In Situ Assembly of Nanomaterials and Molecules for the Signal Enhancement of Electrochemical Biosensors
Abstract
:1. Introduction
2. In Situ Assembly of Nanomaterials for Signal Amplification
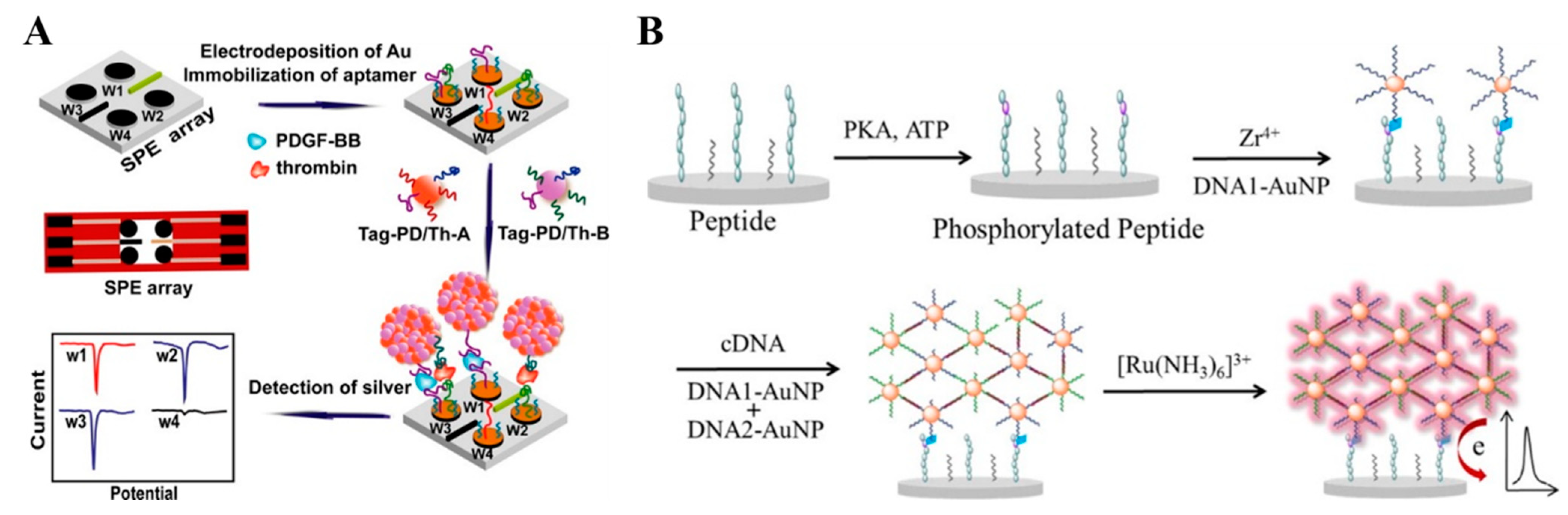
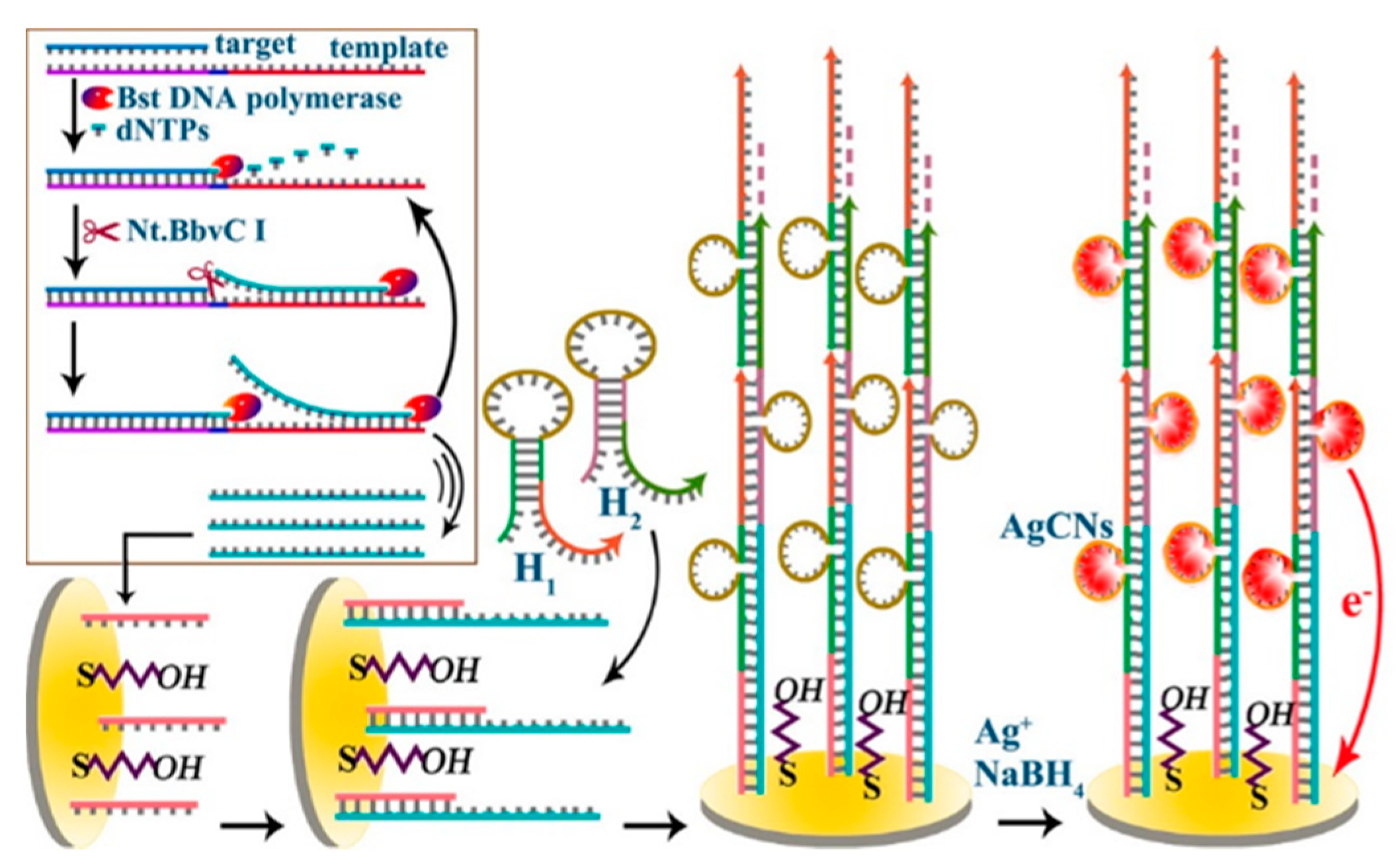
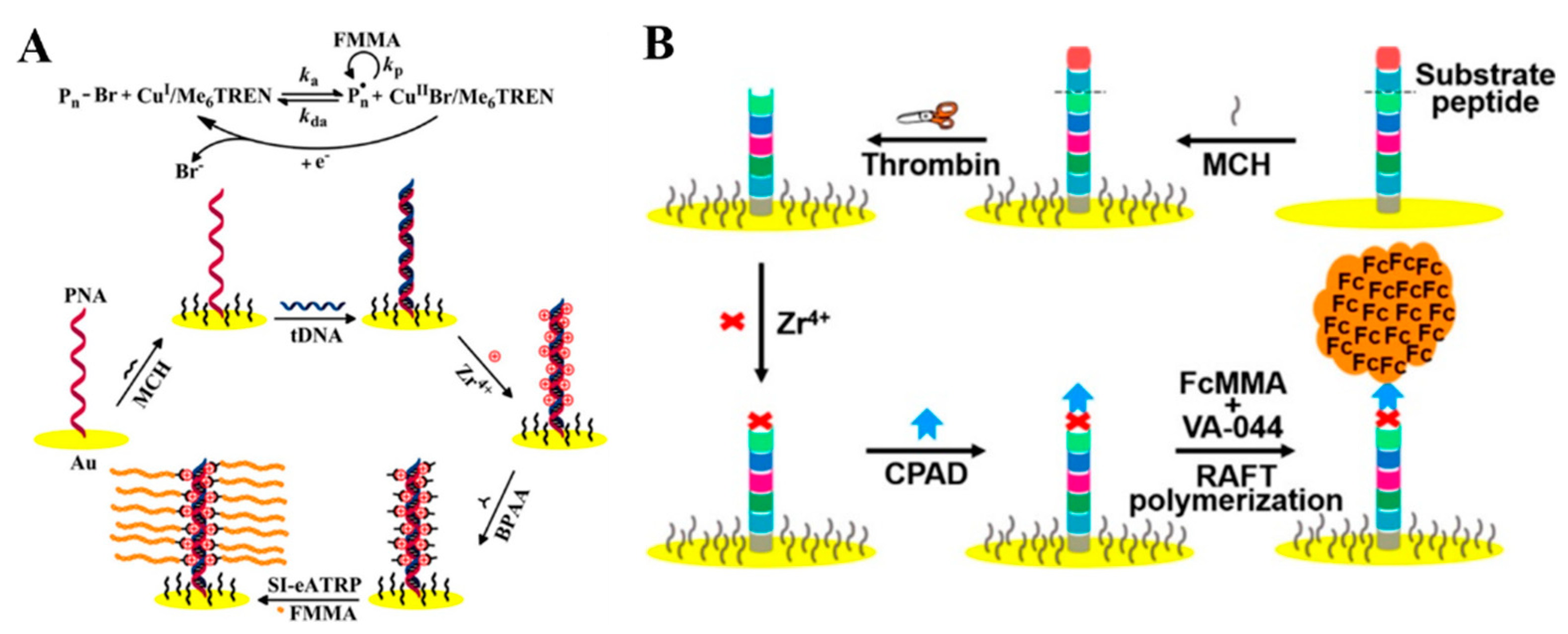
2.1. DNA Hybridization
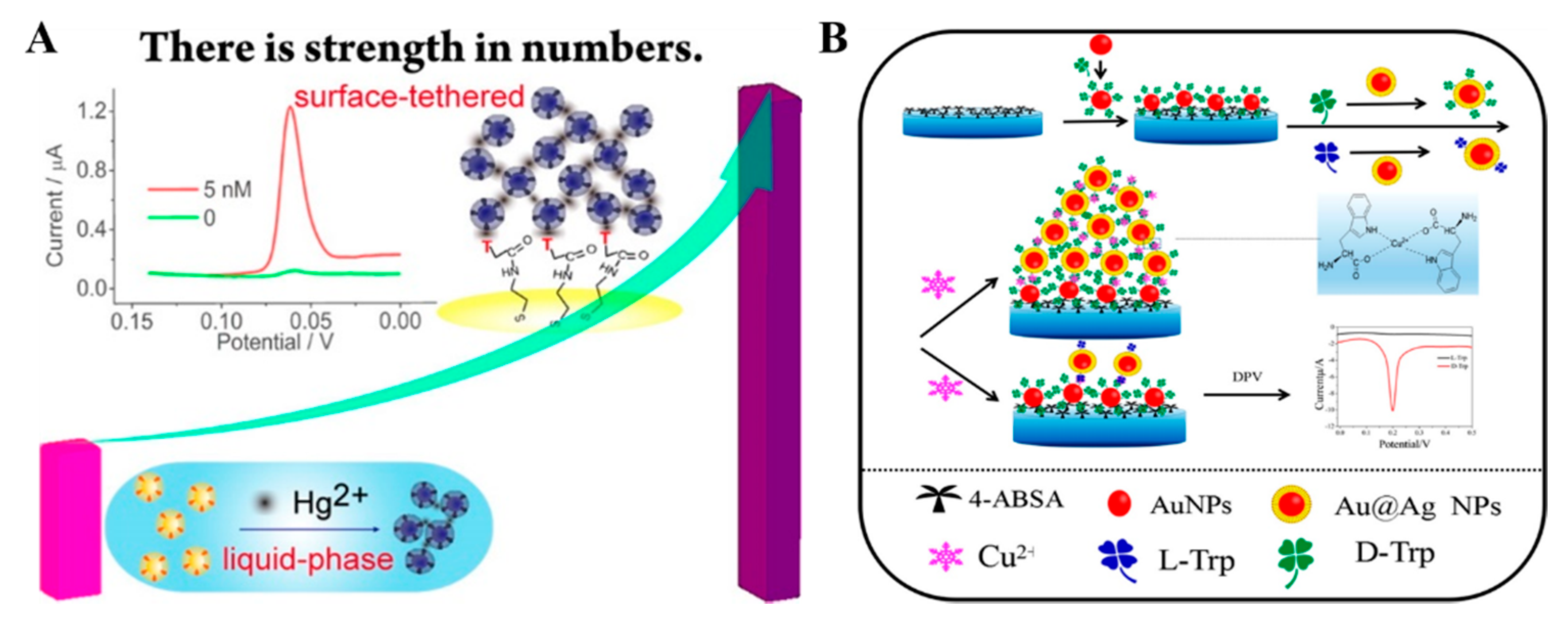
2.2. Metal Ion–Ligand Coordination
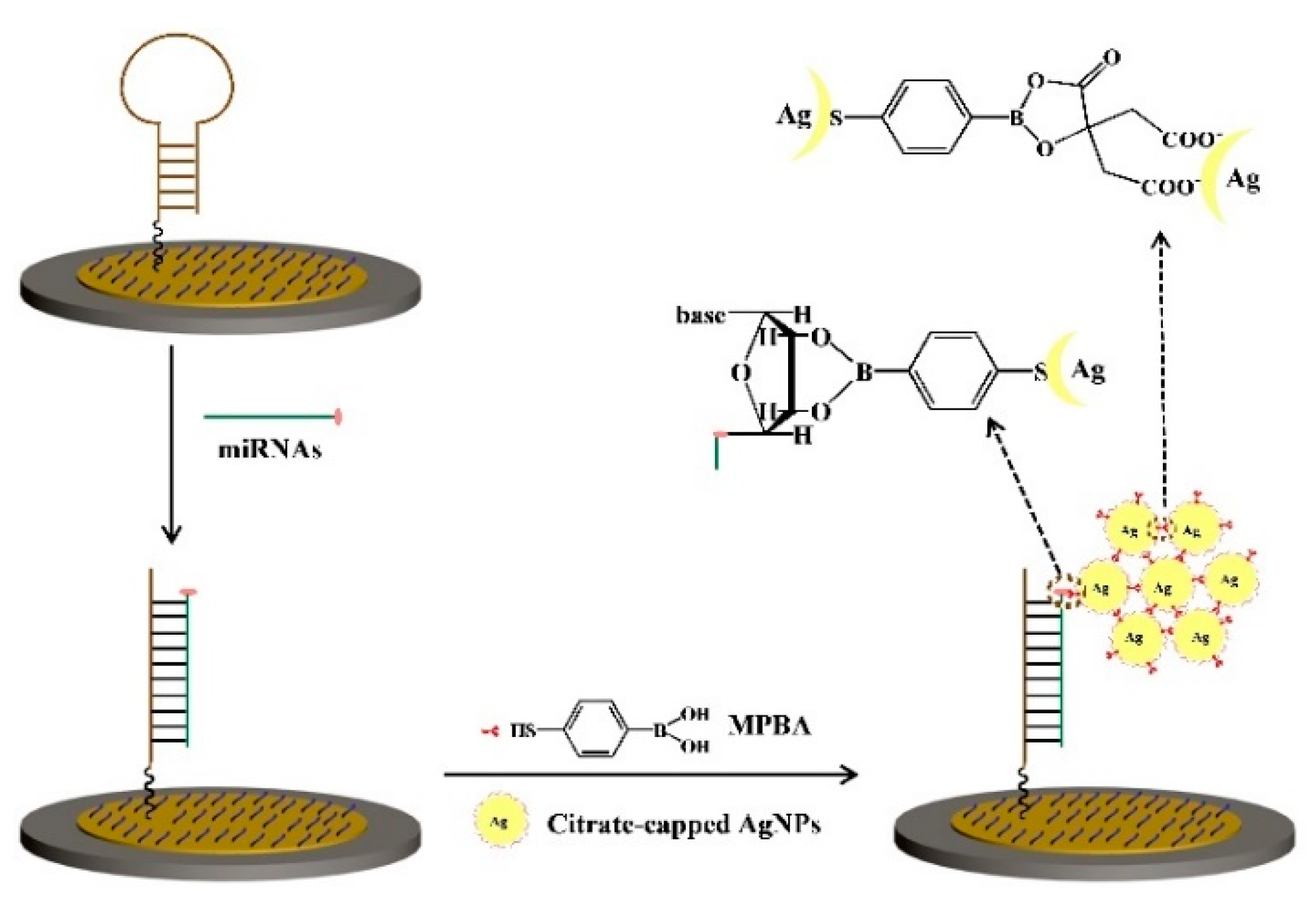
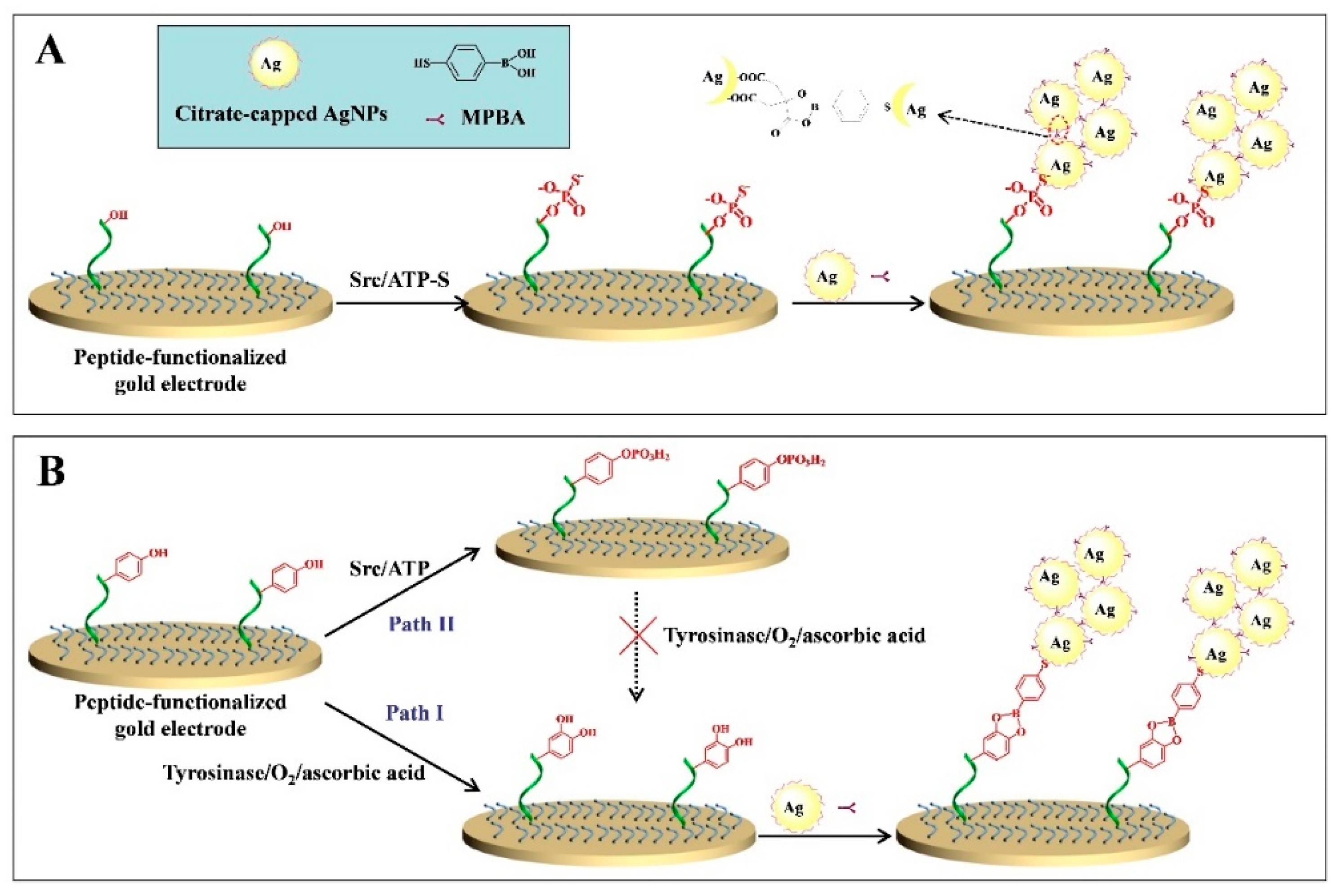
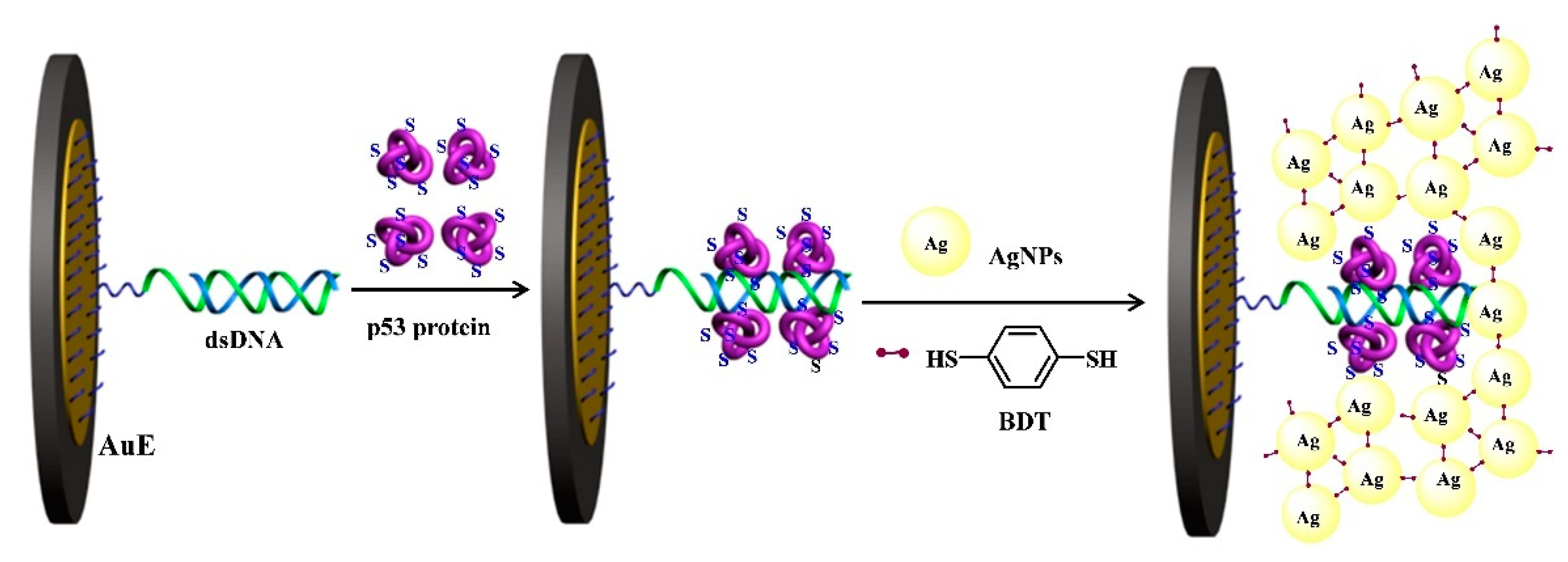
2.3. Metal–Thiol and Boronate Ester Interactions
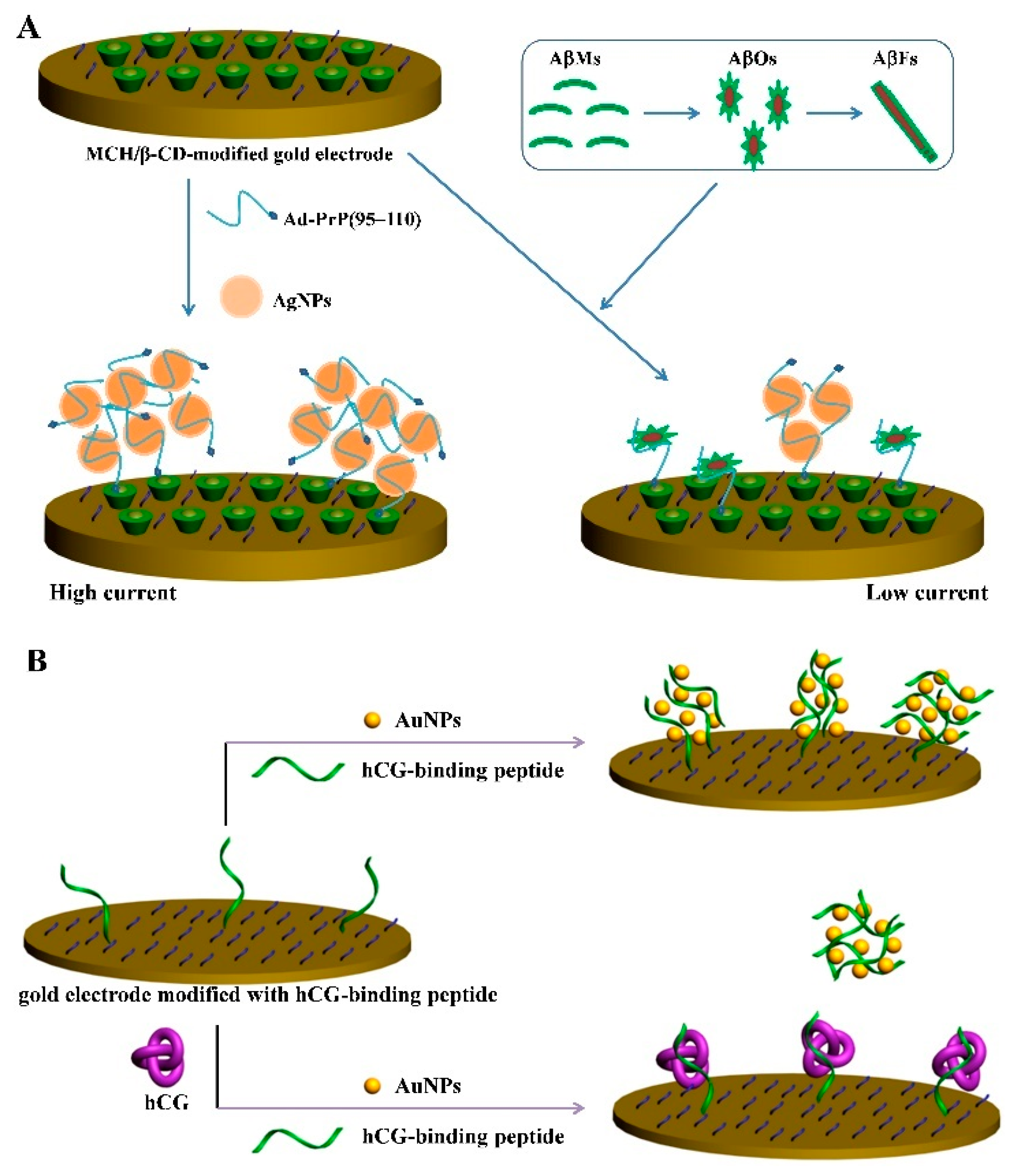
2.4. Peptide-Induced Assembly of Nanoparticles
2.5. SA–Biotin Interaction
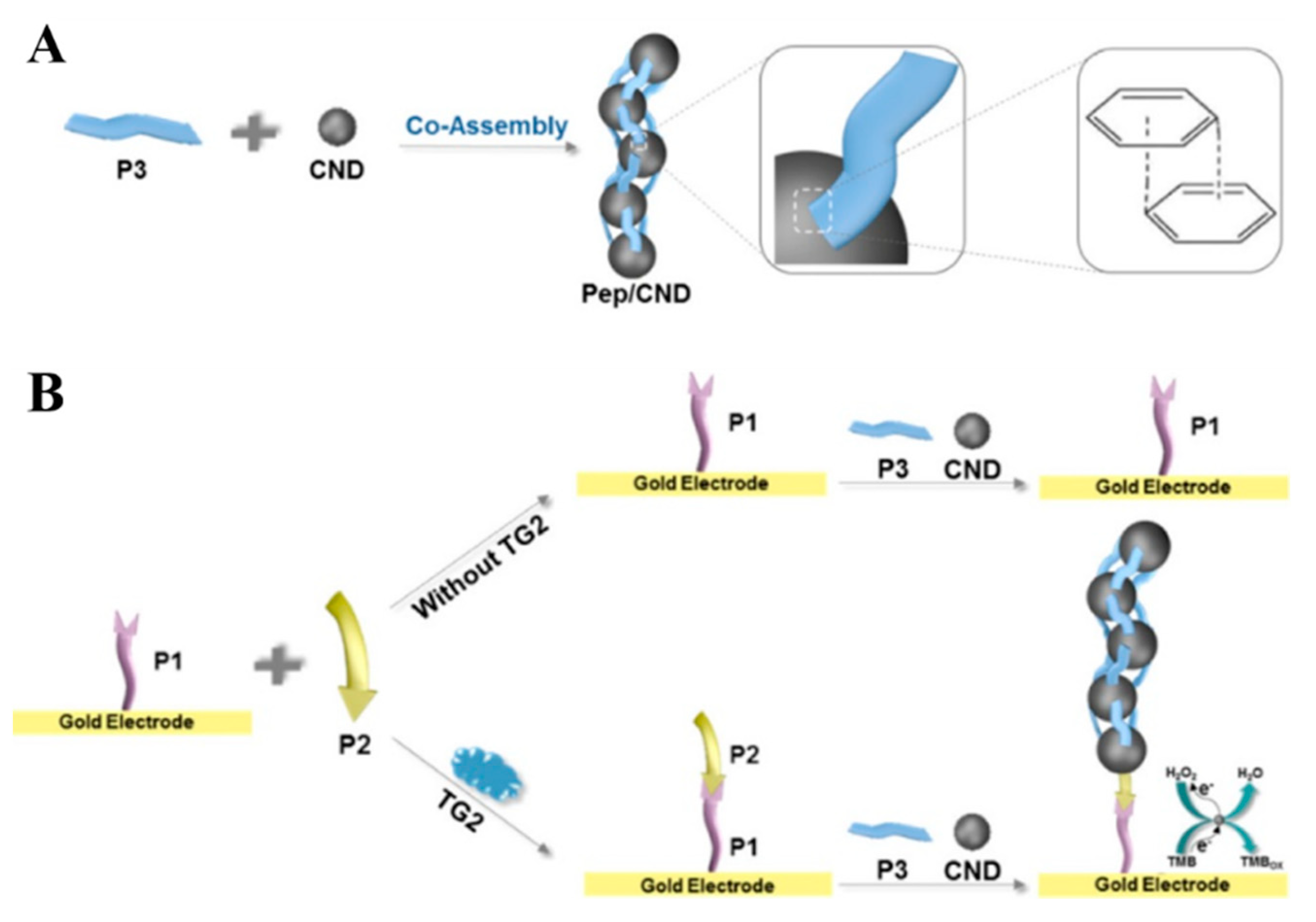
3. In Situ Assembly of Small Molecules and Biomolecules
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Singh, P.; Parent, K.L.; Buttry, D.A. Electrochemical solid-state phase transformations of silver nanoparticles. J. Am. Chem. Soc. 2012, 134, 5610–5617. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.-X.; Xiong, F.; Liu, L.-L. Electrochemical biosensors with silver nanoparticles as signal labels. Int. J. Electrochem. Sci. 2020, 15, 3869. [Google Scholar] [CrossRef]
- Xia, W.; Zhang, P.; Fu, W.; Hu, L.; Wang, Y. Aggregation/dispersion-mediated peroxidase-like activity of MoS2 quantum dots for colorimetric pyrophosphate detection. Chem. Commun. 2019, 55, 2039–2042. [Google Scholar] [CrossRef] [PubMed]
- Deng, S.Q.; Zou, H.Y.; Lan, J.; Huang, C.Z. Aggregation-induced superior peroxidase-like activity of Cu2−xSe nanoparticles for melamine detection. Anal. Methods 2016, 8, 7516–7521. [Google Scholar] [CrossRef]
- Ni, P.; Dai, H.; Wang, Y.; Sun, Y.; Shi, Y.; Hu, J.; Li, Z. Visual detection of melamine based on the peroxidase-like activity enhancement of bare gold nanoparticles. Biosens. Bioelectron. 2014, 60, 286–291. [Google Scholar] [CrossRef]
- Zhao, Y.; Huang, Y.; Zhu, H.; Zhu, Q.; Xia, Y. Three-in-one: Sensing, self-assembly, and cascade catalysis of cyclodextrin modified gold nanoparticles. J. Am. Chem. Soc. 2016, 138, 16645–16654. [Google Scholar] [CrossRef]
- Chen, Y.; Zhang, W.; Zhao, Z.; Cai, Y.; Gong, J.; Kwok, R.T.K.; Lam, J.W.Y.; Sung, H.H.Y.; Williams, I.D.; Tang, B.Z. An easily accessible ionic aggregation-induced emission luminogen with hydrogen-bonding-switchable emission and wash-free imaging ability. Angew. Chem. Int. Ed. 2018, 57, 5011–5015. [Google Scholar] [CrossRef]
