Recent Developments in Rice Molecular Breeding for Tolerance to Heavy Metal Toxicity
Abstract
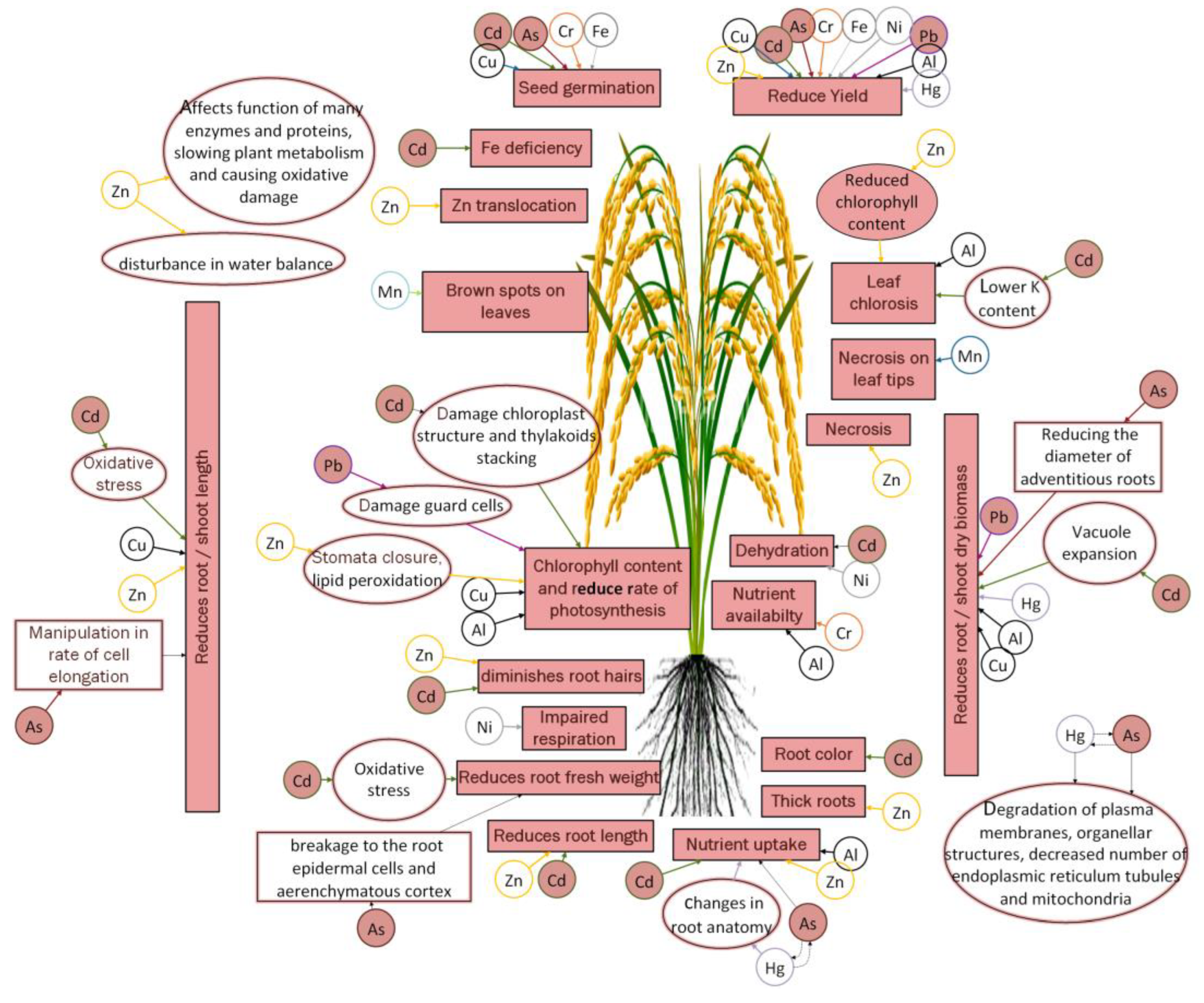
:1. Introduction
| Heavy Metal | Dose/Concentration (mg/kg) | Yield Loss (%) | Reference |
|---|---|---|---|
| Pb | 400–1200 | 46–69 | [25] |
| As | 24.5 | 40 | [26] |
| Cd | 50–150 | 34–63 | [27] |
| Fe | 385–1197 | 16–78 | [28] |
| Cu | 100–1000 | 10–90 | [29] |
| Zn | - | 20–40 | [30] |
| Ni | 40–100 | 54–70 | [31] |
| Al | - | 30–60 | [32] |
| Cr | 200–400 | 30–48 | [32] |
| Hg | 4–5 | 50–70 | [33] |
2. Natural and Anthropogenic Sources of HM Toxicity in Rice
3. Progress in Molecular Breeding to Develop HMT Tolerance in Rice
3.1. Role of HM Transporters (HMTs) and Tolerant Proteins (HMPs)
3.2. Role of microRNAs (miRNAs)
3.3. QTL and Fine Gene Mapping
3.4. HM Tolerant Transgenic Rice
3.5. CRISPR/Cas Genome Editing
4. Breeding Tools for Improving HM Toxicity Tolerance in Rice
4.1. Physiological Screening
4.2. Mutation Breeding
4.3. Molecular Gene Mapping
4.4. Gene Silencing
5. Challenges and Limitations in Breeding for HM-Tolerant Rice
6. Conclusions and Future Prospects
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tong, M.; Liu, X.; Guan, J.; Lin, Y.; Zhou, A.; Qiao, K. Novel biofortification candidate: MTP1 increases microelement contents and decreases toxic heavy metal accumulation in grains. Chemosphere 2023, 318, 137967. [Google Scholar] [CrossRef]
- Singh, A.; Sharma, A.; Verma, R.K.; Chopade, R.L.; Pandit, P.P.; Nagar, V.; Aseri, V.; Choudhary, S.K.; Awasthi, G.; Awasthi, K.K. Heavy metal contamination of water and their toxic effect on living organisms. In The Toxicity of Environmental Pollutants; Dorta, D., De Oliveira, D.P., Eds.; IntechOpen: London, UK, 2022. [Google Scholar]
- Zhang, W.; Liu, M.; Li, C. Soil heavy metal contamination assessment in the Hun-Taizi River watershed, China. Sci. Rep. 2020, 10, 8730. [Google Scholar] [CrossRef]
- Shen, E.; Wang, X.; Lu, Z.; Zhou, F.; Ma, W.; Cui, Z.; Li, Z.; Li, C.; Lin, Y. Overexpression of a beta-1, 6-glucanase gene GluM in transgenic rice confers high resistance to rice blast, sheath blight and false smut. Pest Manag. Sci. 2023, 79, 7394. [Google Scholar] [CrossRef] [PubMed]
- Sodhi, K.K.; Mishra, L.C.; Singh, C.K.; Kumar, M. Perspective on the heavy metal pollution and recent remediation strategies. Curr. Res. Microb. Sci. 2022, 3, 100166. [Google Scholar] [CrossRef] [PubMed]
- Malinowska, E.; Jankowski, K. The effect of different doses of sewage sludge and liming on total cobalt content and its speciation in soil. Agronomy 2020, 10, 1550. [Google Scholar] [CrossRef]
- Bing, H.; Qiu, S.; Tian, X.; Li, J.; Zhu, H.; Wu, Y.; Zhang, G. Trace metal contamination in soils from mountain regions across China: Spatial distribution, sources, and potential drivers. Soil Ecol. Lett. 2021, 3, 189–206. [Google Scholar] [CrossRef]
- Tóth, G.; Hermann, T.; Da Silva, M.; Montanarella, L. Heavy metals in agricultural soils of the European Union with implications for food safety. Environ. Int. 2016, 88, 299–309. [Google Scholar] [CrossRef] [PubMed]
- Rai, P.K. Phytoremediation of Emerging Contaminants in Wetlands; CRC Press: Boca Raton, FL, USA, 2018. [Google Scholar]
- Giri, S.; Singh, A.K.; Mahato, M.K. Monte Carlo simulation-based probabilistic health risk assessment of metals in groundwater via ingestion pathway in the mining areas of Singhbhum copper belt, India. Int. J. Environ. Health Res. 2020, 30, 447–460. [Google Scholar] [CrossRef]
- Giri, S.; Mahato, M.K.; Bhattacharjee, S.; Singh, A.K. Development of a new noncarcinogenic heavy metal pollution index for quality ranking of vegetable, rice, and milk. Ecol. Indic. 2020, 113, 106214. [Google Scholar] [CrossRef]
- Ugulu, I.; Akhter, P.; Khan, Z.I.; Akhtar, M.; Ahmad, K. Trace metal accumulation in pepper (Capsicum annuum L.) grown using organic fertilizers and health risk assessment from consumption. Food Res. Int. 2021, 140, 109992. [Google Scholar] [CrossRef]
- Jin, T.; Nordberg, M.; Frech, W.; Dumont, X.; Bernard, A.; Ye, T.-T.; Kong, Q.; Wang, Z.; Li, P.; Lundström, N.-G. Cadmium biomonitoring and renal dysfunction among a population environmentally exposed to cadmium from smelting in China (ChinaCad). Biometals 2002, 15, 397–410. [Google Scholar] [CrossRef] [PubMed]
- Mayor, D.J.; Gray, N.B.; Elver-Evans, J.; Midwood, A.J.; Thornton, B. Metal-macrofauna interactions determine microbial community structure and function in copper contaminated sediments. PLoS ONE 2013, 8, e64940. [Google Scholar] [CrossRef] [PubMed]
