Use of Leaf and Fruit Morphometric Analysis to Identify and Classify White Mulberry (Morus alba L.) Genotypes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Leaf Morphometric Determinations
2.2. Fruit Morphometric Determinations
2.3. Data Analysis
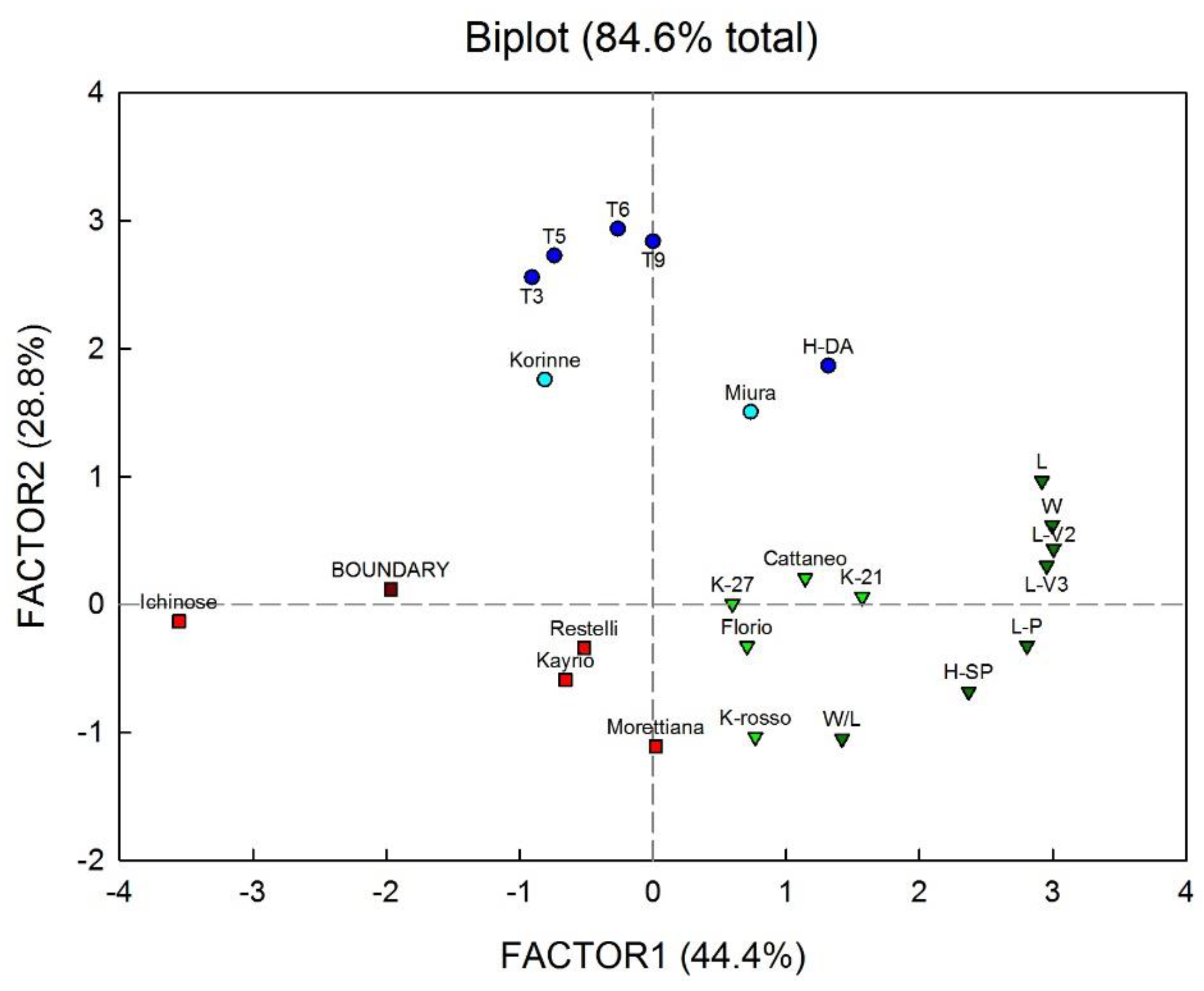
3. Results and Discussion
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Datta, R.K. Mulberry cultivation and utilization in India. In Mulberry for Animal Production; Food and Agriculture Organization of the United Nations: Rome, Italy, 2002; ISBN 92-5-104568-2. [Google Scholar]
- Dandin, S.B.; Rajan, M.V. Microsporogenesis in Hexaploid Morus serrata Roxb. Cytologia 1989, 54, 747–751. [Google Scholar]
- Khurana, P.; Checker, V.G. The advent of genomics in mulberry and perspectives for productivity enhancement. Plant Cell Rep. 2011, 30, 825–838. [Google Scholar] [CrossRef] [PubMed]
- Vijayan, K. The emerging role of genomic tools in mulberry (Morus) genetic improvement. Tree Genet. Genomes 2010, 6, 613–625. [Google Scholar] [CrossRef]
- Cappellozza, L.; Coradazzi, A.T.; Tornadore, N. Studies on the phenotypic variability of seven cultivars of Morus alba L. and three of Morus multicaulis P. (Moracee). Part 1. Sericologia 1995, 35, 257–270. [Google Scholar]
- Cappellozza, L.; Coradazzi, A.T.; Cappellozza, S.; Baldan, B.; Mariani, P. Studies on the phenotypic variability of seven cultivars of Morus alba L. and three of Morus multicaulis P. (Moracee). Part 2. Sericologia 1996, 36, 91–102. [Google Scholar]