- Feng, Q.; Li, Y.; Wang, L.; Li, C.; Wang, J.; Liu, Y.; Li, K.; Hou, H. Multiple-color aggregation-induced emission (AIE) molecules as chemodosimeters for pH sensing. Chem. Commun. 2016, 52, 3123–3126. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Liu, Y.; Guo, Z.; Lei, B.; Zhuang, J.; Zhang, X.; Liu, Z.; Hu, C. Hydrophobic carbon dots with blue dispersed emission and red aggregation-induced emission. Nat. Commun. 2019, 10, 1789–1800. [Google Scholar] [CrossRef]
- Mou, M.; Wu, Y.; Niu, Q.; Wang, Y.; Yan, Z.; Liao, S. Aggregation-induced emission properties of hydrothermally synthesized Cu-In-S quantum dots. Chem. Commun. 2017, 53, 3357–3360. [Google Scholar] [CrossRef]
- Rosi, N.L.; Mirkin, C.A. Nanostructures in biodiagnostics. Chem. Rev. 2005, 105, 1547–1562. [Google Scholar] [CrossRef]
- Rao, H.; Xue, X.; Luo, M.; Liu, H.; Xue, Z. Recent advances in the development of colorimetric analysis and testing based on aggregation-induced nanozymes. Chin. Chem. Lett. 2021, 32, 25–32. [Google Scholar] [CrossRef]
- Storhoff, J.J.; Elghanian, R.; Mucic, R.C.; Mirkin, C.A.; Letsinger, R.L. One-pot colorimetric differentiation of polynucleotides with single base imperfections using gold nanoparticle probes. J. Am. Chem. Soc. 1998, 120, 1959–1964. [Google Scholar] [CrossRef]
- Perez, J.M.; Simeone, F.J.; Saeki, Y.; Josephson, L.; Weissleder, R. Viral-induced self-assembly of magnetic nanoparticles allows the detection of viral particles in biological media. J. Am. Chem. Soc. 2003, 125, 10192–10193. [Google Scholar] [CrossRef]
- Liu, J.; Lu, Y. Fast colorimetric sensing of adenosine and cocaine based on a general sensor design involving aptamers and nanoparticles. Angew. Chem. Int. Ed. 2005, 45, 90–94. [Google Scholar] [CrossRef]
- Liu, Z.D.; Zhu, H.Y.; Zhao, H.X.; Huang, C.Z. Highly selective colorimetric detection of spermine in biosamples on basis of the non-crosslinking aggregation of ssDNA-capped gold nanoparticles. Talanta 2013, 106, 255–260. [Google Scholar] [CrossRef]
- Du, J.; Jiang, L.; Shao, Q.; Liu, X.; Marks, R.S.; Ma, J.; Chen, X. Colorimetric detection of mercury ions based on plasmonic nanoparticles. Small 2012, 9, 1467–1481. [Google Scholar] [CrossRef]
- Li, Y.H.; Zhang, L.; Huang, J.; Liang, R.P.; Qiu, J.D. Fluorescent graphene quantum dots with a boronic acid appended bipyridinium salt to sense monosaccharides in aqueous solution. Chem. Commun. 2013, 49, 5180–5182. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zeng, L.; Wang, Y.; Chen, T.; Chen, W.; Chen, G.; Li, C.; Chen, J. Electrochemical aptasensor based on multidirectional hybridization chain reaction for detection of tumorous exosomes. Sens. Actuators B Chem. 2021, 332, 129471–129480. [Google Scholar] [CrossRef]
- Luo, C.; Tang, H.; Cheng, W.; Yan, L.; Zhang, D.; Ju, H.; Ding, S. A sensitive electrochemical DNA biosensor for specific detection of Enterobacteriaceae bacteria by Exonuclease III-assisted signal amplification. Biosens. Bioelectron. 2013, 48, 132–137. [Google Scholar] [CrossRef]
