Multi-Environment Trials and Stability Analysis for Yield-Related Traits of Commercial Rice Cultivars
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Field Experimental Design
2.2. Phenotypic Data Collection
2.3. Statistical Analysis
3. Results
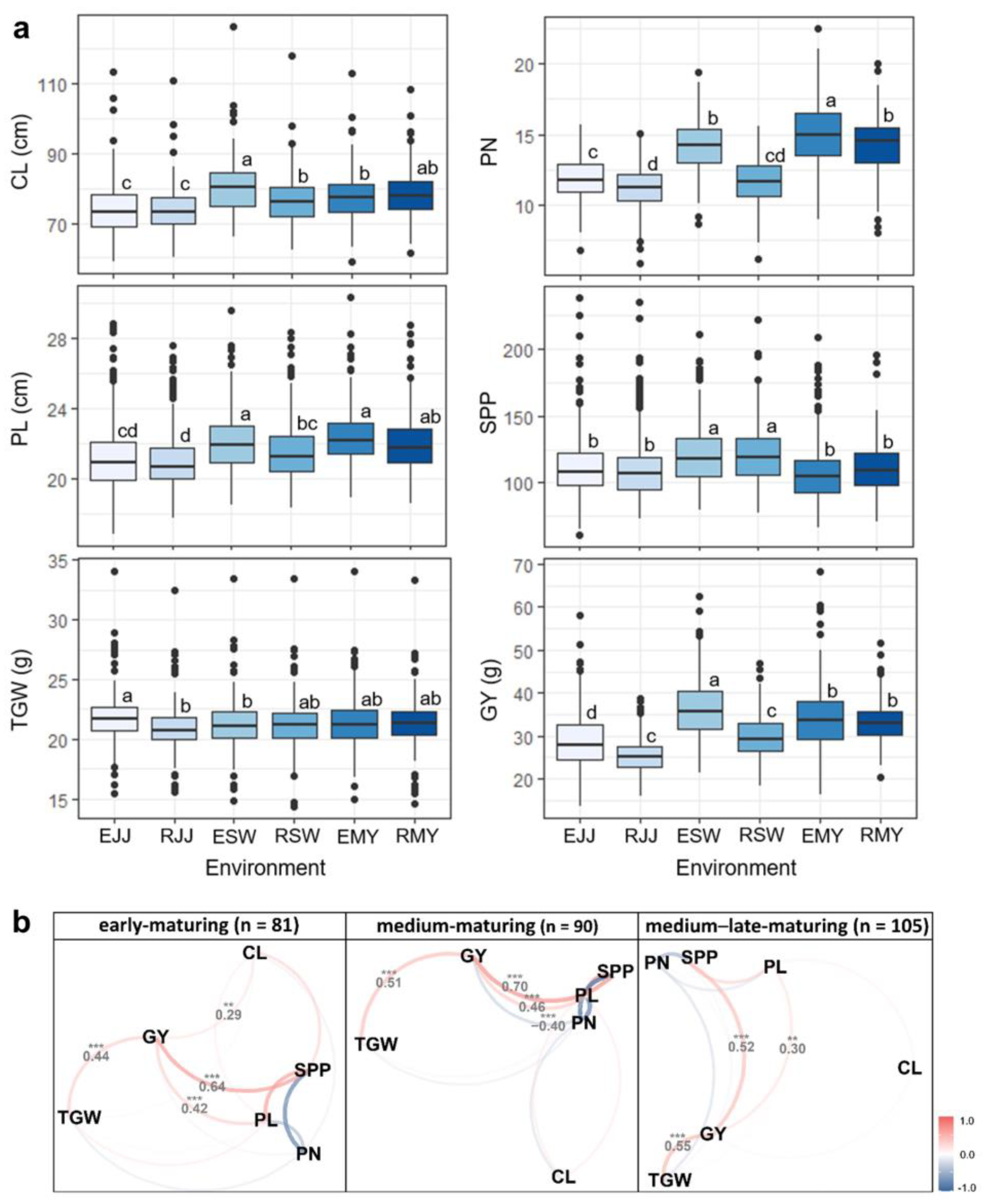
3.1. Descriptive Summary of Yield-Related Traits under Multi-Environments
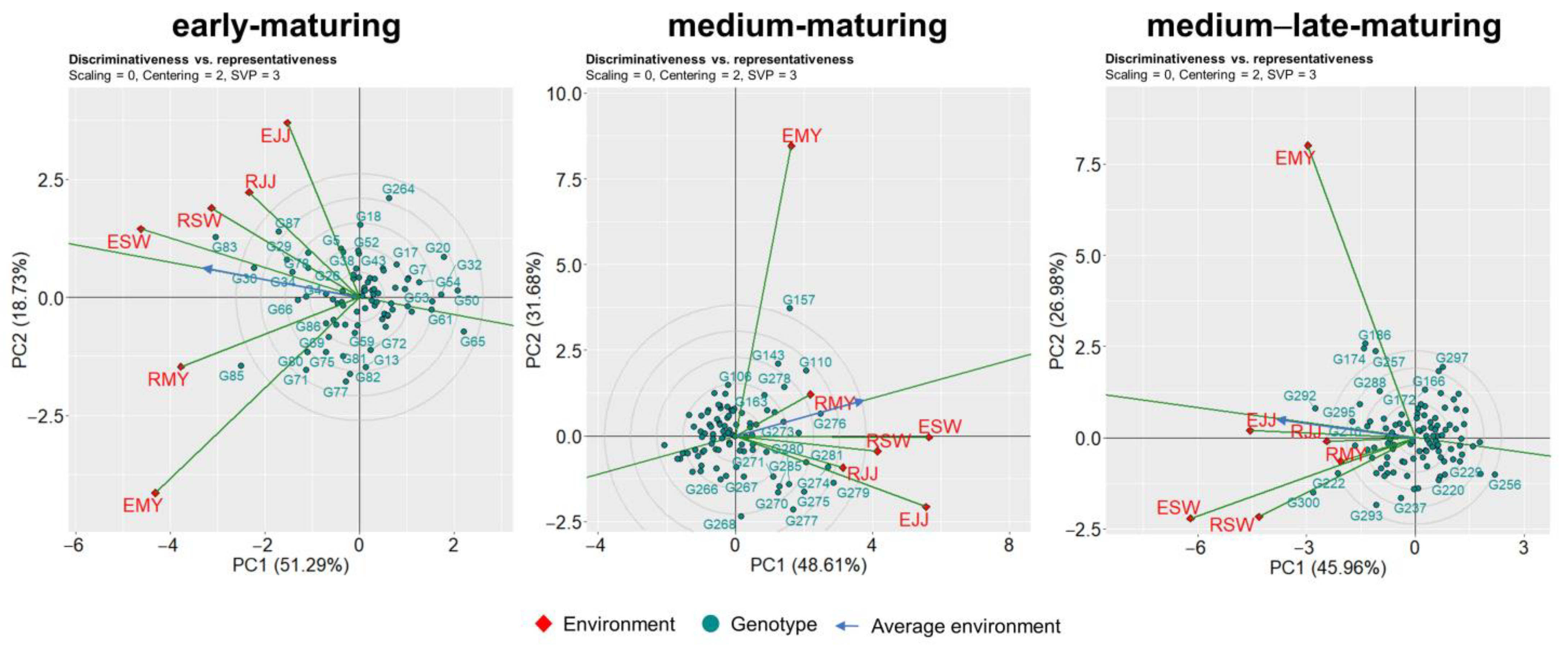
3.2. Mega-Environment Analysis
3.3. Evaluating Idealness of the Environment
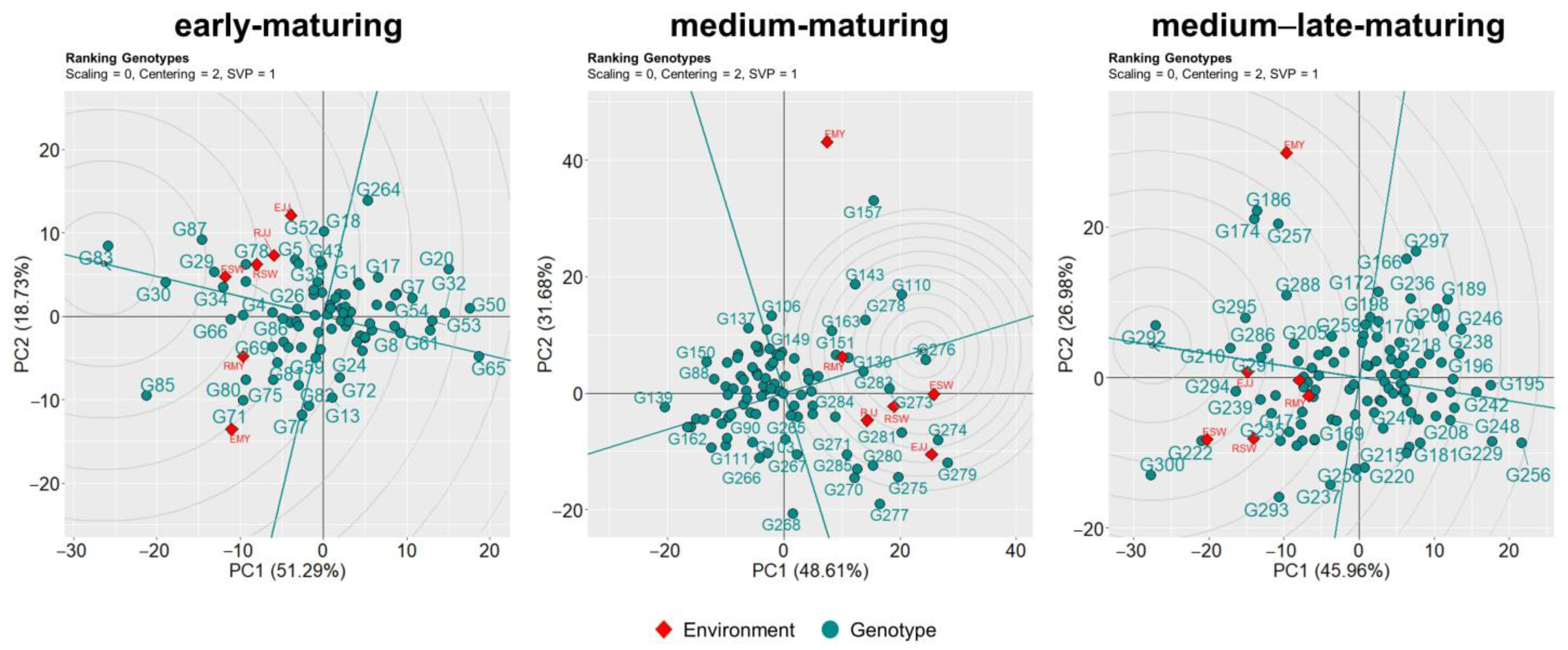
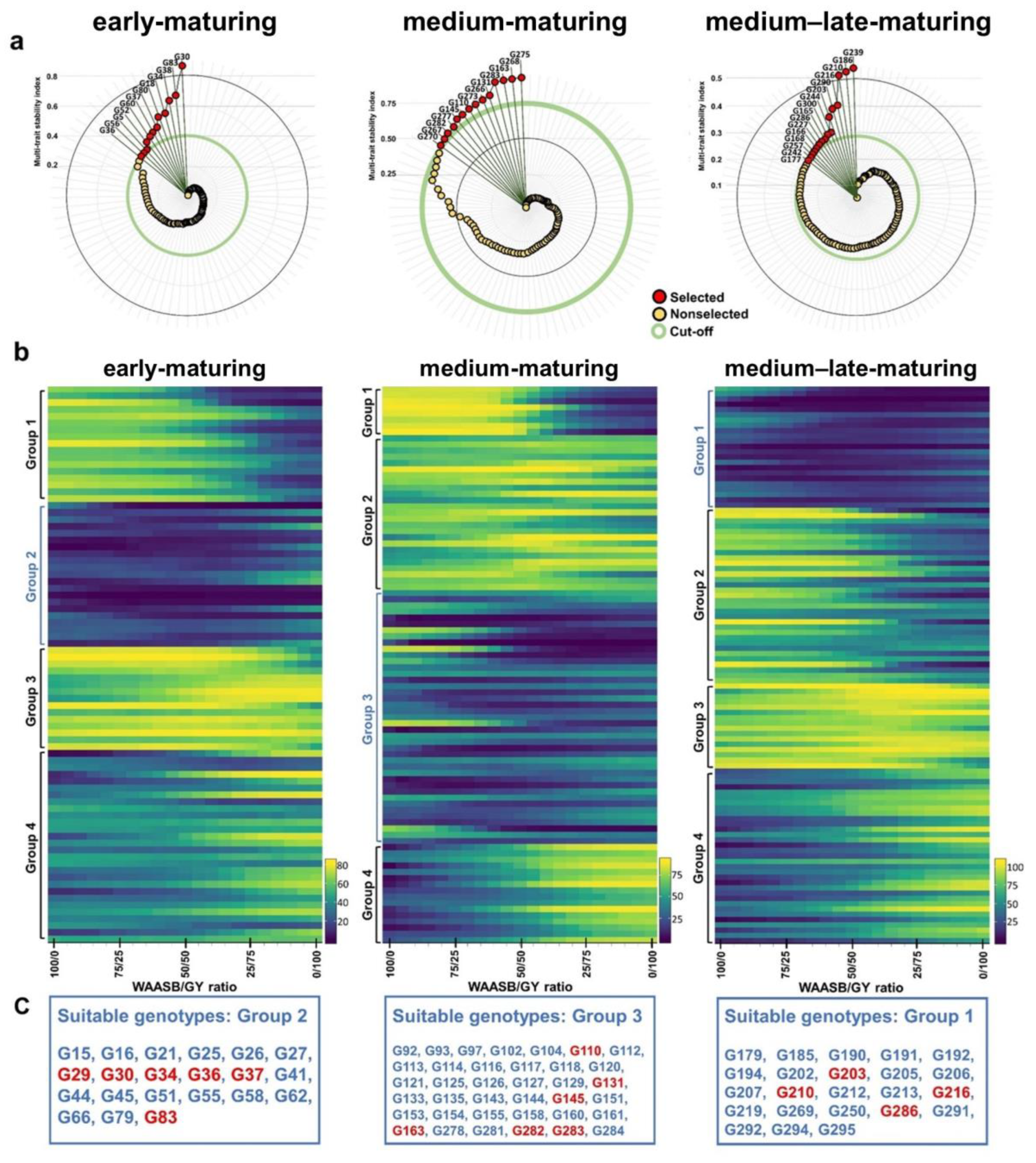
3.4. Ranking Genotypes Based on Performance and Stability
3.5. Superior Genotype Selection Based on Performance and Stability Indices
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Muthayya, S.; Sugimoto, J.D.; Montgomery, S.; Maberly, G.F. An overview of global rice production, supply, trade, and consumption. Ann. New York Acad. Sci. 2014, 1324, 7–14. [Google Scholar] [CrossRef] [PubMed]
- Tester, M.; Langridge, P. Breeding technologies to increase crop production in a changing world. Science 2010, 327, 818–822. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, T.; Matsuoka, M. Identifying and exploiting grain yield genes in rice. Curr. Opin. Plant Biol. 2008, 11, 209–214. [Google Scholar] [CrossRef]