- Kazberuk, W.; Szulc, W.; Rutkowska, B. Use bottom sediment to agriculture—Effect on plant and heavy metal content in soil. Agronomy 2021, 11, 1077. [Google Scholar] [CrossRef]
- Slaveykova, V.I.; Cheloni, G. Preface: Special issue on environmental toxicology of trace metals. Environments 2018, 5, 138. [Google Scholar] [CrossRef]
- Ayejoto, D.A.; Agbasi, J.C.; Egbueri, J.C.; Echefu, K.I. Assessment of oral and dermal health risk exposures associated with contaminated water resources: An update in Ojoto area, southeast Nigeria. Int. J. Environ. Anal. Chem. 2022, 1–21. [Google Scholar] [CrossRef]
- Mu, T.; Zhou, T.; Li, Z.; Hu, P.; Luo, Y.; Christie, P.; Wu, L. Prediction models for rice cadmium accumulation in Chinese paddy fields and the implications in deducing soil thresholds based on food safety standards. Environ. Pollut. 2020, 258, 113879. [Google Scholar] [CrossRef] [PubMed]
- Hussain, B.; Ashraf, M.N.; Abbas, A.; Li, J.; Farooq, M. Cadmium stress in paddy fields: Effects of soil conditions and remediation strategies. Sci. Total Environ. 2021, 754, 142188. [Google Scholar] [CrossRef]
- Sharma, L.K.; McCray, J.M.; Morgan, K. Plant Essential Nutrients and Their Role: SS-AGR-463/AG462, 5/2022. EDIS 2022, 2022, 3. [Google Scholar] [CrossRef]
- Yu, H.-Y.; Liu, C.; Zhu, J.; Li, F.; Deng, D.-M.; Wang, Q.; Liu, C. Cadmium availability in rice paddy fields from a mining area: The effects of soil properties highlighting iron fractions and pH value. Environ. Pollut. 2016, 209, 38–45. [Google Scholar] [CrossRef]
- Özyiğit, İ.İ.; Abakirova, A.; Hocaoğlu-Özyiğit, A.; Kurmanbekova, G.; Chekirov, K.; Yalcin, B.; Yalçin, İ.E. Cadmium stress in barley seedlings: Accumulation, growth, anatomy and physiology. Int. J. Life Sci. Biotechnol. 2021, 4, 204–223. [Google Scholar] [CrossRef]
- Slamet-Loedin, I.H.; Johnson-Beebout, S.E.; Impa, S.; Tsakirpaloglou, N. Enriching rice with Zn and Fe while minimizing Cd risk. Front. Plant Sci. 2015, 6, 121. [Google Scholar] [CrossRef]
- Sageena, G.; Khatana, K.; Nagar, J.K. Biomonitoring of heavy metals contamination in soil ecosystem. In Hazardous and Trace Materials in Soil and Plants; Elsevier: Amsterdam, The Netherlands, 2022; pp. 313–325. [Google Scholar]
- Ashraf, U.; Kanu, A.S.; Deng, Q.; Mo, Z.; Pan, S.; Tian, H.; Tang, X. Lead (Pb) Toxicity; Physio-Biochemical Mechanisms, Grain Yield, Quality, and Pb Distribution Proportions in Scented Rice. Front. Plant Sci. 2017, 8, 259. [Google Scholar] [CrossRef] [PubMed]
- Muehe, E.M.; Wang, T.; Kerl, C.F.; Planer-Friedrich, B.; Fendorf, S. Rice production threatened by coupled stresses of climate and soil arsenic. Nat. Commun. 2019, 10, 4985. [Google Scholar] [CrossRef] [PubMed]
- Kanu, A.S.; Ashraf, U.; Mo, Z.; Fuseini, I.; Mansaray, L.R.; Duan, M.; Pan, S.; Tang, X. Cadmium Uptake and Distribution in Fragrant Rice Genotypes and Related Consequences on Yield and Grain Quality Traits. J. Chem. 2017, 2017, 1405878. [Google Scholar] [CrossRef]
- Audebert, A.; Fofana, M. Rice Yield Gap due to Iron Toxicity in West Africa. J. Agron. Crop Sci. 2009, 195, 66–76. [Google Scholar] [CrossRef]
- Xu, J.; Yang, L.; Wang, Z.; Dong, G.; Huang, J.; Wang, Y. Toxicity of copper on rice growth and accumulation of copper in rice grain in copper contaminated soil. Chemosphere 2007, 62, 602–607. [Google Scholar] [CrossRef]
- Panhwar, Q.A.; Naher, U.A.; Radziah, O.; Shamshuddin, J.; Razi, I.M. Eliminating aluminum toxicity in an acid sulfate soil for rice cultivation using plant growth promoting bacteria. Molecules 2015, 20, 3628–3646. [Google Scholar] [CrossRef] [PubMed]
- Aziz, H.; Sabir, M.; Ahmad, H.R.; Aziz, T.; Zia-ur-Rehman, M.; Hakeem, K.R.; Ozturk, M. Alleviating Effect of Calcium on Nickel Toxicity in Rice. Clean Soil Air Water 2015, 43, 901–909. [Google Scholar] [CrossRef]
- AbdElgawad, H.; Mohammed, A.E.; van Dijk, J.R.; Beemster, G.T.S.; Alotaibi, M.O.; Saleh, A.M. The impact of chromium toxicity on the yield and quality of rice grains produced under ambient and elevated levels of CO2. Front. Plant Sci. 2023, 14, 1019859. [Google Scholar] [CrossRef]
- Hillary, V.E.; Ceasar, S.A. Prime editing in plants and mammalian cells: Mechanism, achievements, limitations, and future prospects. BioEssays 2022, 44, e2200032. [Google Scholar] [CrossRef]
- Xu, D.; Shen, Z.; Dou, C.; Dou, Z.; Li, Y.; Gao, Y.; Sun, Q. Effects of soil properties on heavy metal bioavailability and accumulation in crop grains under different farmland use patterns. Sci. Rep. 2022, 12, 9211. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Ding, L.; Xia, Y.; Wang, F.; Zhu, C. Emerging roles of microRNAs in plant heavy metal tolerance and homeostasis. J. Agric. Food Chem. 2020, 68, 1958–1965. [Google Scholar] [CrossRef] [PubMed]