- Li, M.; Yin, F.; Song, L.; Mao, X.; Li, F.; Fan, C.; Zuo, X.; Xia, Q. Nucleic acid tests for clinical translation. Chem. Rev. 2021, 121, 10469–10558. [Google Scholar] [CrossRef]
- Manikandan, V.S.; Adhikari, B.; Chen, A. Nanomaterial based electrochemical sensors for the safety and quality control of food and beverages. Analyst 2018, 143, 4537–4554. [Google Scholar] [CrossRef]
- Chen, A.; Chatterjee, S. Nanomaterials based electrochemical sensors for biomedical applications. Chem. Soc. Rev. 2013, 42, 5425–5438. [Google Scholar] [CrossRef]
- Zhu, C.; Yang, G.; Li, H.; Du, D.; Lin, Y. Electrochemical sensors and biosensors based on nanomaterials and nanostructures. Anal. Chem. 2015, 87, 230–249. [Google Scholar] [CrossRef]
- Sun, X.; Jian, Y.; Wang, H.; Ge, S.; Yan, M.; Yu, J. Ultrasensitive microfluidic paper-based electrochemical biosensor based on molecularly imprinted film and boronate affinity sandwich assay for glycoprotein detection. ACS Appl. Mater. Interfaces 2019, 11, 16198–16206. [Google Scholar] [CrossRef]
- Chen, J.; Yu, C.; Gao, R.; Geng, Y.; Zhao, Y.; Niu, Y.; Zhang, L.; Yu, Y.; He, J. A palladium-platinum bimetal nanodendritic melamine network for signal amplification in voltammetric sensing of DNA. Microchim. Acta 2018, 185, 138–146. [Google Scholar] [CrossRef]
- Li, H.; Sun, Z.; Zhong, W.; Hao, N.; Xu, D.; Chen, H.Y. Ultrasensitive electrochemical detection for DNA arrays based on silver nanoparticle aggregates. Anal. Chem. 2010, 82, 5477–5483. [Google Scholar] [CrossRef]
- Wei, T.; Dong, T.; Wang, Z.; Bao, J.; Tu, W.; Dai, Z. Aggregation of individual sensing units for signal accumulation: Conversion of liquid-phase colorimetric assay into enhanced surface-tethered electrochemical analysis. J. Am. Chem. Soc. 2015, 137, 8880–8883. [Google Scholar] [CrossRef]
- Li, H.; Qiang, W.; Vuki, M.; Xu, D.; Chen, H.-Y. Fluorescence enhancement of silver nanoparticle hybrid probes and ultrasensitive detection of IgE. Anal. Chem. 2011, 83, 8945–8952. [Google Scholar] [CrossRef]
- Hyeon, J.E.; Kang, D.H.; Han, S.O. Signal amplification by a self-assembled biosensor system designed on the principle of dockerin–cohesin interactions in a cellulosome complex. Analyst 2014, 139, 4790–4793. [Google Scholar] [CrossRef]
- Chen, C.; Liu, Y.; Zheng, Z.; Zhou, G.; Ji, X.; Wang, H.; He, Z. A new colorimetric platform for ultrasensitive detection of protein and cancer cells based on the assembly of nucleic acids and proteins. Anal. Chim. Acta 2015, 880, 1–7. [Google Scholar] [CrossRef]