- Hall, A.J.; Richards, R.A. Prognosis for genetic improvement of yield potential and water-limited yield of major grain crops. Field Crop. Res. 2013, 143, 18–33. [Google Scholar] [CrossRef] [Green Version]
- Li, R.; Li, M.; Ashraf, U.; Liu, S.; Zhang, J. Exploring the relationships between yield and yield-related traits for rice varieties released in China from 1978 to 2017. Front. Plant Sci. 2019, 10, 543. [Google Scholar] [CrossRef] [Green Version]
- Peng, S.; Cassman, K.G.; Virmani, S.; Sheehy, J.; Khush, G. Yield potential trends of tropical rice since the release of IR8 and the challenge of increasing rice yield potential. Crop. Sci. 1999, 39, 1552–1559. [Google Scholar] [CrossRef] [Green Version]
- Kang, M.S. Using genotype-by-environment interaction for crop cultivar development. Adv. Agron. 1997, 62, 199–252. [Google Scholar]
- Malosetti, M.; Ribaut, J.-M.; van Eeuwijk, F.A. The statistical analysis of multi-environment data: Modeling genotype-by-environment interaction and its genetic basis. Front. Physiol. 2013, 4, 44. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eberhart, S.T.; Russell, W. Stability parameters for comparing varieties 1. Crop. Sci. 1966, 6, 36–40. [Google Scholar] [CrossRef] [Green Version]
- Gauch, H., Jr. Statistical Analysis of Regional Yield Trials: AMMI Analysis of Factorial Designs; Elsevier Science Publishers: Amsterdam, The Netherlands, 1992. [Google Scholar]
- Mooers, C. The agronomic placement of varieties. Agron. J. 1921, 13, 337–352. [Google Scholar] [CrossRef]
- Yan, W.; Hunt, L.A.; Sheng, Q.; Szlavnics, Z. Cultivar evaluation and mega-environment investigation based on the GGE biplot. Crop. Sci. 2000, 40, 597–605. [Google Scholar] [CrossRef]
- Olivoto, T.; Lúcio, A.D.; da Silva, J.A.; Marchioro, V.S.; de Souza, V.Q.; Jost, E. Mean performance and stability in multi-environment trials I: Combining features of AMMI and BLUP techniques. Agron. J. 2019, 111, 2949–2960. [Google Scholar] [CrossRef]
- Olivoto, T.; Lúcio, A.D.; da Silva, J.A.; Sari, B.G.; Diel, M.I. Mean performance and stability in multi-environment trials II: Selection based on multiple traits. Agron. J. 2019, 111, 2961–2969. [Google Scholar] [CrossRef]
