang, H.; Wang, T.; Zhang, X.; Chen, H.; Yu, H.; et al. Clinical and immunological features of severe and moderate coronavirus disease 2019. J. Clin. Investig. 2020, 130, 2620–2629. [Google Scholar] [CrossRef]
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef] [PubMed]
- Schulte-Schrepping, J.; Reusch, N.; Paclik, D.; Bassler, K.; Schlickeiser, S.; Zhang, B.; Kramer, B.; Krammer, T.; Brumhard, S.; Bonaguro, L.; et al. Severe COVID-19 Is Marked by a Dysregulated Myeloid Cell Compartment. Cell 2020, 182, 1419–1440.e1423. [Google Scholar] [CrossRef] [PubMed]
- Galati, D.; Zanotta, S.; Capitelli, L.; Bocchino, M. A bird’s eye view on the role of dendritic cells in SARS-CoV-2 infection: Perspectives for immune-based vaccines. Allergy 2022, 77, 100–110. [Google Scholar] [CrossRef]
- Vanderheiden, A.; Ralfs, P.; Chirkova, T.; Upadhyay, A.A.; Zimmerman, M.G.; Bedoya, S.; Aoued, H.; Tharp, G.M.; Pellegrini, K.L.; Manfredi, C.; et al. Type I and Type III Interferons Restrict SARS-CoV-2 Infection of Human Airway Epithelial Cultures. J. Virol. 2020, 94, 1110–1128. [Google Scholar] [CrossRef]
- Zhou, R.; To, K.K.; Wong, Y.C.; Liu, L.; Zhou, B.; Li, X.; Huang, H.; Mo, Y.; Luk, T.Y.; Lau, T.T.; et al. Acute SARS-CoV-2 Infection Impairs Dendritic Cell and T Cell Responses. Immunity 2020, 53, 864–877.e865. [Google Scholar] [CrossRef]
- Alamri, A.; Fisk, D.; Upreti, D.; Kung, S.K.P. A Missing Link: Engagements of Dendritic Cells in the Pathogenesis of SARS-CoV-2 Infections. Int. J. Mol. Sci. 2021, 22, 1118. [Google Scholar] [CrossRef] [PubMed]
- Buttenschon, J.; Mattner, J. The interplay between dendritic cells and CD8 T lymphocytes is a crucial component of SARS-CoV-2 immunity. Cell Mol. Immunol. 2021, 18, 247–249. [Google Scholar] [CrossRef]
- Grabowska, J.; Leopold, V.; Olesek, K.; Nijen Twilhaar, M.K.; Affandi, A.J.; Brouwer, M.C.; Jongerius, I.; Verschoor, A.; van Kooten, C.; van Kooyk, Y.; et al. Platelets interact with CD169(+) macrophages and cDC1 and enhance liposome-induced CD8(+) T cell responses. Front. Immunol. 2023, 14, 1290272. [Google Scholar] [CrossRef]
- Herzog, S.; Fragkou, P.C.; Arneth, B.M.; Mkhlof, S.; Skevaki, C. Myeloid CD169/Siglec1: An immunoregulatory biomarker in viral disease. Front. Med. 2022, 9, 979373. [Google Scholar] [CrossRef] [PubMed]
- Chung, M.; Bernheim, A.; Mei, X.; Zhang, N.; Huang, M.; Zeng, X.; Cui, J.; Xu, W.; Yang, Y.; Fayad, Z.A.; et al. CT Imaging Features of 2019 Novel Coronavirus (2019-nCoV). Radiology 2020, 295, 202–207. [Google Scholar] [CrossRef] [PubMed]
- Bourgoin, P.; Biechele, G.; Ait Belkacem, I.; Morange, P.E.; Malergue, F. Role of the interferons in CD64 and CD169 expressions in whole blood: Relevance in the balance between viral- or bacterial-oriented immune responses. Immun. Inflamm. Dis. 2020, 8, 106–123. [Google Scholar] [CrossRef] [PubMed]
- Rempel, H.; Calosing, C.; Sun, B.; Pulliam, L. Sialoadhesin expressed on IFN-induced monocytes binds HIV-1 and enhances infectivity. PLoS ONE 2008, 3, e1967. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Y.S.; Cheng, Y.; Lin, Q.S.; Wu, A.L.; Yu, J.; Li, C.; Sun, Y.; Zhong, R.Q.; Wu, L.J. Increased expression of Siglec-1 on peripheral blood monocytes and its role in mononuclear cell reactivity to autoantigen in rheumatoid arthritis. Rheumatology 2014, 53, 250–259. [Google Scholar] [CrossRef] [PubMed]
