Furthest-Pair-Based Decision Trees: Experimental Results on Big Data Classification
Abstract
1. Introduction
2. Related Work
3. Furthest-Pair-Based Decision Trees (FPDT)
3.1. Methods
- Accumulate the classes’ probabilities by adding the parent’s probabilities to its children’s; we call this method decision tree 0 (DT0).
- Accumulate the probabilities by adding the parent’s probabilities to its children’s; and weighting these probabilities by the level of the node, assuming that the more we go deeper in the tree, the more likely we reach to a similar example(s), this is done by multiplying the tree level by the classes’ probabilities at a particular node; we call this method decision tree 1 (DT1), and is shown in Algorithm 1 as (Dtype = 1).
- Accumulate the classes’ probabilities by adding the parent’s probabilities to its children's; and weighting these probabilities exponentially, for the same reason in 2, but with higher weight. This is done by multiplying the classes’ probabilities by 2 to the power of the tree’s level at a particular node; we call this method decision tree 2 (DT2), and is shown in Algorithm 1 as (Dtype = 2).
- No accumulation of the probabilities, we use just the probabilities found in a leaf node; we call this method decision tree 3 (DT3), and is shown in Algorithm 1 as (Dtype ≠ 3).
- Similar to DT0 (normal accumulation), but the algorithm continues to cluster until there is only one or a number of similar examples in a leaf-node, this is done even if all the examples of a current node belong to the same class. While DT0–DT3 stop the recursive clustering when all the examples of the current node are belonging to the same class, and consider the current class as a leaf-node, we call this method decision tree 4 (DT4), and is shown in Algorithm 1 as (Dtype = 4).
| Algorithm 1. Training Phase (DT building) of FPDT. |
| Input: Numerical training dataset DATA with n FVs and d features, and DT type (Dtype) Output: A root pointer (RootN) to the resultant DT. 1- Create a DT Node → RootN 2- RootN.Examples ← FVs//all indexes of FVs from the training set 3- (P1, P2) ← Procedure Furthest(DATA ← RootN.Examples, n)//hill climbing algorithm [1] 4- if EN(P1) > EN(P2) swap(P1, P2) 5- RootN.P1 ← P1 6- RootN.P2 ← P2 7- RootN.Left = Null 8- RootN.Right = Null 9- Procedure BuildDT(Node ← RootN) 10- for each FVi in Node, do 11- D1←ED(FVi, Node.P1) 12- D2←ED(FVi, Node.P2) 13- If (D1 < D2) 14- Add index of FVi to Node.Left.Examples 15- else 16- Add index of FVi to Node.Right.Examples 17- end for 18- if (Node.Left.Size == 0 or Node.Right.Size == 0) 19- return //this means a leaf node 20- (P1, P2) ← Furthest(Node.Left.Examples, size(Node.Left.Examples))//work on the left child 21- if (EN(P1) > EN(P2)) then swap(P1, P2) 22- Node.Left.P1 ← P1 23- Node.Left.P2 ← P2 24- Node.Left.ClassP [numclasses] = {0}//initialize the classes’ probabilities to 0; 25- for each i in Node.Left.Examples do 26- Node.Left.ClassP [DATA.Class[i]]++//histogram of classes at Left-Node 27- bool LeftMulticlasses = false//check for single class to prune the tree 28- if there is more than one class at Node.Left.ClassP 29- LeftMulticlasses=true; 30- if (Dtype ==4) //no pruning if chosen 31- LeftMulticlasses=true//even if there is only one class in a node=> cluster it further 32- for each i in numclasses do //calculate probabilities of classes at the left node 33- Node.Left.ClassP [i]= Node.Left.ClassP [i]/ size(Node.Left.Examples) 34- if (Dtype ==1) //increase the probabilities by the increased level 35- for each i in numclasses do 36- Node.Left.ClassP [i]= Node.Left.ClassP [i]* Node.Left.level 37- if (Dtype ==2) //increase the probabilities exponentially by the increased level 38- for each i in numclasses do 39- Node.Left.ClassP [i]= Node.Left.ClassP [i]* 2Node.Left.level 40- if (Dtype != 3)//do accumulation for probabilities, if 3, use just the probabilities in a leaf node 41- for each i in numclasses do 42- Node.Left.ClassP [i] = Node.Left.ClassP [i] + Node.ClassP [i] 43- Node.Left.Left = NULL; 44- Node.Left.Right = NULL; 45- Repeat the previous steps (20–44) on Node.Right 46- if (LeftMulticlasses) 47- BuildDT (Node.Left) 48- if (RightMulticlasses) 49- BuildTree(Node.Right) 50- end Procedure 51- return RootN 52- end Algorithm 1 |
| Algorithm 2. Testing Phase of FPDT. |
| Input: test dataset TESTDATA with n FVs and d features Output: Testing Accuracy (Acc). 1- Acc←0 2- for each FVi in TESTDATA do 3- Procedure GetTreeNode(Node ← RootN, FVi) 4- D1 ← ED(FV[i], Node.P1) 5- D2 ← ED(FV[i], Node.P2) 6- if (D1 < D2 and Node.Left) 7- return GetTreeNode (Node.Left, FVi) 8- else if (D2 ≤ D1 and Node.Right) 9- return GetTreeNode (Node.Right,FVi) 10- else 11- return Node 12- end if 13- end Procedure GetTreeNode 14- class ← argmax(Node.ClassP)// returns the class with the maximum probability 15- if class == Class(FVi) 16- Acc ← Acc+1 17- end for each 18- Acc← Acc/n 19- return Acc 20- end Algorithm 2 |
3.2. Implementation Example
3.3. Data
4. Results and Discussion
- Processor: Intel® Core™ i7-6700 CPU @ 340GHz
- Installed memory (RAM): 16.0 GB
- System type: 64-bit operating system, x64-based processor, MS Windows 10.