- Fu, C.; Jin, S.; Shi, W.; Oh, J.; Cao, H.; Jung, Y.M. Catalyzed deposition of signal reporter for highly sensitive surface-enhanced Raman spectroscopy immunoassay based on tyramine signal amplification strategy. Anal. Chem. 2018, 90, 13159–13162. [Google Scholar] [CrossRef] [Green Version]
- Zahran, M.; Khalifa, Z.; Zahran, M.A.H.; Abdel Azzem, M. Recent advances in silver nanoparticle-based electrochemical sensors for determining organic pollutants in water: A review. Mater. Adv. 2021, 2, 7350–7365. [Google Scholar] [CrossRef]
- Terracciano, M.; Rea, I.; De Stefano, L.; Rendina, I.; Oliviero, G.; Nici, F.; D’Errico, S.; Piccialli, G.; Borbone, N. Synthesis of mixed-sequence oligonucleotides on mesoporous silicon: Chemical strategies and material stability. Nanoscale Res. Lett. 2014, 9, 317–325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Terracciano, M.; De Stefano, L.; Borbone, N.; Politi, J.; Oliviero, G.; Nici, F.; Casalino, M.; Piccialli, G.; Dardano, P.; Varra, M.; et al. Solid phase synthesis of a thrombin binding aptamer on macroporous silica for label free optical quantification of thrombin. RSC Adv. 2016, 6, 86762–86769. [Google Scholar] [CrossRef]
- Cao, X.; Xu, J.; Xia, J.; Zhang, F.; Wang, Z. An electrochemical aptasensor based on the conversion of liquid-phase colorimetric assay into electrochemical analysis for sensitive detection of lysozyme. Sens. Actuators B Chem. 2018, 255, 2136–2142. [Google Scholar] [CrossRef]
- Song, W.; Li, H.; Liang, H.; Qiang, W.; Xu, D. Disposable electrochemical aptasensor array by using in situ DNA hybridization inducing silver nanoparticles aggregate for signal amplification. Anal. Chem. 2014, 86, 2775–2783. [Google Scholar] [CrossRef]
- Wang, Z.; Sun, N.; He, Y.; Liu, Y.; Li, J. DNA assembled gold nanoparticles polymeric network blocks modular highly sensitive electrochemical biosensors for protein kinase activity analysis and inhibition. Anal. Chem. 2014, 86, 6153–6159. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Wang, Y.; Jiang, L.P.; Bi, S.; Zhu, J.J. Cascade amplification-mediated in situ hot-spot assembly for microRNA detection and molecular logic gate operations. Anal. Chem. 2018, 90, 4544–4551. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Wu, H.; Su, Y.; Liang, Y.; Shu, B.; Zhang, C. Electrochemical cloth-based DNA sensors (ECDSs): A new class of electrochemical gene sensors. Anal. Chem. 2020, 92, 7708–7716. [Google Scholar] [CrossRef]
- Mei, C.; Pan, L.; Xu, W.; Xu, H.; Zhang, Y.; Li, Z.; Dong, B.; Ke, X.; McAlinden, C.; Yang, M.; et al. An ultrasensitive reusable aptasensor for noninvasive diabetic retinopathy diagnosis target on tear biomarker. Sens. Actuators B Chem. 2021, 345, 130398–130407. [Google Scholar] [CrossRef]
