Combating Antibiotic Resistance through the Synergistic Effects of Mesoporous Silica-Based Hierarchical Nanocomposites
Abstract
:1. Introduction
2. Materials and Methods
2.1. Reagents and Chemicals
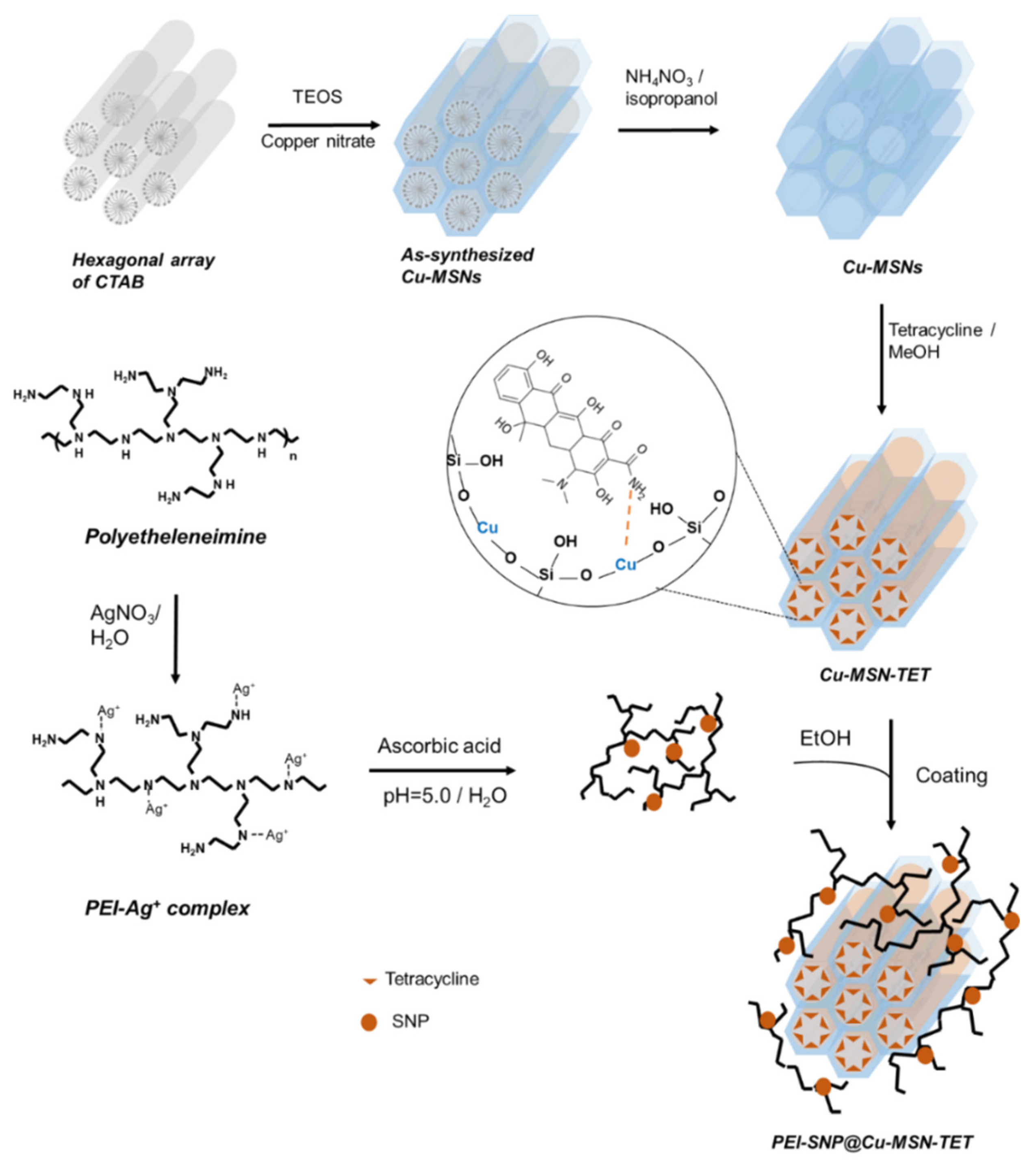
2.2. Fabrication of Cu-MSNs
2.3. Immobilization of TET in Cu-MSNs
2.4. Deposition of PEI-SNP Composite Layer over Cu-MSNs
2.5. Characterizations
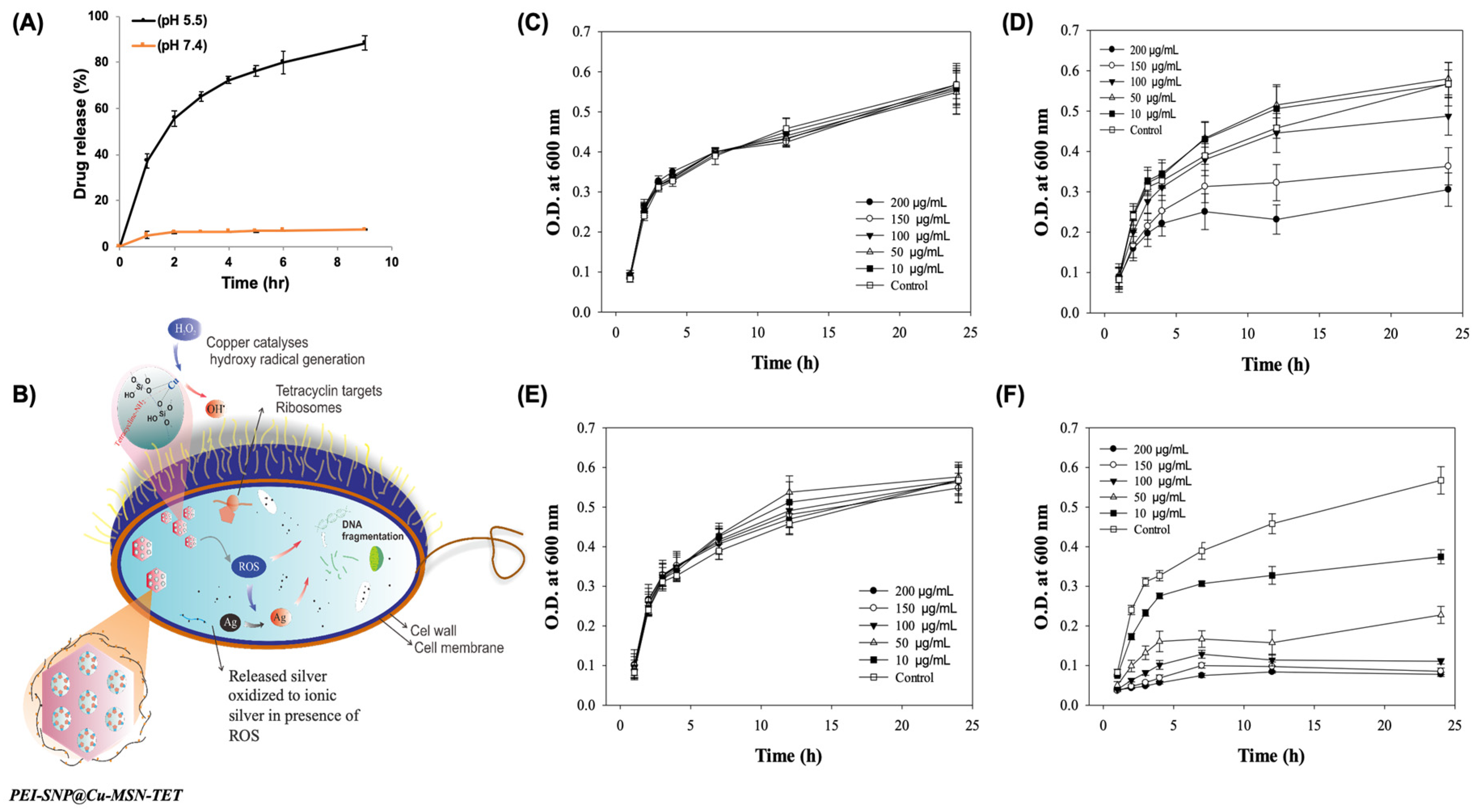
2.6. In Vitro Release
2.7. Biocompatibility
2.8. Antibacterial Efficacy
2.8.1. Bacteria Culture
2.8.2. Bacteria Viability
2.8.3. Bacteria Growth Study
2.8.4. CFU Assay
2.8.5. Determination of ROS
2.8.6. Membrane Permeability Assay
2.9. Statistical Analysis
3. Results and Discussion
3.1. Characterizations
3.2. In vitro TET Release
3.3. Biocompatibility
3.4. Antibacterial Efficacy
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Klein, E.Y.; Van Boeckel, T.P.; Martinez, E.M.; Pant, S.; Gandra, S.; Levin, S.A.; Goossens, H.; Laxminarayan, R. Global increase and geographic convergence in antibiotic consumption between 2000 and 2015. Proc. Natl. Acad. Sci. USA 2018, 115, E3463–E3470. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hwang, A.Y.; Gums, J.G. The emergence and evolution of antimicrobial resistance: Impact on a global scale. Bioorg. Med. Chem. 2016, 24, 6440–6445. [Google Scholar] [CrossRef] [PubMed]
- Roca, I.; Akova, M.; Baquero, F.; Carlet, J.; Cavaleri, M.; Coenen, S.; Cohen, J.; Findlay, D.; Gyssens, I.; Heuer, O.E.; et al. The global threat of antimicrobial resistance: Science for intervention. New Microbes New Infect. 2015, 6, 22–29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Riehemann, K.; Schneider, S.W.; Luger, T.A.; Godin, B.; Ferrari, M.; Fuchs, H. Nanomedicine--challenge and perspectives. Angew. Chem. Int. Ed. 2009, 48, 872–897. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.; Chen, H.; Shi, J. In Vivo Bio-Safety Evaluations and Diagnostic/Therapeutic Applications of Chemically Designed Mesoporous Silica Nanoparticles. Adv. Mater. 2013, 25, 3144–3176. [Google Scholar] [CrossRef] [PubMed]