5. Conclusions
Funding
Conflicts of Interest
References
- Zerbino, P.; Aloini, D.; Dulmin, R.; Mininno, V. Big Data-enabled Customer Relationship Management: A holistic approach. Inf. Process. Manag. 2018, 54, 818–8469. [Google Scholar] [CrossRef]
- LaValle, S.; Lesser, E.; Shockley, R.; Hopkins, M.S.; Kruschwitz, N. Big data, analytics and the path from insights to value. MIT Sloan Manag. Rev. 2011, 52, 21. [Google Scholar]
- Zhang, Q.; Yang, L.T.; Chen, Z.; Li, P. A survey on deep learning for big data. Inf. Fusion 2018, 42, 146–157. [Google Scholar] [CrossRef]
- Bolón-Canedo, V.; Remeseiro, B.; Sechidis, K.; Martinez-Rego, D.; Alonso-Betanzos, A. Algorithmic challenges in Big Data analytics. In Proceedings of the European Symposium on Artificial Neural Networks, Computational Intelligence and Machine Learning, Bruges, Belgium, 26–28 April 2017. [Google Scholar]
- Lv, X. The big data impact and application study on the like ecosystem construction of open internet of things. Clust. Comput. 2018, 1–10. [Google Scholar] [CrossRef]
- Fix, E.; Hodges, J.L., Jr. Discriminatory Analysis-Nonparametric Discrimination: Consistency Properties; USAF School of Aviation Medicine: Randolph Field, TX, USA, 1951. [Google Scholar]
- Cover, T.; Hart, P. Nearest neighbor pattern classification. IEEE Trans. Inf. Theory 1967, 13, 21–27. [Google Scholar] [CrossRef]
- Hassanat, A. Norm-Based Binary Search Trees for Speeding Up KNN Big Data Classification. Computers 2018, 7, 54. [Google Scholar] [CrossRef]
- Hassanat, A.B. Furthest-Pair-Based Binary Search Tree for Speeding Big Data Classification Using K-Nearest Neighbors. Big Data 2018, 6, 225–235. [Google Scholar] [CrossRef]
- Hassanat, B. Two-point-based binary search trees for accelerating big data classification using KNN. PLoS ONE 2018, 13, 14, in press. [Google Scholar]
- Hassanat, B.; Tarawneh, A.S. Fusion of color and statistic features for enhancing content-based image retrieval systems. J. Theor. Appl. Inf. Technol. 2016, 88, 644–655. [Google Scholar]
- Tarawneh, A.S.; Chetverikov, D.; Verma, C.; Hassanat, A.B. Stability and reduction of statistical features for image classification and retrieval: Preliminary results. In Proceedings of the 9th International Conference on Information and Communication Systems, Irbid, Jordan, 3–5 April 2018; pp. 117–121. [Google Scholar]
- Hassanat, A.B. Greedy algorithms for approximating the diameter of machine learning datasets in multidimensional euclidean space. arXiv, 2018; arXiv:1808.03566. [Google Scholar]
- Zhang, S.; Li, X.; Zong, M.; Zhu, X.; Wang, R. Efficient knn classification with different numbers of nearest neighbors. IEEE Trans. Neural Netw. Learn. Syst. 2018, 29, 1774–1785. [Google Scholar] [CrossRef] [PubMed]
- Hassanat, A.B.; Abbadi, M.A.; Altarawneh, G.A.; Alhasanat, A.A. Solving the problem of the K parameter in the KNN classifier using an ensemble learning approach. arXiv, 2014; arXiv:1409.0919. [Google Scholar]
- Wang, F.; Wang, Q.; Nie, F.; Yu, W.; Wang, R. Efficient tree classifiers for large scale datasets. Neurocomputing 2018, 284, 70–79. [Google Scholar] [CrossRef]
- Maillo, J.; Triguero, I.; Herrera, F. A mapreduce-based k-nearest neighbor approach for big data classification. In Proceedings of the 13th IEEE International Symposium on Parallel and Distributed Processing with Application, Helsinki, Finland, 20–22 August 2015; pp. 167–172. [Google Scholar]
- Maillo, J.; Ramírez, S.; Triguero, I.; Herrera, F. kNN-IS: An Iterative Spark-based design of the k-Nearest Neighbors classifier for big data. Knowl.-Based Syst. 2017, 117, 3–15. [Google Scholar] [CrossRef]
- Deng, Z.; Zhu, X.; Cheng, D.; Zong, M.; Zhang, S. Efficient kNN classification algorithm for big data. Neurocomputing 2016, 195, 143–148. [Google Scholar] [CrossRef]
- Gallego, A.J.; Calvo-Zaragoza, J.; Valero-Mas, J.J.; Rico-Juan, J.R. Clustering-based k-nearest neighbor classification for large-scale data with neural codes representation. Pattern Recognit. 2018, 74, 531–543. [Google Scholar] [CrossRef]
- Bentley, J.L. Multidimensional binary search trees used for associative searching. Commun. ACM 1975, 18, 509–517. [Google Scholar] [CrossRef]
- Uhlmann, J.K. Satisfying general proximity/similarity queries with metric trees. Inf. Process. Lett. 1991, 40, 175–179. [Google Scholar] [CrossRef]
- Beygelzimer, A.; Kakade, S.; Langford, J. Cover trees for nearest neighbor. In Proceedings of the 23rd International Conference on Machine Learning, Pittsburgh, PA, USA, 25–29 June 2006; pp. 97–104. [Google Scholar]
- Kibriya, A.M.; Frank, E. An empirical comparison of exact nearest neighbour algorithms. In Proceedings of the 11th European Conference on Principles and Practice of Knowledge Discovery in Databases, Warsaw, Poland, 17–21 September 2007; pp. 140–151. [Google Scholar]
- Cislak, A.; Grabowski, S. Experimental evaluation of selected tree structures for exact and approximate k-nearest neighbor classification. In Proceedings of the Ederated Conference on Computer Science and Information Systems, Warsaw, Poland, 7–10 September 2014; pp. 93–100. [Google Scholar]
- Fan, R.-E. LIBSVM Data: Classification, Regression, and Multi-label. 2011. Available online: https://www.csie.ntu.edu.tw/~cjlin/libsvmtools/datasets/ (accessed on 1 March 2018).