- Sinacori, A.; Reale, S.; Botindari, M.; Alfonzetti, T.; Vitale, F. Nota preliminare sulla determinazione di cultivar di Morus alba L. mediante PCR e RAPD. Italus Hortus 2006, 26, 327–330. [Google Scholar]
- Srivastava, P.P.; Vijayan, K.; Awasthi, A.K.; Saratchandra, B. Genetic analysis of Morus alba through RAPD and ISSR markers. Indian J. Biotech. 2004, 3, 527–532. [Google Scholar]
- Thomas, D. Embryological observations on unpollinated ovary culture of mulberry (Morus alba L.). Acta Biol. Crac. Ser. Bot. 2004, 46, 87–94. [Google Scholar]
- Pavek, D.S.; Lamboy, W.F.; Garvey, E.J. Selecting in situ conservation sites for grape genetic resources in the USA. Genet. Resour. Crop. Evol. 2003, 50, 165–173. [Google Scholar] [CrossRef]
- Garcia-Muñoz, S.; Muñoz-Organero, G.; De Andrés, M.T.; Cabello, F. Ampelography—An old technique with future uses: The case of minor varieties of Vitis vinifera L. from the Balearic Islands. J. Int. Sci. Vigne Vin 2011, 45, 125–137. [Google Scholar] [CrossRef]
- Chomé, P.; Sotes, V.; Benayas, F.; Cayuela, M.; Hernandez, M.; Cabello, F.; Ortiz, J.; Rodriguez-Torres, I.; Chaves, J. Variedades De Vid: Registro De Variedades Comerciales; MAPA: Madrid, Spain, 2003. [Google Scholar]
- UPOV (International Union for the Protection of New Varieties of Plants). Available online: http://www.upov.int/edocs/mdocs/upov/en/tc/44/tg_50_9_proj_3.pdf (accessed on 3 October 2018).
- IPGRI, UPOV, OIV. Descriptors for Grapevine (Vitis spp.); International Union for the Protection of New Varieties of Plants: Geneva, Switzerland; Office International de la Vigne et du Vin: Paris, France; International Plant Genetic Resources Institute: Rome, Italy, 1997; ISBN 92-9043-352-3.