- Liong, M.; Lu, J.; Kovochich, M.; Xia, T.; Ruehm, S.G.; Nel, A.E.; Tamanoi, F.; Zink, J.I. Multifunctional Inorganic Nanoparticles for Imaging, Targeting, and Drug Delivery. ACS Nano 2008, 2, 889–896. [Google Scholar] [CrossRef] [Green Version]
- Azzopardi, E.A.; Ferguson, E.L.; Thomas, D.W. The enhanced permeability retention effect: A new paradigm for drug targeting in infection. J. Antimicrob. Chemother. 2013, 68, 257–274. [Google Scholar] [CrossRef] [Green Version]
- Kuthati, Y.; Kankala, R.K.; Busa, P.; Lin, S.-X.; Deng, J.-P.; Mou, C.-Y.; Lee, C.-H. Phototherapeutic spectrum expansion through synergistic effect of mesoporous silica trio-nanohybrids against antibiotic-resistant gram-negative bacterium. J. Photochem. Photobiol. B 2017, 169, 124–133. [Google Scholar] [CrossRef]
- Prasad, L.K.; O’Mary, H.; Cui, Z. Nanomedicine delivers promising treatments for rheumatoid arthritis. Nanomedicine 2015, 10, 2063–2074. [Google Scholar] [CrossRef] [Green Version]
- Wu, S.-H.; Mou, C.-Y.; Lin, H.-P. Synthesis of mesoporous silica nanoparticles. Chem. Soc. Rev. 2013, 42, 3862–3875. [Google Scholar] [CrossRef]
- Mekaru, H.; Lu, J.; Tamanoi, F. Development of mesoporous silica-based nanoparticles with controlled release capability for cancer therapy. Adv. Drug Deliv. Rev. 2015, 95, 40–49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, C.-G.; Han, Y.-H.; Zhang, J.-T.; Kankala, R.K.; Wang, S.-B.; Chen, A.-Z. Rerouting engineered metal-dependent shapes of mesoporous silica nanocontainers to biodegradable Janus-type (sphero-ellipsoid) nanoreactors for chemodynamic therapy. Chem. Eng. J. 2019, 370, 1188–1199. [Google Scholar] [CrossRef]
- Kankala, R.K.; Liu, C.-G.; Yang, D.-Y.; Wang, S.-B.; Chen, A.-Z. Ultrasmall Platinum Nanoparticles Enable Deep Tumor Penetration and Synergistic Therapeutic Abilities through Free Radical Species-assisted Catalysis to Combat Cancer Multidrug Resistance. Chem. Eng. J. 2020, 383, 123138. [Google Scholar] [CrossRef]
- Tang, F.; Li, L.; Chen, D. Mesoporous Silica Nanoparticles: Synthesis, Biocompatibility and Drug Delivery. Adv. Mater. 2012, 24, 1504–1534. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Che, R.; Elzatahry, A.A.; Zhao, D. Direct Imaging Au Nanoparticle Migration Inside Mesoporous Silica Channels. ACS Nano 2014, 8, 10455–10460. [Google Scholar] [CrossRef]