- Yuan, Y.H.; Wu, Y.D.; Chi, B.Z.; Wen, S.H.; Liang, R.P.; Qiu, J.D. Simultaneously electrochemical detection of microRNAs based on multifunctional magnetic nanoparticles probe coupling with hybridization chain reaction. Biosens. Bioelectron. 2017, 97, 325–331. [Google Scholar] [CrossRef]
- Miao, P.; Tang, Y.; Yin, J. MicroRNA detection based on analyte triggered nanoparticle localization on a tetrahedral DNA modified electrode followed by hybridization chain reaction dual amplification. Chem. Commun. 2015, 51, 15629–15632. [Google Scholar] [CrossRef]
- Li, Z.; Miao, X.; Xing, K.; Peng, X.; Zhu, A.; Ling, L. Ultrasensitive electrochemical sensor for Hg(2+) by using hybridization chain reaction coupled with Ag@Au core-shell nanoparticles. Biosens. Bioelectron. 2016, 80, 339–343. [Google Scholar] [CrossRef]
- Chen, X.; Wang, Y.; Zhang, J.; Zhang, Y. DNA concatemer-silver nanoparticles as a signal probe for electrochemical prostate-specific antigen detection. Analyst 2019, 144, 6313–6320. [Google Scholar] [CrossRef]
- Li, Z.; Miao, X.; Xing, K.; Zhu, A.; Ling, L. Enhanced electrochemical recognition of double-stranded DNA by using hybridization chain reaction and positively charged gold nanoparticles. Biosens. Bioelectron. 2015, 74, 687–690. [Google Scholar] [CrossRef] [PubMed]
- Dong, H.; Jin, S.; Ju, H.; Hao, K.; Xu, L.P.; Lu, H.; Zhang, X. Trace and label-free microRNA detection using oligonucleotide encapsulated silver nanoclusters as probes. Anal. Chem. 2012, 84, 8670–8674. [Google Scholar] [CrossRef]
- Chen, Z.; Liu, Y.; Xin, C.; Zhao, J.; Liu, S. A cascade autocatalytic strand displacement amplification and hybridization chain reaction event for label-free and ultrasensitive electrochemical nucleic acid biosensing. Biosens. Bioelectron. 2018, 113, 1–8. [Google Scholar] [CrossRef]
- Yang, C.; Shi, K.; Dou, B.; Xiang, Y.; Chai, Y.; Yuan, R. In situ DNA-templated synthesis of silver nanoclusters for ultrasensitive and label-free electrochemical detection of microRNA. ACS Appl. Mater. Interfaces 2015, 7, 1188–1193. [Google Scholar] [CrossRef]
- Peng, X.; Zhu, J.; Wen, W.; Bao, T.; Zhang, X.; He, H.; Wang, S. Silver nanoclusters-assisted triple-amplified biosensor for ultrasensitive methyltransferase activity detection based on AuNPs/ERGO hybrids and hybridization chain reaction. Biosens. Bioelectron. 2018, 118, 174–180. [Google Scholar] [CrossRef]
- Xie, X.; Chai, Y.; Yuan, Y.; Yuan, R. Dual triggers induced disassembly of DNA polymer decorated silver nanoparticle for ultrasensitive electrochemical Pb2+ detection. Anal. Chim. Acta 2018, 1034, 56–62. [Google Scholar] [CrossRef] [PubMed]