- Lichman, M. University of California, Irvine, School of Information and Computer Sciences, 2013. Available online: http://archive.ics.uci.edu/ml (accessed on 1 March 2018).
- Nalepa, J.; Kawulok, M. Selecting training sets for support vector machines: A review. Artif. Intell. Rev. 2018, 1–44. [Google Scholar] [CrossRef]
- Rodríguez-Fdez, I.; Canosa, A.; Mucientes, M.; Bugarín, A. STAC: A web platform for the comparison of algorithms using statistical tests. In Proceedings of the 2015 IEEE International Conference on Fuzzy Systems, Istanbul, Turkey, 2–5 August 2015; pp. 1–8. [Google Scholar]
- Shapiro, S.S.; Wilk, M.B. An analysis of variance test for normality (complete samples). Biometrika 1965, 52, 591–611. [Google Scholar] [CrossRef]
- Levene, H. Robust tests for equality of variances. Contrib. Probab. Stat. 1961, 69, 279–292. [Google Scholar]
- Quinlan, J.R. C4.5: Programs for Machine Learning; Morgan Kaufmann Publishers: San Mateo, CA, USA, 1993. [Google Scholar]
- Quinlan, J.R. Simplifying decision trees. Int. J. Man-Mach. Stud. 1987, 27, 221–234. [Google Scholar] [CrossRef]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Witten, I.H.; Frank, E.; Hall, M.A.; Pal, C.J. Data Mining: Practical Machine Learning Tools and Techniques; Morgan Kaufmann Publishers: San Mateo, CA, USA, 2016. [Google Scholar]
- Hassanat, A.B. On identifying terrorists using their victory signs. Data Sci. J. 2018, 17, 27. [Google Scholar] [CrossRef]
- Hassanat, A.B.; Prasath, V.S.; Al-kasassbeh, M.; Tarawneh, A.S.; Al-shamailh, A.J. Magnetic energy-based feature extraction for low-quality fingerprint images. Signal Image Video Process. 2018, 1–8. [Google Scholar] [CrossRef]
- Hassanat, A.B.; Prasath, V.S.; Al-Mahadeen, B.M.; Alhasanat, S.M.M. Classification and gender recognition from veiled-faces. Int. J. Biom. 2017, 9, 347–364. [Google Scholar] [CrossRef]
- Hassanat, A.B. Dimensionality invariant similarity measure. arXiv, 2014; arXiv:1409.0923. [Google Scholar]
- Alkasassbeh, M.; Altarawneh, G.A.; Hassanat, A. On enhancing the performance of nearest neighbour classifiers using hassanat distance metric. arXiv 2015, arXiv:1501.00687. [Google Scholar]




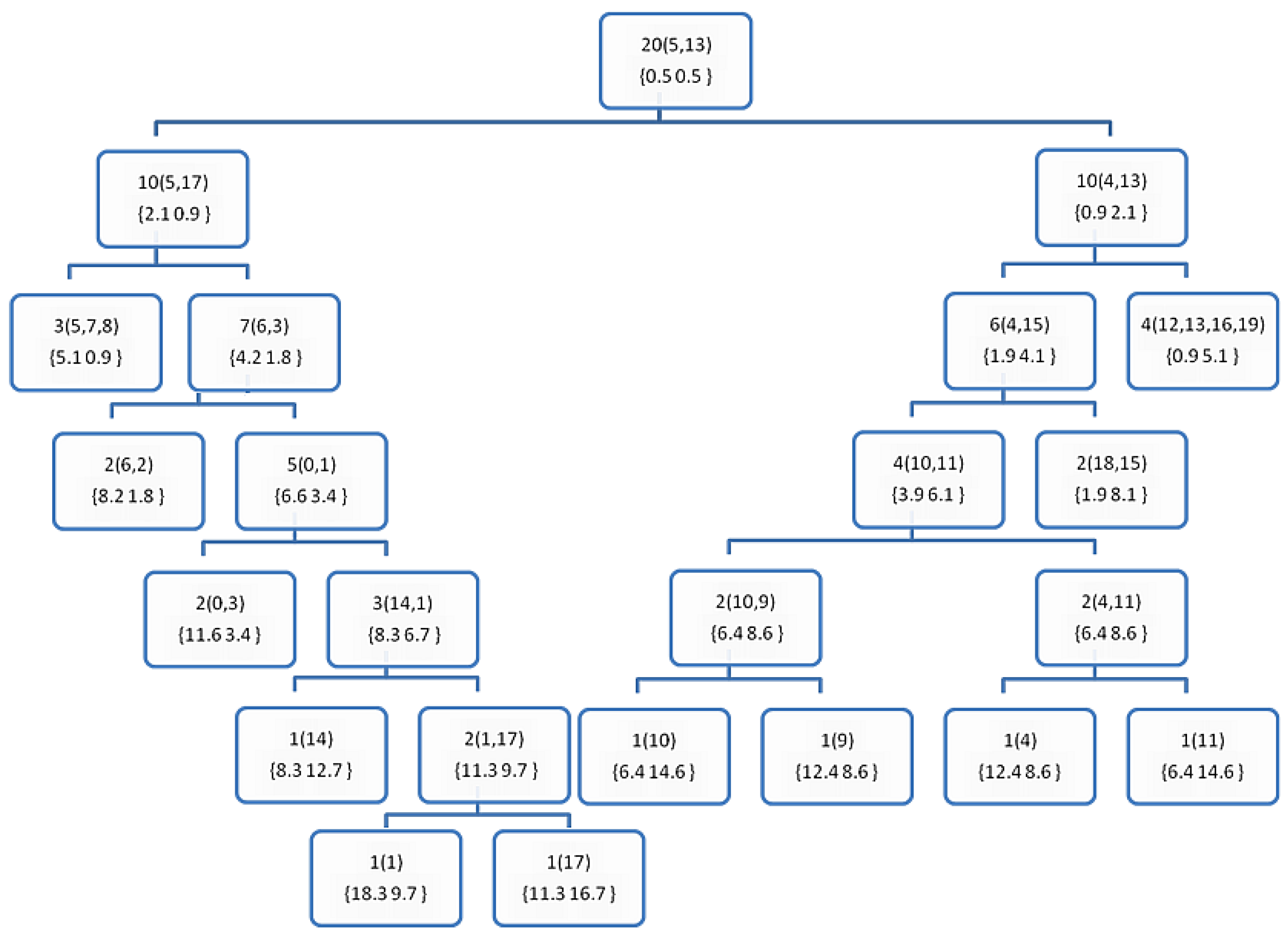



| #Example | X1 | X2 | Class | Euclidean Norms |
|---|---|---|---|---|
| 0 | 4 | 3 | 0 | 5.0 |
| 1 | 2 | 5 | 0 | 5.4 |
| 2 | 2 | 4 | 0 | 4.5 |
| 3 | 4 | 4 | 0 | 5.7 |
| 4 | 3 | 6 | 0 | 6.7 |
| 5 | 1 | 0 | 0 | 1.0 |
| 6 | 1 | 3 | 0 | 3.2 |