- Kaur, R.; Das, S.; Bansal, S.; Singh, G.; Sardar, S.; Dhar, H.; Ram, H. Heavy metal stress in rice: Uptake, transport, signaling, and tolerance mechanisms. Physiol. Plant. 2021, 173, 430–448. [Google Scholar] [CrossRef]
- Wan, Y.; Camara, A.Y.; Yu, Y.; Wang, Q.; Guo, T.; Zhu, L.; Li, H. Cadmium dynamics in soil pore water and uptake by rice: Influences of soil-applied selenite with different water managements. Environ. Pollut. 2018, 240, 523–533. [Google Scholar] [CrossRef]
- Sebastian, A.; Prasad, M.N.V. Cadmium minimization in rice. A review. Agron. Sustain. Dev. 2014, 34, 155–173. [Google Scholar] [CrossRef]
- Jallad, K.N. Heavy metal exposure from ingesting rice and its related potential hazardous health risks to humans. Environ. Sci. Pollut. Res. 2015, 22, 15449–15458. [Google Scholar] [CrossRef]
- Ali, H.; Khan, E.; Ilahi, I. Environmental chemistry and ecotoxicology of hazardous heavy metals: Environmental persistence, toxicity, and bioaccumulation. J. Chem. 2019, 2019, 6730305. [Google Scholar] [CrossRef]
- Zhang, C.; Zou, X.; Yang, H.; Liang, J.; Zhu, T. Bioaccumulation and risk assessment of potentially toxic elements in soil-rice system in karst area, southwest china. Front. Environ. Sci. 2022, 10, 232. [Google Scholar] [CrossRef]
- Norton, G.J.; Adomako, E.E.; Deacon, C.M.; Carey, A.-M.; Price, A.H.; Meharg, A.A. Effect of organic matter amendment, arsenic amendment and water management regime on rice grain arsenic species. Environ. Pollut. 2013, 177, 38–47. [Google Scholar] [CrossRef]
- Lin, H.-J.; Sung, T.-I.; Chen, C.-Y.; Guo, H.-R. Arsenic levels in drinking water and mortality of liver cancer in Taiwan. J. Hazard. Mater. 2013, 262, 1132–1138. [Google Scholar] [CrossRef]
- Si, L.; Xie, Y.; Ma, Q.; Wu, L. The short-term effects of rice straw biochar, nitrogen and phosphorus fertilizer on rice yield and soil properties in a cold waterlogged paddy field. Sustainability 2018, 10, 537. [Google Scholar] [CrossRef]
- Li, Y.; Dong, Z.; Feng, D.; Zhang, X.; Jia, Z.; Fan, Q.; Liu, K. Study on the risk of soil heavy metal pollution in typical developed cities in eastern China. Sci. Rep. 2022, 12, 3855. [Google Scholar] [CrossRef]
- Gong, C.; Wang, S.; Wang, D.; Lu, H.; Dong, H.; Liu, J.; Yan, B.; Wang, L. Ecological and human health risk assessment of heavy metal (loid) s in agricultural soil in hotbed chives hometown of Tangchang, Southwest China. Sci. Rep. 2022, 12, 8563. [Google Scholar] [CrossRef]
- Shao, J.F.; Xia, J.; Yamaji, N.; Shen, R.F.; Ma, J.F. Effective reduction of cadmium accumulation in rice grain by expressing OsHMA3 under the control of the OsHMA2 promoter. J. Exp. Bot. 2018, 69, 2743–2752. [Google Scholar] [CrossRef]
- Colangelo, E.P.; Guerinot, M.L. Put the metal to the petal: Metal uptake and transport throughout plants. Curr. Opin. Plant Biol. 2006, 9, 322–330. [Google Scholar] [CrossRef] [PubMed]
- Greger, M. Metal availability, uptake, transport and accumulation in plants. Heavy Met. Stress Plants Biomol. Ecosyst. 2004, 1–27. [Google Scholar] [CrossRef]
- Thi, K.V.; Lan, P.D.T.; Hang, N.N.T.; Thanh, H.N. Cadmium Immobilization in the Rice–Paddy Soil with Biochar Additive. J. Ecol. Eng. 2022, 23. [Google Scholar]
- Krämer, U.; Talke, I.N.; Hanikenne, M. Transition metal transport. FEBS Lett. 2007, 581, 2263–2272. [Google Scholar] [CrossRef]
- Morel, M.; Crouzet, J.; Gravot, A.; Auroy, P.; Leonhardt, N.; Vavasseur, A.; Richaud, P. AtHMA3, a P1B-ATPase allowing Cd/Zn/co/Pb vacuolar storage in Arabidopsis. Plant Physiol. 2009, 149, 894–904. [Google Scholar] [CrossRef] [PubMed]
- Miyadate, H.; Adachi, S.; Hiraizumi, A.; Tezuka, K.; Nakazawa, N.; Kawamoto, T.; Katou, K.; Kodama, I.; Sakurai, K.; Takahashi, H. OsHMA3, a P1B-type of ATPase affects root-to-shoot cadmium translocation in rice by mediating efflux into vacuoles. New Phytol. 2011, 189, 190–199. [Google Scholar] [CrossRef]
- Cong, W.; Miao, Y.; Xu, L.; Zhang, Y.; Yuan, C.; Wang, J.; Zhuang, T.; Lin, X.; Jiang, L.; Wang, N. Transgenerational memory of gene expression changes induced by heavy metal stress in rice (Oryza sativa L.). BMC Plant Biol. 2019, 19, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Fu, S.; Lu, Y.; Zhang, X.; Yang, G.; Chao, D.; Wang, Z.; Shi, M.; Chen, J.; Chao, D.-Y.; Li, R. The ABC transporter ABCG36 is required for cadmium tolerance in rice. J. Exp. Bot. 2019, 70, 5909–5918. [Google Scholar] [CrossRef] [PubMed]