- Barone, E.; Di Marco, L.; Motisi, A.; Caruso, T. The sicilian olive germplasm and its characterization by using statistical methods. Acta Hort. 1994, 356, 66–69. [Google Scholar] [CrossRef]
- Botta, R.; Schneider, A.; Akkak, A.; Scott, N.S.; Thamas, M.R. Within cultivar grapevine variability studied by morphometrical and molecular marker based techniques. Acta Hort. 2000, 528, 91–96. [Google Scholar] [CrossRef]
- Carroll, J.D. Individual differences and multidimensional scaling. In Multidimensional Scaling: Theory and Applications in the Behavioral Sciences; Shepard, R.N., Romney, A.K., Nerlove, S.B., Eds.; Seminar Press: New York, NY, USA, 1972; Volume 1, pp. 105–155. [Google Scholar]
- Symons, S.J.; Fulcher, R.G. Determination of wheat kernel morphological variation by digital image analysis: II. Variation in cultivars of soft white winter wheats. J. Cereal Sci. 1988, 8, 219–229. [Google Scholar] [CrossRef]
- Camelo-Méndez, G.A.; Camacho-Díaz, B.H.; del Villar-Martínez, A.A.; Arenas-Ocampo, M.L.; Bello-Pérez, L.A.; Jiménez-Aparicio, A.R. Digital image analysis of diverse Mexican rice cultivars. J. Sci. Food Agric. 2012, 92, 2709–2714. [Google Scholar] [CrossRef] [PubMed]
- Asanidze, Z.; Akhalkatsi, M.; Gvritishvili, M. Comparative morphometric study and relationships between the Caucasian species of wild pear (Pyrus spp.) and local cultivars in Georgia. Flora-Morphol. Distrib. Funct. Ecol. Plants 2011, 206, 974–986. [Google Scholar] [CrossRef]
- Menesatti, P.; Costa, C.; Paglia, G.; Pallottino, F.; D’Andrea, S.; Rimatori, V.; Aguzzi, J. Shape-based methodology for multivariate discrimination among Italian hazelnut cultivars. Biosyst. Eng. 2008, 101, 417–424. [Google Scholar] [CrossRef]
- Saddoud, O.; Baraket, G.; Chatti, K.; Trifi, M.; Marrakchi, M.; Salhi-Hannachi, A.; Mars, M. Morphological variability of fig (Ficus carica L.) cultivars. Int. J. Fruit Sci. 2008, 8, 35–51. [Google Scholar] [CrossRef]
- Lo Bianco, R.; Panno, G.; Avellone, G. Characterization of Sicilian olive genotypes by multivariate analysis of leaf and fruit chemical and morphological properties. J. Agric. Sci. 2013, 5, 229–245. [Google Scholar] [CrossRef]
- Beyer, M.; Hahn, R.; Peschel, S.; Harz, M.; Knoche, M. Analysing fruit shape in sweet cherry (Prunus avium L.). Sci. Hortic. 2002, 96, 139–150. [Google Scholar] [CrossRef]
- Piagnani, M.C.; Debellini, C.; LoScalzo, R. Phyllometry and carpometry, chemical and functional characterization of fruits of Sorbus domestica L. (service tree) selections. J. Berry Res. 2012, 2, 7–22. [Google Scholar]