| 7 | 3 | 1 | 0 | 3.2 |
| 8 | 3 | 2 | 0 | 3.6 |
| 9 | 4 | 6 | 0 | 7.2 |
| 10 | 4 | 5 | 1 | 6.4 |
| 11 | 3 | 7 | 1 | 7.6 |
| 12 | 8 | 6 | 1 | 10.0 |
| 13 | 9 | 7 | 1 | 11.4 |
| 14 | 3 | 4 | 1 | 5.0 |
| 15 | 5 | 7 | 1 | 8.6 |
| 16 | 8 | 3 | 1 | 8.5 |
| 17 | 3 | 5 | 1 | 5.8 |
| 18 | 4 | 8 | 1 | 8.9 |
| 19 | 8 | 8 | 1 | 11.3 |
| Dataset | Size | Dimensions | Type | #Class |
|---|---|---|---|---|
| HIGGS | 11,000,000 | 28 | Real | 2 |
| SUSY | 5,000,000 | 18 | Real | 2 |
| Poker | 1,025,010 | 11 | Integers | 10 |
| Covtype | 581,012 | 54 | Integers | 7 |
| Mnist | 70,000 | 784 | Integers | 10 |
| Connect4 | 67,557 | 42 | Integers | 3 |
| Nist | 44,951 | 1024 | Integers | 26 |
| LetRec | 20,000 | 16 | Real | 26 |
| Homus | 15,199 | 1600 | Integers | 32 |
| Gisette | 13,500 | 5000 | Integers | 2 |
| Pendigits | 10,992 | 16 | Integers | 10 |
| Usps | 9298 | 256 | Real | 10 |
| Satimage | 6435 | 36 | Real | 6 |
| Abalone | 4177 | 8 | Real | 3 |
| Climate | 1178 | 18 | Real | 2 |
| German | 1000 | 24 | Integers | 2 |
| Blood | 748 | 4 | Integers | 2 |
| Australian | 690 | 14 | Real | 2 |
| Cancer | 683 | 9 | Integers | 2 |
| Balance | 625 | 4 | Integers | 3 |
| Method | Number of Nodes | Number of Leaves | Maximum Depth | Total Examples in All Leaves | Min Number of Examples in a Leaf | Max Number of Examples in a Leaf |
|---|---|---|---|---|---|---|
| FPBST | 2,045,541 | 1,022,771 | 30 | 1,025,010 | 1 | 3 |
| DT0 | 1,433,617 | 716,809 | 29 | 1,025,010 | 1 | 49 |
| DT1 | 1,433,089 | 716,545 | 29 | 1,025,010 | 1 | 80 |
| DT2 | 1,433,631 | 716,816 | 29 | 1,025,010 | 1 | 71 |
| DT3 | 1,432,131 | 716,066 | 30 | 1,025,010 | 1 | 47 |
| DT4 | 2,045,541 | 1,022,771 | 30 | 1,025,010 | 1 | 3 |
| DT0+ | 1,440,047 | 720,024 | 29 | 1,025,010 | 1 | 99 |
| DT1+ | 1,439,295 | 719,648 | 29 | 1,025,010 | 1 | 42 |
| DT2+ | 1,439,113 | 719,557 | 30 | 1,025,010 | 1 | 56 |
| DT3+ | 1,441,107 | 720,554 | 30 | 1,025,010 | 1 | 44 |
| DT4+ | 2,045,541 | 1,022,771 | 30 | 1,025,010 | 1 | 3 |
| Dataset | FPBST | DT0 | DT1 | DT2 | DT3 | DT4 |
|---|---|---|---|---|---|---|
| Abalone | 0.4990 | 0.5374 | 0.5338 | 0.4906 | 0.5122 | 0.5326 |
| Australian | 0.6435 | 0.6667 | 0.6725 | 0.6203 | 0.6392 | 0.6899 |
| Balance | 0.8258 | 0.8226 | 0.8323 | 0.7855 | 0.8210 | 0.8290 |
| blood | 0.6784 | 0.7662 | 0.7811 | 0.7189 | 0.7135 | 0.7716 |
| Cancer | 0.9618 | 0.9574 | 0.9574 | 0.9544 | 0.9559 | 0.9647 |
| Climate | 0.8722 | 0.9148 | 0.9148 | 0.8537 | 0.8796 | 0.9167 |