- Vergara, B. The flowering response of the rice plant to photoperiod. Int. Rice Res. Instig. Technol. Bull 1976, 8, 75. [Google Scholar]
- Collinson, S.; Ellis, R.; Summerfield, R.; Roberts, E. Durations of the photoperiod-sensitive and photoperiod-insensitive phases of development to flowering in four cultivars of rice (Oryza sativa L.). Ann. Bot. 1992, 70, 339–346. [Google Scholar] [CrossRef]
- Bueno, C.S.; Lafarge, T. Maturity groups and growing seasons as key sources of variation to consider within breeding programs for high yielding rice in the tropics. Euphytica 2017, 213, 1–18. [Google Scholar] [CrossRef]
- Jadhav, S.; Balakrishnan, D.; Shankar V., G.; Beerelli, K.; Chandu, G.; Neelamraju, S. Genotype by environment (G × E) interaction study on yield traits in different maturity groups of Rice. J. Crop. Sci. Biotechnol. 2019, 22, 425–449. [Google Scholar] [CrossRef]
- The Government of the Republic of Korea. 2050 Carbon Neutral Strategy of the Republic of Korea: Towards a Sustainable and Green Society; The Government of the Republic of Korea: Seoul, Republic of Korea, 2020.
- Huang, X.; Kurata, N.; Wang, Z.-X.; Wang, A.; Zhao, Q.; Zhao, Y.; Liu, K.; Lu, H.; Li, W.; Guo, Y. A map of rice genome variation reveals the origin of cultivated rice. Nature 2012, 490, 497–501. [Google Scholar] [CrossRef] [Green Version]
- Lee, C.-M.; Kwon, Y.-H.; Park, H.-M.; Jeung, J.-U.; Park, H.-S.; Baek, M.-K.; Ha, S.-K.; Mo, Y. Days to heading and culm length variation of Korean rice varieties in different environments. Korean Soc. Breed. Sci. 2020, 52, 389–397. [Google Scholar] [CrossRef]
- RDA. Manual for Standard Evaluation Method in Agricultural Experiment and Research; RDA Press: Suwon, Korea, 2012. [Google Scholar]
- Team, R.C. R: A Language and Environment for Statistical Computing; R Core Team: Vienna, Australia, 2013. [Google Scholar]
- Mendiburu, F.D.; Yaseen, M. Agricolae: Statistical Procedures for Agricultural Research, R Package Version 1.4.0.; R Core Team: Vienna, Australia, 2020.
- Kuhn, M.; Jackson, S.; Cimentada, J. Corrr: Correlations in R, R Package Version 0.4.2; R Core Team: Vienna, Australia, 2020.