- Feng, C.; Guo, J.; Li, G.; Ye, B.; Zou, L. Novel multiple strand displacement reaction coupled hybridization chain reaction for label-free and ultrasensitive electrochemical Type b3a2 biosensing. Sens. Actuators B Chem. 2021, 326, 128972–128978. [Google Scholar] [CrossRef]
- Zou, Y.; Zhang, H.; Wang, Z.; Liu, Q.; Liu, Y. A novel ECL method for histone acetyltransferases (HATs) activity analysis by integrating HCR signal amplification and ECL silver clusters. Talanta 2019, 198, 39–44. [Google Scholar] [CrossRef]
- Zhao, J.; Hu, S.; Cao, Y.; Zhang, B.; Li, G. Electrochemical detection of protein based on hybridization chain reaction-assisted formation of copper nanoparticles. Biosens. Bioelectron. 2015, 66, 327–331. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, X.; Zhao, L.; Bao, T.; Wen, W.; Zhang, X.; Wang, S. Integrated amplified aptasensor with in-situ precise preparation of copper nanoclusters for ultrasensitive electrochemical detection of microRNA 21. Biosens. Bioelectron. 2017, 98, 386–391. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Han, L.; Deng, D.; Zhang, Z.; He, H.; Zhang, L.; Luo, L. 4-mercaptobenzoic acid modified silver nanoparticles-enhanced electrochemical sensor for highly sensitive detection of Cu2+. Sens. Actuators B Chem. 2019, 291, 164–169. [Google Scholar] [CrossRef]
- Ejeta, S.Y.; Imae, T. Selective colorimetric and electrochemical detections of Cr(III) pollutant in water on 3-mercaptopropionic acid-functionalized gold plasmon nanoparticles. Anal. Chim. Acta 2021, 1152, 338272–338281. [Google Scholar] [CrossRef]
- Gu, Y.; Jiang, Z.; Ren, D.; Shang, Y.; Hu, Y.; Yi, L. Electrochemiluminescence sensor based on the target recognition-induced aggregation of sensing units for Hg2+ determination. Sens. Actuators B Chem. 2021, 337, 129821–129826. [Google Scholar] [CrossRef]
- Zhao, Y.; Cui, L.; Ke, W.; Zheng, F.; Li, X. Electroactive Au@Ag nanoparticle assembly driven signal amplification for ultrasensitive chiral recognition of d-/l-Trp. ACS Sustain. Chem. Eng. 2019, 7, 5157–5166. [Google Scholar] [CrossRef]
- Wang, N.; Dai, H.; Sai, L.; Ma, H.; Lin, M. Copper ion-assisted gold nanoparticle aggregates for electrochemical signal amplification of lipopolysaccharide sensing. Biosens. Bioelectron. 2019, 126, 529–534. [Google Scholar] [CrossRef] [PubMed]
- Xia, N.; Liu, L.; Chang, Y.; Hao, Y.; Wang, X. 4-mercaptophenylboronic acid-induced in situ formation of silver nanoparticle aggregates as labels on an electrode surface. Electrochem. Commun. 2017, 74, 28–32. [Google Scholar] [CrossRef]