- Lo Bianco, R.; Farina, V.; Indelicato, S.G.; Filizzola, F.; Agozzino, P. Fruit physical, chemical and aromatic attributes of early, intermediate and late apricot cultivars. J. Sci. Food Agric. 2010, 90, 1008–1019. [Google Scholar] [CrossRef] [PubMed]
- Lo Bianco, R.; Farina, V. Fruit quality of Sicilian, commercially affirmed, and newly selected apple varieties: Potential resources for genetic improvement and specialty markets. In Apples: Nutrition, Consumption and Health; Cordoba, S.J., Delgado, F.A., Eds.; Nova Science Publishers: New York, NY, USA, 2012; pp. 193–203. ISBN 978-1-61942-709-9. [Google Scholar]
- Cope, J.S.; Corney, D.; Clark, J.Y.; Remagnino, P.; Wilkin, P. Plant species identification using digital morphometrics: A. review. Expert Syst. Appl. 2012, 39, 7562–7573. [Google Scholar] [CrossRef]
- Vieira, R.G.; Prestes, R.A.; Denardi, F.; Nogueira, A.; Wosiacki, G. Chemical pattern of Brazilian apples: A chemometric approach based on the Fuji and Gala varieties. Food Sci. Technol. 2011, 31, 418–426. [Google Scholar] [CrossRef]
- Garcia, M.G.; Ontivero, M.; Diaz Ricci, J.C.; Castagnaro, A. Morphological traits and high resolution RAPD markers for the identification of the main strawberry varieties cultivated in Argentina. Plant Breed. 2002, 121, 76–80. [Google Scholar] [CrossRef]
- Bishop, C.M. Pattern Recognition and Machine Learning, 1st ed.; Springer-Verlag: New York, NY, USA, 2006; ISBN 978-1-4939-3843-8. [Google Scholar]
- Saini, P.; Chauhan, S.S.; Shabnam, A.A.; Chand, L.; Negi, N. Genetic variability and trait association analysis for agro-morphological markers in mulberry genetic resources from Kashmir, India. Int. J. Curr. Microbiol. App. Sci. 2018, 7, 1799–1812. [Google Scholar] [CrossRef]

| Leaf Trait | Abbreviation | |
|---|---|---|
| Blade length | L |  |
| Blade width | W | |
| Petiole length | L-P | |
| Petiole sinus depth | H-SP | |
| Length of main vein | L-V1 | |
| Length of vein 2 (left) | L-V2 | |
| Length of vein 3 | L-V3 | |
| Length of vein 4 | L-V4 | |
| Length of vein 5 | L-V5 | |
| Length of vein 6 | L-V6 | |
| Length of vein 7 | L-V7 | |
| Length of vein 8 | L-V8 | |
| Length of vein 9 | L-V9 | |
| Length of vein 10 | L-V10 | |
| Height of apical tooth | H-DA | |
| Basal width of apical tooth | W-DA | |
| Petiole sinus angle (concave up) | T1 |  |
| Inner angle V2 (left) | T2 | |
| Inner angle V3 | T3 | |
| Inner angle V4 | T4 | |
| Inner angle V5 | T5 | |
| Inner angle V6 | T6 | |
| Inner angle V7 | T7 | |
| Inner angle V8 | T8 | |
| Inner angle V9 | T9 | |