- Wilk, A.J.; Rustagi, A.; Zhao, N.Q.; Roque, J.; Martinez-Colon, G.J.; McKechnie, J.L.; Ivison, G.T.; Ranganath, T.; Vergara, R.; Hollis, T.; et al. A single-cell atlas of the peripheral immune response in patients with severe COVID-19. Nat. Med. 2020, 26, 1070–1076. [Google Scholar] [CrossRef] [PubMed]
- Wen, W.; Su, W.; Tang, H.; Le, W.; Zhang, X.; Zheng, Y.; Liu, X.; Xie, L.; Li, J.; Ye, J.; et al. Immune cell profiling of COVID-19 patients in the recovery stage by single-cell sequencing. Cell Discov. 2020, 6, 31. [Google Scholar] [CrossRef] [PubMed]
- Ortillon, M.; Coudereau, R.; Cour, M.; Rimmele, T.; Godignon, M.; Gossez, M.; Yonis, H.; Argaud, L.; Lukaszewicz, A.C.; Venet, F.; et al. Monocyte CD169 expression in COVID-19 patients upon intensive care unit admission. Cytom. A 2021, 99, 466–471. [Google Scholar] [CrossRef] [PubMed]
- Laing, A.G.; Lorenc, A.; Del Molino Del Barrio, I.; Das, A.; Fish, M.; Monin, L.; Munoz-Ruiz, M.; McKenzie, D.R.; Hayday, T.S.; Francos-Quijorna, I.; et al. A dynamic COVID-19 immune signature includes associations with poor prognosis. Nat. Med. 2020, 26, 1623–1635. [Google Scholar] [CrossRef]
- Chen, Q.; Yu, B.; Yang, Y.; Huang, J.; Liang, Y.; Zhou, J.; Li, L.; Peng, X.; Cheng, B.; Lin, Y. Immunological and inflammatory profiles during acute and convalescent phases of severe/ critically ill COVID-19 patients. Int. Immunopharmacol. 2021, 97, 107685. [Google Scholar] [CrossRef]
- Hadjadj, J.; Yatim, N.; Barnabei, L.; Corneau, A.; Boussier, J.; Smith, N.; Pere, H.; Charbit, B.; Bondet, V.; Chenevier-Gobeaux, C.; et al. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science 2020, 369, 718–724. [Google Scholar] [CrossRef]
- Matic, S.; Popovic, S.; Djurdjevic, P.; Todorovic, D.; Djordjevic, N.; Mijailovic, Z.; Sazdanovic, P.; Milovanovic, D.; Ruzic Zecevic, D.; Petrovic, M.; et al. SARS-CoV-2 infection induces mixed M1/M2 phenotype in circulating monocytes and alterations in both dendritic cell and monocyte subsets. PLoS ONE 2020, 15, e0241097. [Google Scholar] [CrossRef] [PubMed]
- Zingaropoli, M.A.; Nijhawan, P.; Carraro, A.; Pasculli, P.; Zuccala, P.; Perri, V.; Marocco, R.; Kertusha, B.; Siccardi, G.; Del Borgo, C.; et al. Increased sCD163 and sCD14 Plasmatic Levels and Depletion of Peripheral Blood Pro-Inflammatory Monocytes, Myeloid and Plasmacytoid Dendritic Cells in Patients With Severe COVID-19 Pneumonia. Front. Immunol. 2021, 12, 627548. [Google Scholar] [CrossRef] [PubMed]
- Mantlo, E.; Bukreyeva, N.; Maruyama, J.; Paessler, S.; Huang, C. Antiviral activities of type I interferons to SARS-CoV-2 infection. Antiviral Res. 2020, 179, 104811. [Google Scholar] [CrossRef] [PubMed]