- Ram, H.; Kaur, A.; Gandass, N.; Singh, S.; Deshmukh, R.; Sonah, H.; Sharma, T.R. Molecular characterization and expression dynamics of MTP genes under various spatio-temporal stages and metal stress conditions in rice. PLoS ONE 2019, 14, e0217360. [Google Scholar] [CrossRef] [PubMed]
- Guo, M.; Rupe, M.A.; Dieter, J.A.; Zou, J.; Spielbauer, D.; Duncan, K.E.; Howard, R.J.; Hou, Z.; Simmons, C.R. Cell Number Regulator1 affects plant and organ size in maize: Implications for crop yield enhancement and heterosis. Plant Cell 2010, 22, 1057–1073. [Google Scholar] [CrossRef]
- Qiao, K.; Wang, F.; Liang, S.; Wang, H.; Hu, Z.; Chai, T. Improved Cd, Zn and Mn tolerance and reduced Cd accumulation in grains with wheat-based cell number regulator TaCNR2. Sci. Rep. 2019, 9, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Balk, J.; Pilon, M. Ancient and essential: The assembly of iron–sulfur clusters in plants. Trends Plant Sci. 2011, 16, 218–226. [Google Scholar] [CrossRef] [PubMed]
- Kar, S.; Panda, S.K. Iron homeostasis in rice: Deficit and excess. Proc. Natl. Acad. Sci. India Sect. B Biol. Sci. 2020, 90, 227–235. [Google Scholar] [CrossRef]
- Takahashi, R.; Ishimaru, Y.; Shimo, H.; Ogo, Y.; Senoura, T.; Nishizawa, N.K.; Nakanishi, H. The OsHMA2 transporter is involved in root-to-shoot translocation of Zn and Cd in rice. Plant Cell Environ. 2012, 35, 1948–1957. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Cheng, D.; Chen, Z.; Zhang, M.; Zhang, G.; Jiang, M.; Tan, M. Bioinformatic exploration of the targets of xylem sap miRNAs in maize under cadmium stress. Int. J. Mol. Sci. 2019, 20, 1474. [Google Scholar] [CrossRef]
- Kumar, K.; Mandal, S.N.; Neelam, K.; Reyes, B.G.D.L. MicroRNA-mediated host defense mechanisms against pathogens and herbivores in rice: Balancing gains from genetic resistance with trade-offs to productivity potential. BMC Plant Biol. 2022, 22, 351. [Google Scholar] [CrossRef]
- Sasi, J.M.; Vijaya, K.C.; Kukreja, B.; Budhwar, R.; Shukla, R.N.; Agarwal, M.; Katiyar, A.S. Integrated transcriptomics and miRNAomics provide insights into the complex multi-tiered regulatory networks associated with coleoptile senescence in rice. Front. Plant Sci. 2022, 13, 985402. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Gong, S.; Wang, Y.; Wang, F.; Bao, H.; Sun, J.; Cai, C.; Yi, K.; Chen, Z.; Zhu, C. MicroRNA166 modulates cadmium tolerance and accumulation in rice. Plant Physiol. 2018, 177, 1691–1703. [Google Scholar] [CrossRef] [PubMed]
- Meng, J.G.; Zhang, X.D.; Tan, S.K.; Zhao, K.X.; Yang, Z.M. Genome-wide identification of Cd-responsive NRAMP transporter genes and analyzing expression of NRAMP1 mediated by miR167 in Brassica napus. Biometals 2017, 30, 917–931. [Google Scholar] [CrossRef] [PubMed]
- Wei, L.; Zhang, J.; Wang, C.; Liao, W. Recent progress in the knowledge on the alleviating effect of nitric oxide on heavy metal stress in plants. Plant Physiol. Biochem. 2020, 147, 161–171. [Google Scholar] [CrossRef] [PubMed]
- Anwar, A.; Kim, J.-K. Transgenic breeding approaches for improving abiotic stress tolerance: Recent progress and future perspectives. Int. J. Mol. Sci. 2020, 21, 2695. [Google Scholar] [CrossRef]
- Jahan, N.; Javed, M.A.; Anwar, K.; Tabassum, B.; Parveen, S.; Muzaffar, N. Multiple interval mapping of QTLs and epistasis for iron toxicity tolerance in segregating population of Indica rice. Not. Bot. Horti Agrobot. Cluj-Napoca 2022, 50, 12773. [Google Scholar] [CrossRef]
- Lee, S.-B.; Kim, G.-J.; Shin, J.-D.; Chung, W.; Moon, J.-K.; Choi, G.-H.; Park, Y.-J.; Park, S.-W. Genome-scale profiling and high-throughput analyses unravel the genetic basis of arsenic content variation in rice. Front. Plant Sci. 2022, 13, 2333. [Google Scholar] [CrossRef] [PubMed]
- Adeva, C.; Yun, Y.-T.; Shim, K.-C.; Luong, N.H.; Lee, H.-S.; Kang, J.-W.; Kim, H.-J.; Ahn, S.-N. QTL Mapping of Mineral Element Contents in Rice Using Introgression Lines Derived from an Interspecific Cross. Agronomy 2023, 13, 76. [Google Scholar] [CrossRef]
- Ginting, E.E.; Silalahi, J.; Putra, E.D.L. Analysis of Arsenic in Rice in Medan, North Sumatera Indonesia by Atomic Absorption Spectrophotometer. Orient. J. Chem. 2018, 34, 2651. [Google Scholar] [CrossRef]
- Change, F.C. Unpacking the Burden on Food Safety; FAO—Food and Agriculture Organization of the United Nations: Rome, Italy, 2020. [Google Scholar]
- Jansen, S.; Dera, R.T.S.; MargarethRuth, M.; Nanda, S.I.; Yosy, C.S. Analysis of arsenic in raw and cooked rice by atomic absorption spectrophotometer. In Proceedings of the IOP Conference Series: Earth and Environmental Science, Medan, Indonesia, 19–21 September 2018; p. 012040. [Google Scholar]