| German | 0.6550 | 0.7120 | 0.7050 | 0.6240 | 0.6460 | 0.7110 |
| LetRec | 0.7897 | 0.7143 | 0.7379 | 0.7841 | 0.7935 | 0.7476 |
| Usps | 0.8631 | 0.7758 | 0.8179 | 0.8665 | 0.8614 | 0.8222 |
| Satimage | 0.8672 | 0.8342 | 0.8566 | 0.8594 | 0.8617 | 0.8588 |
| Pendigits | 0.9630 | 0.8779 | 0.9196 | 0.9625 | 0.9678 | 0.9392 |
| Gisette | 0.8907 | 0.8372 | 0.8541 | 0.8867 | 0.8910 | 0.8730 |
| Mnist | 0.8527 | 0.7720 | 0.8055 | 0.8553 | 0.8527 | 0.8135 |
| Homus | 0.4508 | 0.4289 | 0.4481 | 0.4560 | 0.4572 | 0.4386 |
| Nist | 0.4795 | 0.4507 | 0.4684 | 0.4853 | 0.4858 | 0.4605 |
| Connect4 | 0.6222 | 0.6613 | 0.6743 | 0.6216 | 0.6197 | 0.6659 |
| Covtype | 0.9314 | 0.7752 | 0.8318 | 0.9313 | 0.9315 | 0.8533 |
| Poker | 0.5372 | 0.5881 | 0.5889 | 0.5366 | 0.5351 | 0.5870 |
| SUSY | 0.7098 | 0.7547 | 0.7651 | 0.7103 | 0.7093 | 0.7599 |
| HIGGS | 0.5860 | 0.5998 | 0.6062 | 0.5857 | 0.5856 | 0.6010 |
| Average | 0.7339 | 0.7224 | 0.7386 | 0.7294 | 0.7360 | 0.7418 |
| Dataset | FPBST | DT0 | DT1 | DT2 | DT3 | DT4 |
|---|---|---|---|---|---|---|
| Abalone | 196 | 170 | 163 | 164 | 162 | 156 |
| Australian | 45 | 44 | 37 | 39 | 39 | 42 |
| Balance | 15 | 8 | 8 | 9 | 8 | 10 |
| blood | 19 | 13 | 13 | 13 | 13 | 15 |
| Cancer | 29 | 11 | 9 | 8 | 9 | 20 |
| Climate | 29 | 21 | 21 | 20 | 20 | 30 |
| German | 56 | 62 | 61 | 68 | 62 | 70 |
| LetRec | 1595 | 1249 | 1151 | 1142 | 1176 | 1338 |
| Usps | 8096 | 8030 | 8119 | 8084 | 8098 | 9922 |
| Satimage | 860 | 743 | 769 | 771 | 774 | 958 |
| Pendigits | 910 | 574 | 576 | 578 | 581 | 882 |
| Gisette | 76,157 | 60,297 | 58,053 | 59,413 | 58,600 | 72,881 |
| Mnist | 142,712 | 118,787 | 118,678 | 117,771 | 116,226 | 141,527 |
| Homus | 83,337 | 76,153 | 72,626 | 73,118 | 73,821 | 77,758 |
| Nist | 118,746 | 99,569 | 98,802 | 100,926 | 98,987 | 106,223 |
| Connect4 | 6860 | 5832 | 5854 | 5408 | 5323 | 5857 |
| Covtype | 94,265 | 73,359 | 71,481 | 75,018 | 72,736 | 90,682 |
| Poker | 74,536 | 70,164 | 66,805 | 68,613 | 61,009 | 75,957 |
| SUSY | 923,749 | 955,593 | 948,978 | 959,964 | 906,365 | 1,017,843 |
| HIGGS | 2,974,260 | 3,117,121 | 2,855,369 | 2,958,233 | 2,951,326 | 2,939,329 |
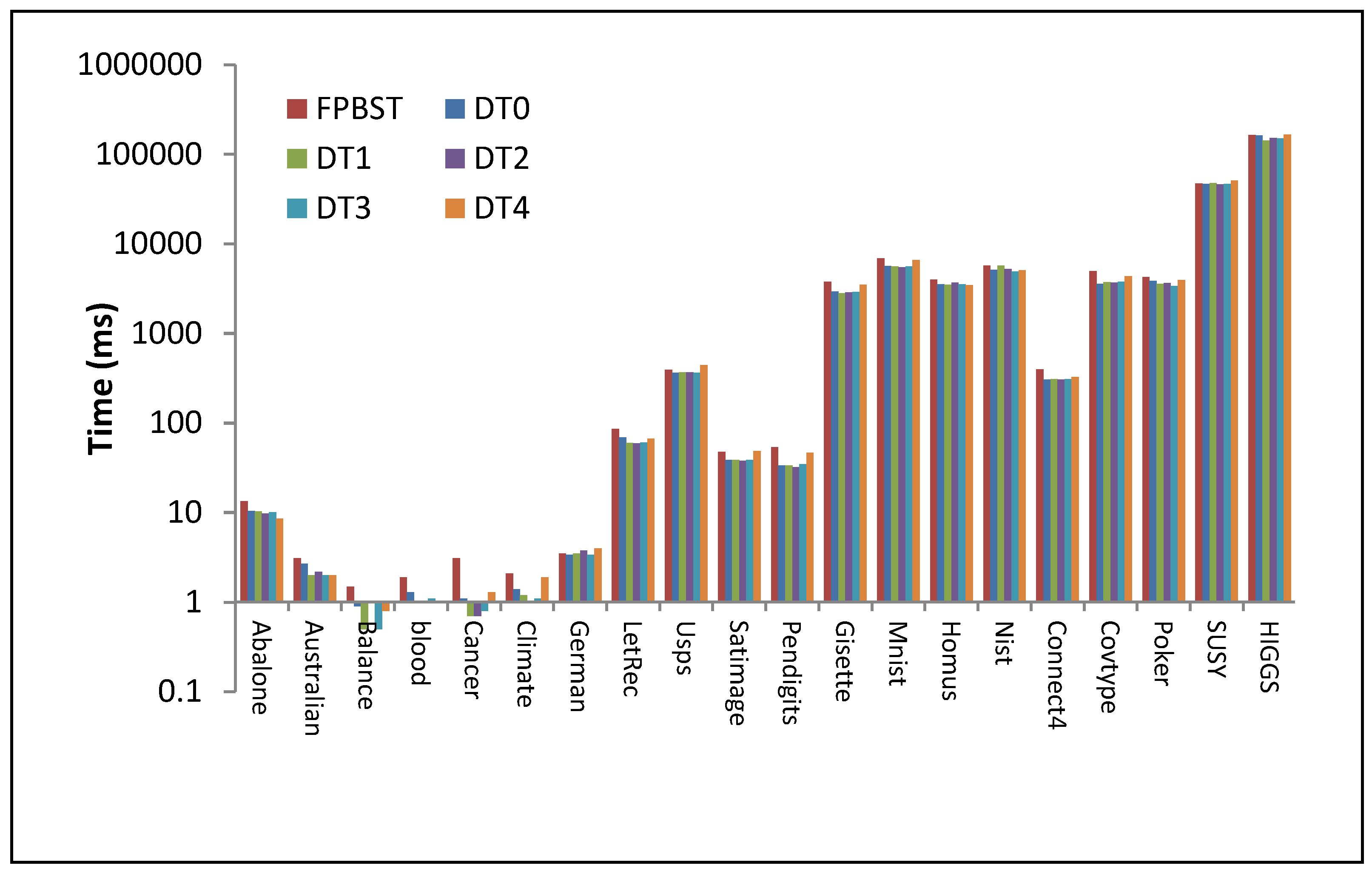
| Dataset | FPBST | DT0 | DT1 | DT2 | DT3 | DT4 |
|---|---|---|---|---|---|---|
| Abalone | 13.5 | 10.5 | 10.3 | 9.8 | 10.1 | 8.6 |
| Australian | 3.1 | 2.7 | 2.0 | 2.2 | 2.0 | 2.0 |
| Balance | 1.5 | 0.9 | 0.5 | 1.0 | 0.5 | 0.8 |