- Braun, K.; Pochert, A.; Linden, M.; Davoudi, M.; Schmidtchen, A.; Nordstrom, R.; Malmsten, M. Membrane interactions of mesoporous silica nanoparticles as carriers of antimicrobial peptides. J. Colloid Interface Sci. 2016, 475, 161–170. [Google Scholar] [CrossRef]
- Wang, Y.; Ding, X.; Chen, Y.; Guo, M.; Zhang, Y.; Guo, X.; Gu, H. Antibiotic-loaded, silver core-embedded mesoporous silica nanovehicles as a synergistic antibacterial agent for the treatment of drug-resistant infections. Biomaterials 2016, 101, 207–216. [Google Scholar] [CrossRef]
- Manzano, M.; Vallet-Regi, M. Ultrasound responsive mesoporous silica nanoparticles for biomedical applications. Chem. Commun. 2019, 55, 2731–2740. [Google Scholar] [CrossRef] [Green Version]
- Teng, Z.; Wang, C.; Tang, Y.; Li, W.; Bao, L.; Zhang, X.; Su, X.; Zhang, F.; Zhang, J.; Wang, S.; et al. Deformable Hollow Periodic Mesoporous Organosilica Nanocapsules for Significantly Improved Cellular Uptake. J. Am. Chem. Soc. 2018, 140, 1385–1393. [Google Scholar] [CrossRef]
- Lee, C.-H.; Lo, L.-W.; Mou, C.-Y.; Yang, C.-S. Synthesis and Characterization of Positive-Charge Functionalized Mesoporous Silica Nanoparticles for Oral Drug Delivery of an Anti-Inflammatory Drug. Adv. Funct. Mater. 2008, 18, 3283–3292. [Google Scholar] [CrossRef]
- Chernousova, S.; Epple, M. Silver as Antibacterial Agent: Ion, Nanoparticle, and Metal. Angew. Chem. Int. Ed. 2013, 52, 1636–1653. [Google Scholar] [CrossRef] [PubMed]
- Franci, G.; Falanga, A.; Galdiero, S.; Palomba, L.; Rai, M.; Morelli, G.; Galdiero, M. Silver nanoparticles as potential antibacterial agents. Molecules 2015, 20, 8856–8874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, D.; Wang, Q.; Yang, T.; Cao, J.; Lin, Q.; Yuan, Z.; Li, L. Polyethyleneimine Capped Silver Nanoclusters as Efficient Antibacterial Agents. Int. J. Environ. Res. Public Health 2016, 13, 334. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rai, M.K.; Deshmukh, S.D.; Ingle, A.P.; Gade, A.K. Silver nanoparticles: The powerful nanoweapon against multidrug-resistant bacteria. J. Appl. Microbiol. 2012, 112, 841–852. [Google Scholar] [CrossRef] [PubMed]