- Murugaiyan, V.; Ali, J.; Mahender, A.; Aslam, U.M.; Jewel, Z.A.; Pang, Y.; Marfori-Nazarea, C.M.; Wu, L.-B.; Frei, M.; Li, Z. Mapping of genomic regions associated with arsenic toxicity stress in a backcross breeding populations of rice (Oryza sativa L.). Rice 2019, 12, 1–14. [Google Scholar] [CrossRef]
- Tyagi, W.; Yumnam, J.S.; Sen, D.; Rai, M. Root transcriptome reveals efficient cell signaling and energy conservation key to aluminum toxicity tolerance in acidic soil adapted rice genotype. Sci. Rep. 2020, 10, 4580. [Google Scholar] [CrossRef] [PubMed]
- Stein, R.J.; Duarte, G.L.; Scheunemann, L.; Spohr, M.G.; de Araújo Júnior, A.T.; Ricachenevsky, F.K.; Rosa, L.M.G.; Zanchin, N.I.T.; dos Santos, R.P.; Fett, J.P. Genotype variation in rice (Oryza sativa L.) tolerance to Fe toxicity might be linked to root cell wall lignification. Front. Plant Sci. 2019, 10, 746. [Google Scholar] [CrossRef] [PubMed]
- Shilin, D.; Chaolei, L.; Lianguang, S.; Shenglong, Y.; Anpeng, Z.; Hongzhen, J.; Banpu, R.; Guonan, F.; Biao, T.; Guoyou, Y. Identification of QTLs for cadmium tolerance during seedling stage and validation of qCDSL1 in rice. Rice Sci. 2021, 28, 81–88. [Google Scholar] [CrossRef]
- Maghrebi, M.; Baldoni, E.; Lucchini, G.; Vigani, G.; Valè, G.; Sacchi, G.A.; Nocito, F.F. Analysis of cadmium root retention for two contrasting rice accessions suggests an important role for OsHMA2. Plants 2021, 10, 806. [Google Scholar] [CrossRef]
- Pan, X.; Li, Y.; Liu, W.; Liu, S.; Min, J.; Xiong, H.; Dong, Z.; Duan, Y.; Yu, Y.; Li, X. QTL mapping and candidate gene analysis of cadmium accumulation in polished rice by genome-wide association study. Sci. Rep. 2020, 10, 1–11. [Google Scholar] [CrossRef]
- Bollinedi, H.; Yadav, A.K.; Vinod, K.; Gopala Krishnan, S.; Bhowmick, P.K.; Nagarajan, M.; Neeraja, C.; Ellur, R.K.; Singh, A.K. Genome-wide association study reveals novel marker-trait associations (MTAs) governing the localization of Fe and Zn in the rice grain. Front. Genet. 2020, 11, 213. [Google Scholar] [CrossRef]
- Utami, D.D.; Rosdianti, I.; Chrisnawati, L.; Subardi, S.; Nurani, S.; Suwarno, S. Identification of iron tolerant candidate loci in rice determined through genome-wide association study. Indones. J. Agric. Sci. 2020, 21, 17–29. [Google Scholar] [CrossRef]
- Liu, H.; Long, S.-X.; Pinson, S.R.; Tang, Z.; Guerinot, M.L.; Salt, D.E.; Zhao, F.-J.; Huang, X.-Y. Univariate and multivariate QTL analyses reveal covariance among mineral elements in the rice ionome. Front. Genet. 2021, 12, 638555. [Google Scholar] [CrossRef]
- Sim, J.-E.; Oh, S.-D.; Kang, K.; Shin, Y.-M.; Yun, D.-W.; Baek, S.-H.; Choi, Y.-E.; Park, S.-U.; Kim, J.-K. Metabolite Profiling to Evaluate Metabolic Changes in Genetically Modified Protopanaxadiol-Enriched Rice. Plants 2023, 12, 758. [Google Scholar] [CrossRef]
- Islam, T.; Manna, M.; Reddy, M.K. Glutathione peroxidase of Pennisetum glaucum (PgGPx) is a functional Cd2+ dependent peroxiredoxin that enhances tolerance against salinity and drought stress. PLoS ONE 2015, 10, e0143344. [Google Scholar] [CrossRef]
- Shri, M.; Dave, R.; Diwedi, S.; Shukla, D.; Kesari, R.; Tripathi, R.D.; Trivedi, P.K.; Chakrabarty, D. Heterologous expression of Ceratophyllum demersum phytochelatin synthase, CdPCS1, in rice leads to lower arsenic accumulation in grain. Sci. Rep. 2014, 4, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, A.; Mustafiz, A.; Pareek, A.; Sopory, S.K.; Singla-Pareek, S.L. Glyoxalase III enhances salinity tolerance through reactive oxygen species scavenging and reduced glycation. Physiol. Plant. 2022, 174, e13693. [Google Scholar] [CrossRef] [PubMed]
- Islam, T.; Reddy, M. Evaluation of Cd2+ stress tolerance in transgenic rice overexpressing PgGPx gene that maintains cellular ion and reactive oxygen species homeostasis. PLoS ONE 2022, 17, e0273974. [Google Scholar] [CrossRef]
- Wu, Y.; Yang, Z.; How, J.; Xu, H.; Chen, L.; Li, K. Overexpression of a peroxidase gene (AtPrx64) of Arabidopsis thaliana in tobacco improves plant’s tolerance to aluminum stress. Plant Mol. Biol. 2017, 95, 157–168. [Google Scholar] [CrossRef] [PubMed]
- Kidwai, M.; Dhar, Y.V.; Gautam, N.; Tiwari, M.; Ahmad, I.Z.; Asif, M.H.; Chakrabarty, D. Oryza sativa class III peroxidase (OsPRX38) overexpression in Arabidopsis thaliana reduces arsenic accumulation due to apoplastic lignification. J. Hazard. Mater. 2019, 362, 383–393. [Google Scholar] [CrossRef]