| blood | 1.9 | 1.3 | 1.0 | 1.0 | 1.1 | 1.0 |
| Cancer | 3.1 | 1.1 | 0.7 | 0.7 | 0.8 | 1.3 |
| Climate | 2.1 | 1.4 | 1.2 | 1.0 | 1.1 | 1.9 |
| German | 3.5 | 3.4 | 3.5 | 3.8 | 3.4 | 4.0 |
| LetRec | 85.2 | 69.4 | 60.2 | 59.8 | 60.9 | 67.0 |
| Usps | 390.3 | 361.4 | 365.4 | 366.3 | 361.2 | 441.9 |
| Satimage | 47.9 | 38.7 | 38.7 | 38.0 | 38.8 | 48.8 |
| Pendigits | 53.9 | 33.6 | 33.6 | 32.3 | 34.8 | 46.8 |
| Gisette | 3752 | 2920 | 2804 | 2865 | 2884 | 3480 |
| Mnist | 6869 | 5631 | 5563 | 5474 | 5571 | 6591 |
| Homus | 3965 | 3520 | 3492 | 3695 | 3540 | 3443 |
| Nist | 5686 | 5088 | 5728 | 5236 | 4893 | 5058 |
| Connect4 | 395 | 304 | 308 | 304 | 308 | 324 |
| Covtype | 4935 | 3561 | 3719 | 3662 | 3764 | 4326 |
| Poker | 4243 | 3834 | 3546 | 3641 | 3381 | 3924 |
| SUSY | 46,906 | 46,521 | 47,801 | 46,027 | 46,430 | 50,604 |
| HIGGS | 164,652 | 161,623 | 142,220 | 151,414 | 149,363 | 164,853 |
| Dataset | DT0 Speed | DT1 Speed | DT2 Speed | DT3 Speed | DT4 Speed | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Train | Test | Train | Test | Train | Test | Train | Test | Train | Test | |
| Abalone | 1.15 | 1.29 | 1.20 | 1.31 | 1.19 | 1.38 | 1.21 | 1.34 | 1.25 | 1.57 |
| Australian | 1.03 | 1.15 | 1.21 | 1.55 | 1.17 | 1.41 | 1.16 | 1.55 | 1.06 | 1.55 |
| Balance | 1.82 | 1.67 | 1.80 | 3.00 | 1.78 | 1.50 | 1.89 | 3.00 | 1.50 | 1.88 |
| blood | 1.40 | 1.46 | 1.44 | 1.90 | 1.44 | 1.90 | 1.47 | 1.73 | 1.22 | 1.90 |
| Cancer | 2.72 | 2.82 | 3.23 | 4.43 | 3.59 | 4.43 | 3.30 | 3.88 | 1.46 | 2.38 |
| Climate | 1.40 | 1.50 | 1.41 | 1.75 | 1.42 | 2.10 | 1.45 | 1.91 | 0.97 | 1.11 |
| German | 0.90 | 1.03 | 0.92 | 1.00 | 0.82 | 0.92 | 0.90 | 1.03 | 0.80 | 0.88 |
| LetRec | 1.28 | 1.23 | 1.39 | 1.42 | 1.40 | 1.42 | 1.36 | 1.40 | 1.19 | 1.27 |
| Usps | 1.01 | 1.08 | 1.00 | 1.07 | 1.00 | 1.07 | 1.00 | 1.08 | 0.82 | 0.88 |
| Satimage | 1.16 | 1.24 | 1.12 | 1.24 | 1.11 | 1.26 | 1.11 | 1.23 | 0.90 | 0.98 |
| Pendigits | 1.59 | 1.60 | 1.58 | 1.60 | 1.57 | 1.67 | 1.57 | 1.55 | 1.03 | 1.15 |
| Gisette | 1.26 | 1.28 | 1.31 | 1.34 | 1.28 | 1.31 | 1.30 | 1.30 | 1.04 | 1.08 |
| Mnist | 1.20 | 1.22 | 1.20 | 1.23 | 1.21 | 1.25 | 1.23 | 1.23 | 1.01 | 1.04 |
| Homus | 1.09 | 1.13 | 1.15 | 1.14 | 1.14 | 1.07 | 1.13 | 1.12 | 1.07 | 1.15 |
| Nist | 1.19 | 1.12 | 1.20 | 0.99 | 1.18 | 1.09 | 1.20 | 1.16 | 1.12 | 1.12 |
| Connect4 | 1.18 | 1.30 | 1.17 | 1.28 | 1.27 | 1.30 | 1.29 | 1.28 | 1.17 | 1.22 |
| Covtype | 1.28 | 1.39 | 1.32 | 1.33 | 1.26 | 1.35 | 1.30 | 1.31 | 1.04 | 1.14 |