- Kalan, L.R.; Pepin, D.M.; Ul-Haq, I.; Miller, S.B.; Hay, M.E.; Precht, R.J. Targeting biofilms of multidrug-resistant bacteria with silver oxynitrate. Int. J. Antimicrob. Agents 2017, 49, 719–726. [Google Scholar] [CrossRef] [PubMed]
- Vankayala, R.; Sagadevan, A.; Vijayaraghavan, P.; Kuo, C.-L.; Hwang, K.C. Metal Nanoparticles Sensitize the Formation of Singlet Oxygen. Angew. Chem. Int. Ed. 2011, 50, 10640–10644. [Google Scholar] [CrossRef] [PubMed]
- Sondi, I.; Salopek-Sondi, B. Silver nanoparticles as antimicrobial agent: A case study on E. coli as a model for Gram-negative bacteria. J. Colloid Interface Sci. 2004, 275, 177–182. [Google Scholar] [CrossRef]
- Choi, O.; Hu, Z. Size Dependent and Reactive Oxygen Species Related Nanosilver Toxicity to Nitrifying Bacteria. Environ. Sci. Technol. 2008, 42, 4583–4588. [Google Scholar] [CrossRef]
- Jung, W.K.; Koo, H.C.; Kim, K.W.; Shin, S.; Kim, S.H.; Park, Y.H. Antibacterial Activity and Mechanism of Action of the Silver Ion in Staphylococcus aureus and Escherichia coli. Appl. Environ. Microbiol. 2008, 74, 2171–2178. [Google Scholar] [CrossRef] [Green Version]
- Lee, H.J.; Lee, S.G.; Oh, E.J.; Chung, H.Y.; Han, S.I.; Kim, E.J.; Seo, S.Y.; Ghim, H.D.; Yeum, J.H.; Choi, J.H. Antimicrobial polyethyleneimine-silver nanoparticles in a stable colloidal dispersion. Colloids Surf. B 2011, 88, 505–511. [Google Scholar] [CrossRef]
- Xu, P.; Liang, J.; Cao, X.; Tang, J.; Gao, J.; Wang, L.; Shao, W.; Gao, Q.; Teng, Z. Facile synthesis of monodisperse of hollow mesoporous SiO2 nanoparticles and in-situ growth of Ag nanoparticles for antibacterial. J. Colloid Interface Sci. 2016, 474, 114–118. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Qi, J.; Zhang, W.; Cai, Q.; Jiang, X. Facile, One-Pot Synthesis, and Antibacterial Activity of Mesoporous Silica Nanoparticles Decorated with Well-Dispersed Silver Nanoparticles. ACS Appl. Mater. Interfaces 2014, 6, 12038–12045. [Google Scholar] [CrossRef] [PubMed]