- Chen, Z.; Pan, Y.; Wang, S.; Ding, Y.; Yang, W.; Zhu, C. Overexpression of a protein disulfide isomerase-like protein from Methanothermobacter thermoautotrophicum enhances mercury tolerance in transgenic rice. Plant Sci. 2012, 197, 10–20. [Google Scholar] [CrossRef]
- Ding, Y.; Qu, A.; Gong, S.; Huang, S.; Lv, B.; Zhu, C. Molecular identification and analysis of Cd-responsive microRNAs in rice. J. Agric. Food Chem. 2013, 61, 11668–11675. [Google Scholar] [CrossRef]
- Tang, L.; Mao, B.; Li, Y.; Lv, Q.; Zhang, L.; Chen, C.; He, H.; Wang, W.; Zeng, X.; Shao, Y. Knockout of OsNramp5 using the CRISPR/Cas9 system produces low Cd-accumulating indica rice without compromising yield. Sci. Rep. 2017, 7, 14438. [Google Scholar] [CrossRef]
- Nieves-Cordones, M.; Mohamed, S.; Tanoi, K.; Kobayashi, N.I.; Takagi, K.; Vernet, A.; Guiderdoni, E.; Périn, C.; Sentenac, H.; Véry, A.A. Production of low-Cs+ rice plants by inactivation of the K+ transporter OsHAK1 with the CRISPR-Cas system. Plant J. 2017, 92, 43–56. [Google Scholar] [CrossRef]
- Takahashi, R.; Ishimaru, Y.; Nakanishi, H.; Nishizawa, N.K. Role of the iron transporter OsNRAMP1 in cadmium uptake and accumulation in rice. Plant Signal Behav. 2011, 6, 1813–1816. [Google Scholar] [CrossRef]
- Huang, X.-Y.; Deng, F.; Yamaji, N.; Pinson, S.R.; Fujii-Kashino, M.; Danku, J.; Douglas, A.; Guerinot, M.L.; Salt, D.E.; Ma, J.F. A heavy metal P-type ATPase OsHMA4 prevents copper accumulation in rice grain. Nat. Commun. 2016, 7, 12138. [Google Scholar] [CrossRef] [PubMed]
- Tian-Yu, G.; Zi-Ai, Q.; Si-Ying, C.; Jing, Y.; Zi-Jun, F.; Jun-Min, W.; Ji-Ming, G. Dual-function DEFENSIN8 mediates phloem cadmium unloading and accumulation in rice grains. Plant Physiol. 2023, 191, 515–527. [Google Scholar] [CrossRef]
- Mehmood, S.S.; Lu, G.; Luo, D.; Hussain, M.A.; Raza, A.; Zafar, Z.; Zhang, X.; Cheng, Y.; Zou, X.; Lv, Y. Integrated analysis of transcriptomics and proteomics provides insights into the molecular regulation of cold response in Brassica napus. Environ. Exp. Bot. 2021, 187, 104480. [Google Scholar] [CrossRef]
- Cao, F.; Dai, H.; Hao, P.-F.; Wu, F. Silicon regulates the expression of vacuolar H+-pyrophosphatase 1 and decreases cadmium accumulation in rice (Oryza sativa L.). Chemosphere 2020, 240, 124907. [Google Scholar] [CrossRef] [PubMed]
- Cao, H.W.; Zhao, Y.N.; Liu, X.S.; Rono, J.K.; Yang, Z.M. A metal chaperone OsHIPP16 detoxifies cadmium by repressing its accumulation in rice crops. Environ. Pollut. 2022, 311, 120058. [Google Scholar] [CrossRef]
- Gostimskaya, I. CRISPR-Cas9: A History of Its Discovery and Ethical Considerations of Its Use in Genome Editing. Biochemistry 2022, 87, 777–788. [Google Scholar] [CrossRef]
- Raza, A.; Su, W.; Hussain, M.A.; Mehmood, S.S.; Zhang, X.; Cheng, Y.; Zou, X.; Lv, Y. Integrated analysis of metabolome and transcriptome reveals insights for cold tolerance in rapeseed (Brassica napus L.). Front. Plant Sci. 2021, 12, 1796. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.D.; Huang, S.; Yamaji, N.; Zhang, W.; Ma, J.F.; Zhao, F.J. OsNRAMP1 transporter contributes to cadmium and manganese uptake in rice. Plant Cell Environ. 2020, 43, 2476–2491. [Google Scholar] [CrossRef]
- Songmei, L.; Jie, J.; Yang, L.; Jun, M.; Shouling, X.; Yuanyuan, T.; Youfa, L.; Qingyao, S.; Jianzhong, H. Characterization and evaluation of OsLCT1 and OsNramp5 mutants generated through CRISPR/Cas9-mediated mutagenesis for breeding low Cd rice. Rice Sci. 2019, 26, 88–97. [Google Scholar] [CrossRef]
- Tang, L.; Dong, J.; Qu, M.; Lv, Q.; Zhang, L.; Peng, C.; Hu, Y.; Li, Y.; Ji, Z.; Mao, B. Knockout of OsNRAMP5 enhances rice tolerance to cadmium toxicity in response to varying external cadmium concentrations via distinct mechanisms. Sci. Total Environ. 2022, 832, 155006. [Google Scholar] [CrossRef]
- Tang, L.; Dong, J.; Tan, L.; Ji, Z.; Li, Y.; Sun, Y.; Chen, C.; Lv, Q.; Mao, B.; Hu, Y. Overexpression of OsLCT2, a low-affinity cation transporter gene, reduces cadmium accumulation in shoots and grains of rice. Rice 2021, 14, 1–15. [Google Scholar] [CrossRef]
- Mishra, N.; Srivastava, A.P.; Esmaeili, N.; Hu, W.; Shen, G. Overexpression of the rice gene OsSIZ1 in Arabidopsis improves drought-, heat-, and salt-tolerance simultaneously. PLoS ONE 2018, 13, e0201716. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.S.; Feng, S.J.; Zhang, B.Q.; Wang, M.Q.; Cao, H.W.; Rono, J.K.; Chen, X.; Yang, Z.M. OsZIP1 functions as a metal efflux transporter limiting excess zinc, copper and cadmium accumulation in rice. BMC Plant Biol. 2019, 19, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Jeong, J.S.; Kim, Y.S.; Redillas, M.C.; Jang, G.; Jung, H.; Bang, S.W.; Choi, Y.D.; Ha, S.H.; Reuzeau, C.; Kim, J.K. OsNAC5 overexpression enlarges root diameter in rice plants leading to enhanced drought tolerance and increased grain yield in the field. Plant Biotechnol. J. 2013, 11, 101–114. [Google Scholar] [CrossRef] [PubMed]