| Inner angle V10 | T10 | |
| Apical tooth angle | T-DA | |
| Petiole vein angle (3rd order, left) | Z1 | |
| Petiole vein angle (3rd order, right) | Z2 | |
| Ratio between blade width and length | W/L | |
| Ratio between apical tooth basal width and height | W-DA/H-DA | |
| Margin index (serrated = 0, smooth = 1) | BOUNDARY |
| Leaf Descriptors | Cattaneo | Florio | Ichinose | Kayrio | Kokusò-21 | Kokusò-27 | Kokusò-R | Korinne | Miura | Morettiana | Restelli | C.V. |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| L | 18.0 ± 0.64 | 15.4 ± 0.57 | 8.10 ± 0.38 | 14.4 ± 0.39 | 19.4 ± 0.77 | 16.9 ± 0.69 | 16.7 ± 0.95 | 15.6 ± 0.37 | 19.0 ± 0.69 | 13.8 ± 0.54 | 15.5 ± 0.54 | 19.7 |
| W | 11.0 ± 0.32 | 11.8 ± 0.43 | 4.81 ± 0.19 | 9.40 ± 0.19 | 13.4 ± 0.56 | 11.6 ± 0.51 | 10.8 ± 0.36 | 10.2 ± 0.29 | 12.6 ± 0.77 | 10.3 ± 0.50 | 10.3 ± 0.35 | 21.1 |
| L-P | 4.87 ± 0.25 | 4.57 ± 0.18 | 1.82 ± 0.11 | 3.37 ± 0.10 | 4.35 ± 0.15 | 3.13 ± 0.23 | 4.41 ± 0.32 | 3.10 ± 0.08 | 4.17 ± 0.21 | 4.07 ± 0.15 | 3.06 ± 0.10 | 24.3 |
| H-SP | 1.64 ± 0.10 | 1.33 ± 0.09 | 0.35 ± 0.04 | 0.77 ± 0.06 | 1.46 ± 0.09 | 1.07 ± 0.05 | 0.97 ± 0.06 | 0.46 ± 0.04 | 0.59 ± 0.10 | 0.82 ± 0.09 | 0.43 ± 0.07 | 48.8 |
| L-V1 | 16.7 ± 0.67 | 14.3 ± 0.52 | 7.73 ± 0.36 | 13.6 ± 0.40 | 17.6 ± 0.78 | 15.9 ± 0.69 | 15.9 ± 0.95 | 15.2 ± 0.37 | 18.1 ± 0.60 | 13.1 ± 0.49 | 15.3 ± 0.53 | 19.0 |
| L-V2 | 10.8 ± 0.34 | 9.04 ± 0.36 | 4.41 ± 0.32 | 7.61 ± 0.21 | 10.3 ± 0.52 | 9.17 ± 0.44 | 9.49 ± 0.44 | 7.91 ± 0.23 | 10.1 ± 0.52 | 7.96 ± 0.38 | 9.12 ± 0.30 | 20.2 |
| L-V3 | 10.4 ± 0.45 | 8.67 ± 0.29 | 4.14 ± 0.28 | 7.78 ± 0.25 | 10.2 ± 0.44 | 8.88 ± 0.43 | 9.38 ± 0.42 | 8.18 ± 0.28 | 9.21 ± 0.47 | 7.87 ± 0.31 | 9.30 ± 0.37 | 19.8 |
| L-V4 | 8.24 ± 0.19 | 6.93 ± 0.26 | 3.22 ± 0.25 | 6.97 ± 0.16 | 8.55 ± 0.36 | 6.21 ± 0.37 | 7.08 ± 0.31 | 6.89 ± 0.36 | 8.04 ± 0.57 | 6.05 ± 0.33 | 7.00 ± 0.23 | 21.0 |
| L-V5 | 7.34 ± 0.29 | 6.59 ± 0.28 | 3.22 ± 0.22 | 7.02 ± 0.17 | 8.58 ± 0.36 | 6.84 ± 0.37 | 6.68 ± 0.31 | 6.44 ± 0.25 | 7.66 ± 0.41 | 6.14 ± 0.28 | 6.73 ± 0.20 | 19.9 |
| L-V6 | 6.10 ± 0.18 | 5.66 ± 0.22 | 2.50 ± 0.24 | 5.52 ± 0.18 | 7.53 ± 0.37 | 6.19 ± 0.31 | 5.75 ± 0.44 | 5.58 ± 0.28 | 6.85 ± 0.38 | 5.23 ± 0.22 | 5.61 ± 0.22 | 22.0 |
| L-V7 | 5.17 ± 0.23 | 5.07 ± 0.23 | 2.01 ± 0.29 | 4.84 ± 0.23 | 6.46 ± 0.33 | 5.57 ± 0.32 | 4.96 ± 0.31 | 5.47 ± 0.28 | 6.36 ± 0.23 | 4.63 ± 0.20 | 4.95 ± 0.21 | 23.2 |
| L-V8 | 3.96 ± 0.17 | 4.16 ± 0.17 | 1.88 ± 0.20 | 3.57 ± 0.19 | 5.26 ± 0.28 | 4.54 ± 0.29 | 3.92 ± 0.34 | 4.59 ± 0.23 | 5.11 ± 0.25 | 3.98 ± 0.15 | 4.02 ± 0.25 | 21.9 |