- Prompetchara, E.; Ketloy, C.; Palaga, T. Immune responses in COVID-19 and potential vaccines: Lessons learned from SARS and MERS epidemic. Asian Pac. J. Allergy Immunol. 2020, 38, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Oosterhoff, D.; Lougheed, S.; van de Ven, R.; Lindenberg, J.; van Cruijsen, H.; Hiddingh, L.; Kroon, J.; van den Eertwegh, A.J.; Hangalapura, B.; Scheper, R.J.; et al. Tumor-mediated inhibition of human dendritic cell differentiation and function is consistently counteracted by combined p38 MAPK and STAT3 inhibition. Oncoimmunology 2012, 1, 649–658. [Google Scholar] [CrossRef]
- Yu, H.; Pardoll, D.; Jove, R. STATs in cancer inflammation and immunity: A leading role for STAT3. Nat. Rev. Cancer 2009, 9, 798–809. [Google Scholar] [CrossRef] [PubMed]
- Oblitas, C.M.; Demelo-Rodriguez, P.; Alvarez-Sala-Walther, L.A.; Rubio-Rivas, M.; Navarro-Romero, F.; Giner Galvan, V.; de Jorge-Huerta, L.; Fonseca Aizpuru, E.; Garcia Garcia, G.M.; Beato Perez, J.L.; et al. Prognostic Value of D-dimer to Lymphocyte Ratio (DLR) in Hospitalized Coronavirus Disease 2019 (COVID-19) Patients: A Validation Study in a National Cohort. Viruses 2024, 16, 335. [Google Scholar] [CrossRef]
- Lopez Reboiro, M.L.; Suarez Fuentetaja, R.; Gutierrez Lopez, R.; Ares Castro-Conde, B.; Sardina Gonzalez, C.; Navarro Menendez, I.; Pereyra Barrionuevo, M.F.; Alves Perez, M.T.; Lopez Castro, J. Role of lupus anticoagulant and von Willebrand factor in chronic reactive endotheliitis in COVID-19. J. Infect. 2021, 82, e27–e28. [Google Scholar] [CrossRef]
| Parameter | |
|---|---|
| Participants, n | 58 |
| Age (yrs) | 68 ± 12.8 |
| Sex (M/F) | 40/18 |
| Smoking habit (either active or former) | 33 (58) |
| Body mass index (kg/m2) | 27.3 [25–30.8] |
| Type II diabetes | 34 (58) |
| Systemic arterial hypertension | 33 (58) |
| Chest CT Chung score | 13 [8–15] |
| paO2/FiO2 | 122.5 [96–170] |
| neutrophils/lymphocytes | 13 [5.6–24.1] |
| C reactive protein (mg/dL) | 4.9 [2.1–9.4] |
| Interleukin-6 (pg/mL) | 34 [17–59] |
| KL-6 (U/mL) | 657 [349–1354] |
| D-Dimer (μg/mL) | 422 [250–937] |
| Lenght of hospitalization (days) | 16 [10.75–24.25] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Galati, D.; Mallardo, D.; Nicastro, C.; Zanotta, S.; Capitelli, L.; Lombardi, C.; Baino, B.; Cavalcanti, E.; Sale, S.; Labonia, F.; et al. The Dysregulation of the Monocyte–Dendritic Cell Interplay Is Associated with In-Hospital Mortality in COVID-19 Pneumonia. J. Clin. Med. 2024, 13, 2481. https://doi.org/10.3390/jcm13092481
Galati D, Mallardo D, Nicastro C, Zanotta S, Capitelli L, Lombardi C, Baino B, Cavalcanti E, Sale S, Labonia F, et al. The Dysregulation of the Monocyte–Dendritic Cell Interplay Is Associated with In-Hospital Mortality in COVID-19 Pneumonia. Journal of Clinical Medicine. 2024; 13(9):2481. https://doi.org/10.3390/jcm13092481
Chicago/Turabian StyleGalati, Domenico, Domenico Mallardo, Carmine Nicastro, Serena Zanotta, Ludovica Capitelli, Carmen Lombardi, Bianca Baino, Ernesta Cavalcanti, Silvia Sale, Francesco Labonia, and et al. 2024. "The Dysregulation of the Monocyte–Dendritic Cell Interplay Is Associated with In-Hospital Mortality in COVID-19 Pneumonia" Journal of Clinical Medicine 13, no. 9: 2481. https://doi.org/10.3390/jcm13092481