- Kumagai, S.; Suzuki, T.; Tezuka, K.; Satoh-Nagasawa, N.; Takahashi, H.; Sakurai, K.; Watanabe, A.; Fujimura, T.; Akagi, H. Functional analysis of the C-terminal region of the vacuolar cadmium-transporting rice OsHMA3. FEBS Lett. 2014, 588, 789–794. [Google Scholar] [CrossRef]
- Shen, J.; Lv, B.; Luo, L.; He, J.; Mao, C.; Xi, D.; Ming, F. The NAC-type transcription factor OsNAC2 regulates ABA-dependent genes and abiotic stress tolerance in rice. Sci. Rep. 2017, 7, 40641. [Google Scholar] [CrossRef]
- Qi, D.-L.; Lee, J.-R.; Yang, C.-G.; Lee, M.-C.; Cao, G.-L.; Zhang, J.-G.; Zhou, Q.-Y.; Suh, S.-C.; Zhang, S.-Y.; Han, L.-Z. Detection of QTL for alkali tolerance at the germination stage in japonica rice. Chin. J. OF Rice Sci. 2009, 23, 589. [Google Scholar]
- Chen, J.; Zou, W.; Meng, L.; Fan, X.; Xu, G.; Ye, G. Advances in the uptake and transport mechanisms and QTLs mapping of cadmium in rice. Int. J. Mol. Sci. 2019, 20, 3417. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; Luo, X.; Wang, L.; Wei, Y.; Li, J.; Xie, H.; Zhang, J.; Xie, G. Genome-wide association study reveals the QTLs for seed storability in world rice core collections. Plants 2021, 10, 812. [Google Scholar] [CrossRef]
- Yamazaki, S.; Ueda, Y.; Mukai, A.; Ochiai, K.; Matoh, T. Rice phytochelatin synthases OsPCS1 and OsPCS2 make different contributions to cadmium and arsenic tolerance. Plant Direct 2018, 2, e00034. [Google Scholar] [CrossRef]
- Lu, C.; Zhang, L.; Tang, Z.; Huang, X.-Y.; Ma, J.F.; Zhao, F.-J. Producing cadmium-free Indica rice by overexpressing OsHMA3. Environ. Int. 2019, 126, 619–626. [Google Scholar] [CrossRef]
- Arya, G.C.; Sarkar, S.; Manasherova, E.; Aharoni, A.; Cohen, H. The Plant Cuticle: An Ancient Guardian Barrier Set Against Long-Standing Rivals. Front. Plant Sci. 2021, 12, 663165. [Google Scholar] [CrossRef] [PubMed]
- Rono, J.K.; Le Wang, L.; Wu, X.C.; Cao, H.W.; Zhao, Y.N.; Khan, I.U.; Yang, Z.M. Identification of a new function of metallothionein-like gene OsMT1e for cadmium detoxification and potential phytoremediation. Chemosphere 2021, 265, 129136. [Google Scholar] [CrossRef]
- Zhao, D.-D.; Park, J.-R.; Jang, Y.-H.; Kim, E.-G.; Du, X.-X.; Farooq, M.; Yun, B.-J.; Kim, K.-M. Identification of one major QTL and a novel gene OsIAA17q5 associated with tiller number in rice using QTl analysis. Plants 2022, 11, 538. [Google Scholar] [CrossRef]
- Ogo, Y.; Itai, R.N.; Kobayashi, T.; Aung, M.S.; Nakanishi, H.; Nishizawa, N.K. OsIRO2 is responsible for iron utilization in rice and improves growth and yield in calcareous soil. Plant Mol. Biol. 2011, 75, 593–605. [Google Scholar] [CrossRef] [PubMed]
- Yamaji, N.; Huang, C.F.; Nagao, S.; Yano, M.; Sato, Y.; Nagamura, Y.; Ma, J.F. A zinc finger transcription factor ART1 regulates multiple genes implicated in aluminum tolerance in rice. Plant Cell 2009, 21, 3339–3349. [Google Scholar] [CrossRef]
- Sasaki, A.; Yamaji, N.; Ma, J.F. Overexpression of OsHMA3 enhances Cd tolerance and expression of Zn transporter genes in rice. J. Exp. Bot. 2014, 65, 6013–6021. [Google Scholar] [CrossRef] [PubMed]
- Tao, J.; Lu, L. Advances in genes-encoding transporters for cadmium uptake, translocation, and accumulation in plants. Toxics 2022, 10, 411. [Google Scholar] [CrossRef]
- Li, J.-C.; Guo, J.-B.; Xu, W.-Z.; Ma, M. RNA Interference-mediated Silencing of Phytochelatin Synthase Gene Reduce Cadmium Accumulation in Rice Seeds. J. Integr. Plant Biol. 2007, 49, 1032–1037. [Google Scholar] [CrossRef]
- Zhao, J.; Yang, W.; Zhang, S.; Yang, T.; Liu, Q.; Dong, J.; Fu, H.; Mao, X.; Liu, B. Genome-wide association study and candidate gene analysis of rice cadmium accumulation in grain in a diverse rice collection. Rice 2018, 11, 1–15. [Google Scholar] [CrossRef]
- Vinocur, B.; Altman, A. Recent advances in engineering plant tolerance to abiotic stress: Achievements and limitations. Curr. Opin. Biotechnol. 2005, 16, 123–132. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Parihar, P.; Singh, R.; Singh, V.P.; Prasad, S.M. Heavy Metal Tolerance in Plants: Role of Transcriptomics, Proteomics, Metabolomics, and Ionomics. Front Plant Sci. 2016, 6, 1143. [Google Scholar] [CrossRef] [PubMed]