- Bates, D.; Mächler, M.; Bolker, B.; Walker, S. Fitting linear mixed-effects models using lme4. arXiv Prepr. 2014, arXiv:1406.5823. [Google Scholar]
- Yan, W.; Kang, M.S. GGE Biplot Analysis: A Graphical Tool for Breeders, Geneticists, and Agronomists; CRC Press: Boca Raton, FL, USA, 2002. [Google Scholar]
- Nataraj, V.; Bhartiya, A.; Singh, C.P.; Devi, H.N.; Deshmukh, M.P.; Verghese, P.; Singh, K.; Mehtre, S.P.; Kumari, V.; Maranna, S. WAASB-based stability analysis and simultaneous selection for grain yield and early maturity in soybean. Agron. J. 2021, 113, 3089–3099. [Google Scholar] [CrossRef]
- Rocha, J.R.D.A.S.D.C.; Machado, J.C.; Carneiro, P.C.S. Multitrait index based on factor analysis and ideotype-design: Proposal and application on elephant grass breeding for bioenergy. Gcb Bioenergy 2018, 10, 52–60. [Google Scholar] [CrossRef]
- Olivoto, T.; Lúcio, A.D.C. metan: An R package for multi-environment trial analysis. Methods Ecol. Evol. 2020, 11, 783–789. [Google Scholar] [CrossRef]
- Mo, Y.; Jeong, J.-M.; Kim, B.-K.; Kwon, S.-W.; Jeung, J.-U. Utilization of elite Korean japonica rice varieties for association mapping of heading time, culm length, and amylose and protein content. Korean J. Crop. Sci. 2020, 65, 1–21. [Google Scholar]
- Mo, Y.; Lee, C.-M.; Park, H.-M.; Ha, S.-K.; Kim, M.-J.; Kwak, J.; Lee, H.-S.; Lee, J.-H.; Jeung, J.-U. Hd1 Allele Types and Their Associations with Major Agronomic Traits in Korean Rice Cultivars. Plants 2021, 10, 2408. [Google Scholar] [CrossRef]
- Yan, W.; Tinker, N.A. Biplot analysis of multi-environment trial data: Principles and applications. Can. J. Plant Sci. 2006, 86, 623–645. [Google Scholar] [CrossRef] [Green Version]
- Khan, M.M.H.; Rafii, M.Y.; Ramlee, S.I.; Jusoh, M.; Al Mamun, M. AMMI and GGE biplot analysis for yield performance and stability assessment of selected Bambara groundnut (Vigna subterranea L. Verdc.) genotypes under the multi-environmental trials (METs). Sci. Rep. 2021, 11, 22791. [Google Scholar] [CrossRef]
- Yan, W.; Kang, M.S.; Ma, B.; Woods, S.; Cornelius, P.L. GGE biplot vs. AMMI analysis of genotype-by-environment data. Crop. Sci. 2007, 47, 643–653. [Google Scholar] [CrossRef] [Green Version]
- Sharifi, P.; Aminpanah, H.; Erfani, R.; Mohaddesi, A.; Abbasian, A. Evaluation of genotype× environment interaction in rice based on AMMI model in Iran. Rice Sci. 2017, 24, 173–180. [Google Scholar] [CrossRef]
- Enyew, M.; Feyissa, T.; Geleta, M.; Tesfaye, K.; Hammenhag, C.; Carlsson, A.S. Genotype by environment interaction, correlation, AMMI, GGE biplot and cluster analysis for grain yield and other agronomic traits in sorghum (Sorghum bicolor L. Moench). PLoS ONE 2021, 16, e0258211. [Google Scholar] [CrossRef]
- Pour-Aboughadareh, A.; Barati, A.; Koohkan, S.A.; Jabari, M.; Marzoghian, A.; Gholipoor, A.; Shahbazi-Homonloo, K.; Zali, H.; Poodineh, O.; Kheirgo, M. Dissection of genotype-by-environment interaction and yield stability analysis in barley using AMMI model and stability statistics. Bull. Natl. Res. Cent. 2022, 46, 19. [Google Scholar] [CrossRef]
- Huang, X.; Jang, S.; Kim, B.; Piao, Z.; Redona, E.; Koh, H.-J. Evaluating Genotype× Environment Interactions of Yield Traits and Adaptability in Rice Cultivars Grown under Temperate, Subtropical and Tropical Environments. Agriculture 2021, 11, 558. [Google Scholar] [CrossRef]