- Kuthati, Y.; Kankala, R.K.; Lin, S.-X.; Weng, C.-F.; Lee, C.-H. pH-Triggered Controllable Release of Silver–Indole-3 Acetic Acid Complexes from Mesoporous Silica Nanoparticles (IBN-4) for Effectively Killing Malignant Bacteria. Mol. Pharm. 2015, 12, 2289–2304. [Google Scholar] [CrossRef] [PubMed]
- Kankala, R.K.; Liu, C.-G.; Chen, A.-Z.; Wang, S.-B.; Xu, P.-Y.; Mende, L.K.; Liu, C.-L.; Lee, C.-H.; Hu, Y.-F. Overcoming Multidrug Resistance through the Synergistic Effects of Hierarchical pH-Sensitive, ROS-Generating Nanoreactors. ACS Biomater. Sci. Eng. 2017, 3, 2431. [Google Scholar] [CrossRef]
- Yuan, Z.; Cai, N.; Du, Y.; He, Y.; Yeung, E.S. Sensitive and selective detection of copper ions with highly stable polyethyleneimine-protected silver nanoclusters. Anal. Chem. 2014, 86, 419–426. [Google Scholar] [CrossRef]
- Mosmann, T. Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. J. Immunol. Methods 1983, 65, 55–63. [Google Scholar] [CrossRef]
- Helander, I.M.; Alakomi, H.-L.; Latva-Kala, K.; Koski, P. Polyethyleneimine is an effective permeabilizer of Gram-negative bacteria. Microbiology 1997, 140, 3193. [Google Scholar] [CrossRef] [Green Version]
- Paschoal, M.A.; Tonon, C.C.; Spolidório, D.M.P.; Bagnato, V.S.; Giusti, J.S.M.; Santos-Pinto, L. Photodynamic potential of curcumin and blue LED against Streptococcus mutans in a planktonic culture. Photodiagn. Photodyn. Ther. 2013, 10, 313–319. [Google Scholar] [CrossRef]
- Aranda, A.; Sequedo, L.; Tolosa, L.; Quintas, G.; Burello, E.; Castell, J.V.; Gombau, L. Dichloro-dihydro-fluorescein diacetate (DCFH-DA) assay: A quantitative method for oxidative stress assessment of nanoparticle-treated cells. Toxicol. Vitro 2013, 27, 954–963. [Google Scholar] [CrossRef]
- Chakotiya, A.S.; Tanwar, A.; Narula, A.; Sharma, R.K. Alternative to antibiotics against Pseudomonas aeruginosa: Effects of Glycyrrhiza glabra on membrane permeability and inhibition of efflux activity and biofilm formation in Pseudomonas aeruginosa and its in vitro time-kill activity. Microb. Pathog. 2016, 98, 98–105. [Google Scholar] [CrossRef]
- Kasibhatla, S.; Amarante-Mendes, G.P.; Finucane, D.; Brunner, T.; Bossy-Wetzel, E.; Green, D.R. Propidium Iodide (PI) Uptake Assay to Detect Apoptosis. CSH Protoc. 2006, 2006. [Google Scholar] [CrossRef] [PubMed]