| Poker | 1.06 | 1.11 | 1.12 | 1.20 | 1.09 | 1.17 | 1.22 | 1.25 | 0.98 | 1.08 |
| SUSY | 0.97 | 1.01 | 0.97 | 0.98 | 0.96 | 1.02 | 1.02 | 1.01 | 0.91 | 0.93 |
| HIGGS | 0.95 | 1.02 | 1.04 | 1.16 | 1.01 | 1.09 | 1.01 | 1.10 | 1.01 | 1.00 |
| Average | 1.28 | 1.33 | 1.34 | 1.55 | 1.34 | 1.48 | 1.36 | 1.52 | 1.08 | 1.27 |
| Dataset | FPBST | DT+ | DT+ Speed | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Acc. | Train | Test | DT | Acc. | Train | Test | Train | Test | |
| Abalone | 0.4990 | 196 | 14 | 0+ | 0.5441 | 206 | 12 | 0.95 | 1.13 |
| Australian | 0.6435 | 45 | 3 | 4+ | 0.6957 | 48 | 3 | 0.94 | 1.03 |
| Balance | 0.8258 | 15 | 2 | 1+ | 0.8468 | 10 | 1.2 | 1.50 | 1.25 |
| blood | 0.6784 | 19 | 2 | 1+ | 0.7635 | 19 | 1.5 | 0.98 | 1.27 |
| Cancer | 0.9618 | 29 | 3 | 4+ | 0.9574 | 32 | 2.9 | 0.92 | 1.07 |
| Climate | 0.8722 | 29 | 2 | 4+ | 0.9148 | 32 | 2.1 | 0.91 | 1.00 |
| German | 0.6550 | 56 | 4 | 0+ | 0.7100 | 52 | 3.4 | 1.08 | 1.03 |
| LetRec | 0.7897 | 1595 | 85 | 3+ | 0.7884 | 1268 | 68.6 | 1.26 | 1.24 |
| Usps | 0.8631 | 8096 | 390 | 2+ | 0.8694 | 6456 | 306.3 | 1.25 | 1.27 |
| Satimage | 0.8672 | 860 | 48 | 3+ | 0.8652 | 670 | 34.8 | 1.28 | 1.38 |
| Pendigits | 0.9630 | 910 | 54 | 3+ | 0.9642 | 583 | 31.7 | 1.56 | 1.70 |
| Gisette | 0.8907 | 76,157 | 3752 | 3+ | 0.8904 | 57,101 | 2762.8 | 1.33 | 1.36 |
| Mnist | 0.8527 | 142,712 | 6869 | 2+ | 0.8523 | 118,015 | 5493.6 | 1.21 | 1.25 |
| Homus | 0.4508 | 83,337 | 4023 | 3+ | 0.4468 | 80,330 | 3554 | 1.04 | 1.13 |
| Nist | 0.4795 | 118,746 | 5686 | 3+ | 0.4828 | 108,402 | 4775 | 1.10 | 1.19 |
| Connect4 | 0.6222 | 6860 | 395 | 1+ | 0.6723 | 5735 | 298 | 1.20 | 1.33 |
| Covtype | 0.9314 | 94,265 | 4935 | 3+ | 0.9312 | 74,155 | 3728 | 1.27 | 1.32 |
| Poker | 0.5372 | 74,536 | 4243 | 1+ | 0.5889 | 66,029 | 3570 | 1.13 | 1.19 |
| SUSY | 0.7098 | 923,749 | 46,906 | 1+ | 0.7639 | 864,713 | 44,285 | 1.07 | 1.06 |
| HIGGS | 0.5860 | 2,974,260 | 164,652 | 1+ | 0.6061 | 2,854,760 | 141,130 | 1.04 | 1.17 |
| Average | 0.7339 | 225,324 | 12,103 | 0.7577 | 211,931 | 10,503 | 1.15 | 1.22 | |
| Dataset | Statistic | P-Value | Result |
|---|---|---|---|
| FPBST | 0.93589 | 0.2003 | Null hypothesis is accepted |
| DT+ | 0.92717 | 0.13622 | Null hypothesis is accepted |
| Statistic | P-Value | Result |
|---|---|---|
| 0.45998 | 0.50174 | Null hypothesis is accepted |
| T Statistic | P-Value | Result |
|---|---|---|
| –3.8335 | 0.00112 | Ho is rejected |