| L-V9 | 3.21 ± 0.21 | 3.31 ± 0.18 | 1.48 ± 0.20 | 2.87 ± 0.23 | 4.25 ± 0.23 | 3.45 ± 0.23 | 3.00 ± 0.21 | 4.39 ± 0.18 | 4.63 ± 0.25 | 3.19 ± 0.24 | 3.13 ± 0.19 | 25.8 |
| L-V10 | 2.28 ± 0.15 | 2.56 ± 0.17 | 0.95 ± 0.18 | 2.10 ± 0.11 | 3.39 ± 0.19 | 2.82 ± 0.21 | 2.36 ± 0.28 | 3.48 ± 0.20 | 3.60 ± 0.22 | 2.41 ± 0.15 | 2.21 ± 0.17 | 29.6 |
| H-DA | 1.24 ± 0.12 | 0.80 ± 0.05 | 0.74 ± 0.06 | 1.17 ± 0.09 | 1.24 ± 0.08 | 1.12 ± 0.05 | 1.46 ± 0.11 | 1.61 ± 0.09 | 1.65 ± 0.12 | 1.04 ± 0.05 | 0.92 ± 0.06 | 25.7 |
| W-DA | 0.92 ± 0.06 | 0.64 ± 0.04 | 0.44 ± 0.05 | 0.67 ± 0.04 | 0.75 ± 0.04 | 0.59 ± 0.03 | 0.76 ± 0.02 | 0.82 ± 0.04 | 0.88 ± 0.05 | 0.67 ± 0.03 | 0.62 ± 0.04 | 19.7 |
| T1 | 117 ± 5.19 | 115 ± 3.65 | 150 ± 7.65 | 139 ± 3.97 | 115 ± 3.06 | 121 ± 3.09 | 125 ± 2.52 | 158 ± 1.67 | 97 ± 10.7 | 134 ± 4.27 | 135 ± 12.5 | 13.7 |
| T2 | 40.1 ± 1.24 | 37.3 ± 1.30 | 41.5 ± 3.59 | 43.1 ± 1.61 | 48.7 ± 2.36 | 41.3 ± 1.94 | 36.3 ± 1.80 | 55.7 ± 2.02 | 46.8 ± 1.06 | 32.9 ± 1.29 | 45.0 ± 1.69 | 14.9 |
| T3 | 42.6 ± 1.47 | 34.5 ± 1.35 | 44.3 ± 2.55 | 42.9 ± 1.70 | 41.8 ± 1.24 | 42.3 ± 1.82 | 35.5 ± 1.69 | 52.4 ± 1.95 | 47.5 ± 1.45 | 30.8 ± 1.44 | 46.4 ± 1.82 | 14.9 |
| T4 | 40.7 ± 1.81 | 44.7 ± 1.24 | 44.0 ± 1.64 | 37.3 ± 1.26 | 44.9 ± 1.39 | 44.2 ± 1.15 | 32.0 ± 1.67 | 52.4 ± 1.88 | 50.1 ± 1.09 | 38.8 ± 1.62 | 38.2 ± 0.96 | 13.8 |
| T5 | 42.7 ± 1.75 | 44.6 ± 1.31 | 45.5 ± 1.60 | 35.6 ± 0.97 | 39.5 ± 1.26 | 40.5 ± 1.36 | 30.4 ± 1.26 | 53.3 ± 1.24 | 49.5 ± 1.06 | 38.9 ± 1.09 | 38.2 ± 1.27 | 15.4 |
| T6 | 40.4 ± 0.75 | 42.8 ± 1.09 | 41.6 ± 2.27 | 37.1 ± 1.24 | 42.3 ± 1.57 | 39.1 ± 1.20 | 31.9 ± 1.88 | 52.6 ± 1.36 | 48.7 ± 1.18 | 37.0 ± 1.02 | 37.2 ± 1.23 | 14.0 |
| T7 | 42.2 ± 1.01 | 43.0 ± 1.41 | 40.6 ± 2.58 | 36.7 ± 1.39 | 40.1 ± 1.13 | 40.8 ± 1.26 | 35.2 ± 1.44 | 49.1 ± 1.72 | 49.2 ± 1.07 | 38.3 ± 1.10 | 38.0 ± 1.09 | 11.0 |
| T8 | 43.3 ± 1.91 | 43.8 ± 1.12 | 42.6 ± 2.77 | 38.0 ± 1.49 | 42.5 ± 1.99 | 43.5 ± 1.27 | 35.8 ± 1.68 | 48.6 ± 1.34 | 51.0 ± 1.55 | 40.4 ± 2.05 | 37.5 ± 1.03 | 10.7 |
| T9 | 43.4 ± 1.39 | 45.0 ± 1.23 | 43.6 ± 2.13 | 38.0 ± 1.80 | 42.0 ± 1.12 | 45.1 ± 1.36 | 36.8 ± 1.73 | 48.9 ± 1.21 | 49.3 ± 1.32 | 37.7 ± 0.82 | 38.6 ± 0.97 | 10.3 |
| T10 | 45.6 ± 2.46 | 43.5 ± 1.21 | 40.5 ± 2.98 | 36.9 ± 1.08 | 43.0 ± 1.50 | 43.1 ± 1.41 | 39.0 ± 2.02 | 49.8 ± 1.15 | 49.0 ± 1.28 | 41.1 ± 1.65 | 41.6 ± 1.35 | 9.11 |
| T-DA | 52.0 ± 4.13 | 50.1 ± 3.80 | 33.1 ± 3.63 | 31.3 ± 1.93 | 37.9 ± 4.01 | 27.8 ± 2.32 | 38.7 ± 2.48 | 23.0 ± 0.88 | 24.9 ± 2.22 | 34.4 ± 3.46 | 42.1 ± 2.83 | 26.3 |