- Mock, J.J.; Barbic, M.; Smith, D.R.; Schultz, D.A.; Schultz, S. Shape effects in plasmon resonance of individual colloidal silver nanoparticles. J. Chem. Phys. 2002, 116, 6755. [Google Scholar] [CrossRef]
- Lin, C.H.; Cheng, S.H.; Liao, W.N.; Wei, P.R.; Sung, P.J.; Weng, C.F.; Lee, C.H. Mesoporous silica nanoparticles for the improved anticancer efficacy of cis-platin. Int. J. Pharm. 2012, 429, 138–147. [Google Scholar] [CrossRef] [PubMed]
- Kumar Trivedi, M. Spectroscopic Characterization of Chloramphenicol and Tetracycline: An Impact of Biofield Treatment. Pharm. Anal. Acta 2015, 6. [Google Scholar] [CrossRef]
- Luo, H.W.; Chen, J.J.; Sheng, G.P.; Su, J.H.; Wei, S.Q.; Yu, H.Q. Experimental and theoretical approaches for the surface interaction between copper and activated sludge microorganisms at molecular scale. Sci. Rep. 2014, 4, 7078. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| Sample | BET Surface Area (m²/g) | Pore Volume (cm³/g) | Pore Size (nm) | Particle Size (nm) | Zeta potential (mV) |
|---|---|---|---|---|---|
| Pristine Cu-MSNs | 436 | 0.81 | 2.18 | 140.5 | 32.8 ± 1.3 |
| Cu-MSN-ext | 1317 | 1.89 | 2.76 | 187.1 | −17.5 ± 1.1 |
| Cu-MSN-TET | 1133 | 1.65 | 2.26 | 246.5 | −30.8 ± 1.5 |
| PEI-SNP@Cu-MSN-TET | 445 | 0.95 | 256.8 | 32.0 ± 0.7 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kankala, R.K.; Lin, W.-Z.; Lee, C.-H. Combating Antibiotic Resistance through the Synergistic Effects of Mesoporous Silica-Based Hierarchical Nanocomposites. Nanomaterials 2020, 10, 597. https://doi.org/10.3390/nano10030597
Kankala RK, Lin W-Z, Lee C-H. Combating Antibiotic Resistance through the Synergistic Effects of Mesoporous Silica-Based Hierarchical Nanocomposites. Nanomaterials. 2020; 10(3):597. https://doi.org/10.3390/nano10030597
Chicago/Turabian StyleKankala, Ranjith Kumar, Wei-Zhi Lin, and Chia-Hung Lee. 2020. "Combating Antibiotic Resistance through the Synergistic Effects of Mesoporous Silica-Based Hierarchical Nanocomposites" Nanomaterials 10, no. 3: 597. https://doi.org/10.3390/nano10030597