- Shephard, A.M.; Zambre, A.M.; Snell-Rood, E.C. Evaluating costs of heavy metal tolerance in a widely distributed, invasive butterfly. Evol. Appl. 2021, 14, 1390–1402. [Google Scholar] [CrossRef] [PubMed]
- Lam, P.K.S. Interpopulation differences in acute response of Brotia hainanensis (Gastropoda, Prosobranchia) to cadmium: Genetic or environmental variance? Environ. Pollut. 1996, 94, 1–7. [Google Scholar] [CrossRef]
- Sahrawat, K.Á. Iron toxicity in wetland rice and the role of other nutrients. J. Plant Nutr. 2005, 27, 1471–1504. [Google Scholar] [CrossRef]

| Metalloid | Targeted Gene(s) | Molecular Functions | Role in HM Toxicity Tolerance | References |
|---|---|---|---|---|
| As, Si | OsLsi | Silicon (Si) transporter | Low arsenic uptake | [33] |
| Cd | OsABCG36 | G-type ATP-binding cassette (ABC) transporter | Cadmium sequestration and toxicity tolerance | [55] |
| Cs | OsHAK1 | High-Affinity Potassium (K+) Transporter | Low cesium accumulation in root and shoots | [94] |
| As, Cd | OsNRAMP1 | Cadmium, Iron, and manganese uptake/transporter | Low arsenic and cadmium content in grains | [103] |
| Cd, Pb, Mn, Fe | OsNRAMP5 | Major transporter for metal uptake | Low cadmium content in grains | [104] |
| Cd | OsLCT1 | Low-Affinity Cation Transporter | Low cadmium uptake | [104] |
| Cd | OsLCT2 | Low-Affinity Cation Transporter | Low cadmium accumulation in grains | [105] |
| SR | Genes/QTL | Metalloids | Mechanism Involved | Ref |
|---|---|---|---|---|
| 1 | OsNRAMP5 | Fe and Mn, as well as Cd and Zn | Overexpression of OsNRAMP5 to increase tolerance to Cd toxicity | [93] |
| 2 | OsHMA3 | Cd | Overexpression of OsHMA3 to increase tolerance to Cd toxicity | [116] |
| 3 | OsABCG31 | Cd and Pb | Overexpression of OsABCG31 enhanced their tolerance to Cd and Pd toxicity | [117] |
| 4 | OsLCT1 | Al | Overexpression of OsLCT1 to increase tolerance to Al toxicity | [106] |
| 5 | OsSIZ | Cd | Overexpression of OsSIZ1 enhanced tolerance to Cd toxicity. | [107] |
| 6 | OsZIP1 | Zn | Overexpression of OsZIP1 can enhance tolerance to Zn toxicity | [108] |
| 7 | OsNAC5 | Cd and Pb | Overexpression of OsNAC5 enhanced their tolerance to Cd and Pb toxicity | [109] |
| 8 | OsMT1e | Cd and Zn | OsMT1e encodes a metallothionein protein involved in metal detoxification | [118] |
| 9 | OsNAC2 | Cd | OsNAC2 is a transcription factor that is involved in regulating the expression of genes involved in stress responses in rice | [111] |
| 10 | qNRG2-1 | Ni | qNRG2-1, a QTL on chromosome 2 is associated with Ni tolerance in rice associated with the expression of the OsNramp5 gene | [119] |
| 11 | qCdt1 | Cd | qCdt1-2, a QTL on chromosome 1 is associated with Cd tolerance with overexpression of OsHMA3 | [110] |
| 12 | RM219 | As | accumulation of As | [112] |
| 13 | qPC1 | Cd | qPC1 QTL was associated with lower Cd accumulation in rice grains | [113] |
| 14 | OsIRO2 | Cd | OsIRO2 is a transcription factor regulating the expression of genes involved in Fe homeostasis in rice | [120] |
| 15 | qGCD7 | Cd | This QTL has been found to be important for rice tolerance to Cd toxicity | [121] |
| 16 | qHLR1 | Cd and Pb | This QTL has been found to be important for rice tolerance to both Cd and Pb toxicity | [122] |
| 17 | qCST11 | Pb | This QTL is associated with Cd and Pb tolerance in rice identified through a GWAS | [80] |
| 18 | OsIRT1 | Fe | This gene encodes an Fe transporter involved in Cd uptake in rice. It was found to be upregulated under Cd stress | [123] |
| 19 | qMRS6.1 | Mn | This QTL is associated with manganese (Mn) tolerance in rice. It was identified through a GWAS | [114] |
| 20 | OsPCS1 | Cd | This gene encodes phytochelatin synthase involved in the synthesis of phytochelatins, a group of peptides that bind to HMs and detoxify them in plants. It was found to be upregulated under Cd stress in rice. | [115] |
| 21 | qHTSF4.1 | As | The QTL is associated with Cd and As tolerance in rice | [116] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Haider, Z.; Ahmad, I.; Zia, S.; Gan, Y. Recent Developments in Rice Molecular Breeding for Tolerance to Heavy Metal Toxicity. Agriculture 2023, 13, 944. https://doi.org/10.3390/agriculture13050944
Haider Z, Ahmad I, Zia S, Gan Y. Recent Developments in Rice Molecular Breeding for Tolerance to Heavy Metal Toxicity. Agriculture. 2023; 13(5):944. https://doi.org/10.3390/agriculture13050944
Chicago/Turabian StyleHaider, Zulqarnain, Irshan Ahmad, Samta Zia, and Yinbo Gan. 2023. "Recent Developments in Rice Molecular Breeding for Tolerance to Heavy Metal Toxicity" Agriculture 13, no. 5: 944. https://doi.org/10.3390/agriculture13050944