- Balakrishnan, D.; Subrahmanyam, D.; Badri, J.; Raju, A.K.; Rao, Y.V.; Beerelli, K.; Mesapogu, S.; Surapaneni, M.; Ponnuswamy, R.; Padmavathi, G. Genotype× environment interactions of yield traits in backcross introgression lines derived from Oryza sativa cv. Swarna/Oryza nivara. Front. Plant Sci. 2016, 7, 1530. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Das, A.; Parihar, A.K.; Saxena, D.; Singh, D.; Singha, K.; Kushwaha, K.; Chand, R.; Bal, R.; Chandra, S.; Gupta, S. Deciphering genotype-by-environment interaction for targeting test environments and rust resistant genotypes in field pea (Pisum sativum L.). Front. Plant Sci. 2019, 10, 825. [Google Scholar] [CrossRef] [PubMed]
- de Leon, N.; Jannink, J.L.; Edwards, J.W.; Kaeppler, S.M. Introduction to a special issue on genotype by environment interaction. Crop. Sci. 2016, 56, 2081–2089. [Google Scholar] [CrossRef] [Green Version]
- Yan, W.; Cornelius, P.L.; Crossa, J.; Hunt, L. Two types of GGE biplots for analyzing multi-environment trial data. Crop. Sci. 2001, 41, 656–663. [Google Scholar] [CrossRef] [Green Version]
- Islam, S.S.; Anothai, J.; Nualsri, C.; Soonsuwon, W. Analysis of genotype-environment interaction and yield stability of Thai upland rice ('Oryza sativa'L.) genotypes using AMMI model. Aust. J. Crop. Sci. 2020, 14, 362–370. [Google Scholar] [CrossRef]
- Tumuhimbise, R.; Melis, R.; Shanahan, P.; Kawuki, R. Genotype × environment interaction effects on early fresh storage root yield and related traits in cassava. Crop. J. 2014, 2, 329–337. [Google Scholar] [CrossRef] [Green Version]
- Becker, H.; Leon, J. Stability analysis in plant breeding. Plant Breed. 1988, 101, 1–23. [Google Scholar] [CrossRef]
- Lin, C.-S.; Binns, M.R.; Lefkovitch, L.P. Stability analysis: Where do we stand? 1. Crop. Sci. 1986, 26, 894–900. [Google Scholar] [CrossRef] [Green Version]
- Pour-Aboughadareh, A.; Sanjani, S.; Nikkhah-Chamanabad, H.; Mehrvar, M.R.; Asadi, A.; Amini, A. Identification of salt-tolerant barley genotypes using multiple-traits index and yield performance at the early growth and maturity stages. Bull. Natl. Res. Cent. 2021, 45, 117. [Google Scholar] [CrossRef]
- Sharifi, P.; Erfani, A.; Abbasian, A.; Mohaddesi, A. Stability of some of rice genotypes based on WAASB and MTSI indices. Iran. J. Genet. Plant Breed. 2020, 9, 1–11. [Google Scholar]
- Heredia, M.C.; Kant, J.; Prodhan, M.; Dixit, S.; Wissuwa, M. Breeding rice for a changing climate by improving adaptations to water saving technologies. Theor. Appl. Genet. 2021, 135, 17–33. [Google Scholar] [CrossRef] [PubMed]
- Wassmann, R.; Papen, H.; Rennenberg, H. Methane emission from rice paddies and possible mitigation strategies. Chemosphere 1993, 26, 201–217. [Google Scholar] [CrossRef]
- Yagi, K.; Tsuruta, H.; Minami, K. Possible options for mitigating methane emission from rice cultivation. Nutr. Cycl. Agroecosystems 1997, 49, 213–220. [Google Scholar] [CrossRef]
- Mahmood, T.; Ahmed, T.; Trethowan, R. Genotype × Environment × Management (GEM) Reciprocity and Crop Productivity; FAO: Rome, Italy, 2022. [Google Scholar]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, S.Y.; Lee, H.-S.; Lee, C.-M.; Ha, S.-K.; Park, H.-M.; Lee, S.-M.; Kwon, Y.; Jeung, J.-U.; Mo, Y. Multi-Environment Trials and Stability Analysis for Yield-Related Traits of Commercial Rice Cultivars. Agriculture 2023, 13, 256. https://doi.org/10.3390/agriculture13020256
Lee SY, Lee H-S, Lee C-M, Ha S-K, Park H-M, Lee S-M, Kwon Y, Jeung J-U, Mo Y. Multi-Environment Trials and Stability Analysis for Yield-Related Traits of Commercial Rice Cultivars. Agriculture. 2023; 13(2):256. https://doi.org/10.3390/agriculture13020256
Chicago/Turabian StyleLee, Seung Young, Hyun-Sook Lee, Chang-Min Lee, Su-Kyung Ha, Hyang-Mi Park, So-Myeong Lee, Youngho Kwon, Ji-Ung Jeung, and Youngjun Mo. 2023. "Multi-Environment Trials and Stability Analysis for Yield-Related Traits of Commercial Rice Cultivars" Agriculture 13, no. 2: 256. https://doi.org/10.3390/agriculture13020256