- Liu, L.; Sun, T.; Ren, H. Electrochemical detection of hydrogen peroxide by inhibiting the p-benzenediboronic acid-triggered assembly of citrate-capped Au/Ag nanoparticles on electrode surface. Materials 2017, 10, 40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xia, N.; Wu, D.; Sun, T.; Wang, Y.; Ren, X.; Zhao, F.; Liu, L.; Yi, X. Magnetic bead-based electrochemical and colorimetric methods for the detection of poly(ADP-ribose) polymerase-1 with boronic acid derivatives as the signal probes. Sens. Actuators B Chem. 2021, 327, 128913–128920. [Google Scholar] [CrossRef]
- Xia, N.; Wu, D.; Yu, H.; Sun, W.; Yi, X.; Liu, L. Magnetic bead-based electrochemical and colorimetric assays of circulating tumor cells with boronic acid derivatives as the recognition elements and signal probes. Talanta 2021, 221, 121640–121647. [Google Scholar] [CrossRef]
- Shinde, S.; Kim, D.-Y.; Saratale, R.G.; Syed, A.; Ameen, F.; Ghodake, G. A spectral probe for detection of aluminum (III) ions using surface functionalized gold nanoparticles. Nanomaterials 2017, 7, 287. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ali, A.; Hussain, F.; Attacha, S.; Kalsoom, A.; Qureshi, W.A.; Shakeel, M.; Militky, J.; Tomkova, B.; Kremenakova, D. Development of novel antimicrobial and antiviral green synthesized silver nanocomposites for the visual detection of Fe3+ ions. Nanomaterials 2021, 11, 2076. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Chang, Y.; Xia, N.; Peng, P.; Zhang, L.; Jiang, M.; Zhang, J.; Liu, L. Simple, sensitive and label-free electrochemical detection of microRNAs based on the in situ formation of silver nanoparticles aggregates for signal amplification. Biosens. Bioelectron. 2017, 94, 235–242. [Google Scholar] [CrossRef]
- Liu, L.; Cheng, C.; Chang, Y.; Ma, H.; Hao, Y. Two sensitive electrochemical strategies for the detection of protein kinase activity based on the 4-mercaptophenylboronic acid-induced in situ assembly of silver nanoparticles. Sens. Actuators B Chem. 2017, 248, 178–186. [Google Scholar] [CrossRef]
- Xia, N.; Cheng, C.; Liu, L.; Peng, P.; Liu, C.; Chen, J. Electrochemical glycoprotein aptasensors based on the in-situ aggregation of silver nanoparticles induced by 4-mercaptophenylboronic acid. Microchim. Acta 2017, 184, 4393–4400. [Google Scholar] [CrossRef]
- Hou, L.; Huang, Y.; Hou, W.; Yan, Y.; Liu, J.; Xia, N. Modification-free amperometric biosensor for the detection of wild-type p53 protein based on the in situ formation of silver nanoparticle networks for signal amplification. Int. J. Biol. Macromol. 2020, 158, 580–586. [Google Scholar] [CrossRef] [PubMed]
- Sun, K.; Chang, Y.; Zhou, B.; Wang, X.; Liu, L. Gold nanoparticles-based electrochemical method for the detection of protein kinase with a peptide-like inhibitor as the bioreceptor. Int. J. Nanomed. 2017, 12, 1905–1915. [Google Scholar] [CrossRef] [Green Version]