| Method | Training Time | Testing Time | Model Size | Accuracy |
|---|---|---|---|---|
| FPBST | long | long | large | moderate |
| DT0 | short | short | small | low |
| DT1 | short | short | small | moderate |
| DT2 | short | short | small | low |
| DT3 | short | short | small | moderate |
| DT4 | long | long | large | high |
| DT0+ | short | short | small | high |
| DT1+ | short | short | small | high |
| DT2+ | short | short | small | moderate |
| DT3+ | short | short | small | high |
| DT4+ | long | long | large | high |
| Dataset | J48 | REPTree | RF | DT0+ | DT1+ | DT3+ |
|---|---|---|---|---|---|---|
| Abalone | 0.5281 | 0.5286 | 0.5430 | 0.5439 | 0.5403 | 0.5002 |
| Australian | 0.8522 | 0.8478 | 0.8754 | 0.6783 | 0.6899 | 0.6479 |
| Balance | 0.7664 | 0.7648 | 0.8176 | 0.8339 | 0.8323 | 0.8371 |
| blood | 0.7781 | 0.7741 | 0.7273 | 0.7716 | 0.7689 | 0.7297 |
| Cancer | 0.9605 | 0.9531 | 0.9707 | 0.9691 | 0.9691 | 0.9574 |
| Climate | 0.9259 | 0.9148 | 0.9370 | 0.9148 | 0.9093 | 0.8630 |
| German | 0.7390 | 0.7390 | 0.7630 | 0.7100 | 0.6990 | 0.6640 |
| LetRec | 0.8825 | 0.8424 | 0.9645 | 0.7137 | 0.7463 | 0.7943 |
| Usps | 0.8943 | 0.8780 | 0.9607 | 0.7823 | 0.8200 | 0.8626 |
| Satimage | 0.8623 | 0.8595 | 0.9197 | 0.8381 | 0.8569 | 0.8619 |
| Pendigits | 0.9637 | 0.9518 | 0.9916 | 0.8802 | 0.9181 | 0.9663 |
| Gisette * | - | - | - | - | - | - |
| Mnist * | - | - | - | - | - | - |
| Homus * | - | - | - | - | - | - |
| Nist * | - | - | - | - | - | - |
| Connect4 * | - | - | - | - | - | - |
| Covtype * | - | - | - | - | - | - |
| Poker * | - | - | - | - | - | - |
| SUSY * | - | - | - | - | - | - |
| HIGGS * | - | - | - | - | - | - |
© 2018 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hassanat, A.B.A. Furthest-Pair-Based Decision Trees: Experimental Results on Big Data Classification. Information 2018, 9, 284. https://doi.org/10.3390/info9110284
Hassanat ABA. Furthest-Pair-Based Decision Trees: Experimental Results on Big Data Classification. Information. 2018; 9(11):284. https://doi.org/10.3390/info9110284
Chicago/Turabian StyleHassanat, Ahmad B. A. 2018. "Furthest-Pair-Based Decision Trees: Experimental Results on Big Data Classification" Information 9, no. 11: 284. https://doi.org/10.3390/info9110284
APA StyleHassanat, A. B. A. (2018). Furthest-Pair-Based Decision Trees: Experimental Results on Big Data Classification. Information, 9(11), 284. https://doi.org/10.3390/info9110284