| Z1 | 116 ± 3.17 | 128 ± 2.29 | 129 ± 3.89 | 125 ± 2.57 | 117 ± 1.68 | 124 ± 1.64 | 126 ± 3.15 | 128 ± 2.48 | 121 ± 5.12 | 128 ± 3.67 | 109 ± 2.07 | 5.29 |
| Z2 | 120 ± 2.14 | 126 ± 1.69 | 129 ± 3.94 | 132 ±1.36 | 121 ± 1.50 | 123 ± 1.32 | 130 ± 2.44 | 129 ± 2.61 | 119 ± 4.54 | 132 ± 2.85 | 110 ± 3.04 | 5.38 |
| W/L | 0.62 ± 0.01 | 0.77 ± 0.01 | 0.60 ± 0.01 | 0.66 ± 0.02 | 0.69 ± 0.01 | 0.69 ± 0.01 | 0.66 ± 0.02 | 0.66 ± 0.01 | 0.65 ± 0.02 | 0.75 ± 0.01 | 0.67 ± 0.01 | 7.47 |
| W-DA/H-DA | 0.88 ± 0.09 | 0.88 ± 0.07 | 0.59 ± 0.05 | 0.59 ± 0.04 | 0.73 ± 0.09 | 0.57 ± 0.05 | 0.55 ± 0.03 | 0.52 ± 0.02 | 0.57 ± 0.04 | 0.67 ± 0.04 | 0.72 ± 0.05 | 19.5 |
| BOUNDARY | 0.90 ± 0.01 | 0.87 ± 0.01 | 0.94 ± 0.02 | 0.91 ± 0.01 | 0.85 ± 0.01 | 0.78 ± 0.01 | 0.88 ± 0.02 | 0.93 ± 0.01 | 0.86 ± 0.01 | 0.87 ± 0.01 | 0.94 ± 0.01 | 5.26 |
| Cultivars | H (cm) | W (cm) | L-ped (cm) | TSS (°Brix) | Weight (g) | W/H (g/cm) |
|---|---|---|---|---|---|---|
| Cattaneo | 2.68 ± 0.08 | 1.54 ± 0.04 | 0.90 ± 0.05 | 13.9 ± 1.13 | 3.13 ± 0.29 | 0.58 ± 0.02 |
| Florio | 2.28 ± 0.07 | 1.19 ± 0.03 | 1.01 ± 0.04 | 16.1 ± 0.50 | 1.38 ± 0.08 | 0.52 ± 0.01 |
| Giazzola | 1.21 ± 0.08 | 0.89 ± 0.06 | 0.76 ± 0.06 | 12.1 ± 0.16 | 0.42 ± 0.11 | 0.73 ± 0.02 |
| Kayrio | 1.55 ± 0.03 | 1.03 ± 0.02 | 0.67 ± 0.04 | 8.01 ± 0.32 | 0.80 ± 0.03 | 0.67 ± 0.01 |
| Kokusò-21 | 2.66 ± 0.08 | 1.32 ± 0.03 | 0.99 ± 0.05 | 17.1 ± 0.86 | 2.27 ± 0.14 | 0.50 ± 0.01 |
| Kokusò Rosso | 1.50 ± 0.06 | 0.96 ± 0.03 | 0.56 ± 0.04 | 17.5 ± 1.73 | 0.71 ± 0.06 | 0.65 ± 0.03 |
| Korinne | 1.39 ± 0.06 | 1.11 ± 0.07 | 0.96 ± 0.12 | 20.4 ± 1.35 | 0.76 ± 0.12 | 0.80 ± 0.03 |
| Miura | 1.51 ± 0.05 | 1.13 ± 0.03 | 1.20 ± 0.07 | 25.7 ± 0.92 | 0.76 ± 0.07 | 0.76 ± 0.04 |
| Spagna Rosso | 2.57 ± 0.08 | 1.56 ± 0.03 | 0.73 ± 0.05 | 17.0 ± 0.57 | 3.11 ± 0.25 | 0.61 ± 0.01 |
| C.V. | 31.4 | 20.1 | 23.0 | 30.5 | 72.5 | 16.1 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lo Bianco, R.; Mirabella, F. Use of Leaf and Fruit Morphometric Analysis to Identify and Classify White Mulberry (Morus alba L.) Genotypes. Agriculture 2018, 8, 157. https://doi.org/10.3390/agriculture8100157
Lo Bianco R, Mirabella F. Use of Leaf and Fruit Morphometric Analysis to Identify and Classify White Mulberry (Morus alba L.) Genotypes. Agriculture. 2018; 8(10):157. https://doi.org/10.3390/agriculture8100157
Chicago/Turabian StyleLo Bianco, Riccardo, and Fabio Mirabella. 2018. "Use of Leaf and Fruit Morphometric Analysis to Identify and Classify White Mulberry (Morus alba L.) Genotypes" Agriculture 8, no. 10: 157. https://doi.org/10.3390/agriculture8100157