- Xia, N.; Wang, X.; Wang, X.; Zhou, B. Gold nanoparticle-based colorimetric and electrochemical methods for dipeptidyl peptidase-IV activity assay and inhibitor screening. Materials 2016, 9, 857. [Google Scholar] [CrossRef] [PubMed]
- Xing, Y.; Feng, X.Z.; Zhang, L.; Hou, J.; Han, G.C.; Chen, Z. A sensitive and selective electrochemical biosensor for the determination of beta-amyloid oligomer by inhibiting the peptide-triggered in situ assembly of silver nanoparticles. Int. J. Nanomed. 2017, 12, 3171–3179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Balducci, C.; Beeg, M.; Stravalaci, M.; Bastone, A.; Sclip, A.; Biasini, E.; Tapella, L.; Colombo, L.; Manzoni, C.; Borsello, T.; et al. Synthetic amyloid-beta oligomers impair long-term memory independently of cellular prion protein. Proc. Nat. Acad. Sci. USA 2010, 107, 2295–2300. [Google Scholar] [CrossRef] [Green Version]
- Laurén, J.; Gimbel, D.A.; Nygaard, H.B.; Gilbert, J.W.; Strittmatter, S.M. Cellular prion protein mediates impairment of synaptic plasticity by amyloid-beta oligomers. Nature 2009, 457, 1128–1132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, S.; Yadav, S.P.; Surewicz, W.K. Interaction between human prion protein and amyloid-beta (Abeta) oligomers: Role of N-terminal residues. J. Biol. Chem. 2010, 285, 26377–26383. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xia, N.; Zhou, B.; Huang, N.; Jiang, M.; Zhang, J.; Liu, L. Visual and fluorescent assays for selective detection of beta-amyloid oligomers based on the inner filter effect of gold nanoparticles on the fluorescence of CdTe quantum dots. Biosens. Bioelectron. 2016, 85, 625–632. [Google Scholar] [CrossRef] [PubMed]
- Xia, N.; Wang, X.; Zhou, B.; Wu, Y.; Mao, W.; Liu, L. Electrochemical detection of amyloid-beta oligomers based on the signal amplification of a network of silver nanoparticles. ACS Appl. Mater. Interfaces 2016, 8, 19303–19311. [Google Scholar] [CrossRef] [PubMed]
- Xia, N.; Wang, X.; Yu, J.; Wu, Y.; Cheng, S.; Xing, Y.; Liu, L. Design of electrochemical biosensors with peptide probes as the receptors of targets and the inducers of gold nanoparticles assembly on electrode surface. Sens. Actuators B Chem. 2017, 239, 834–840. [Google Scholar] [CrossRef]
- Xia, N.; Chen, Z.; Liu, Y.; Ren, H.; Liu, L. Peptide aptamer-based biosensor for the detection of human chorionic gonadotropin by converting silver nanoparticles-based colorimetric assay into sensitive electrochemical analysis. Sens. Actuators B Chem. 2017, 243, 784–791. [Google Scholar] [CrossRef]
- Han, Y.; Zhang, Y.; Wu, S.; Jalalah, M.; Alsareii, S.A.; Yin, Y.; Harraz, F.A.; Li, G. Co-assembly of peptides and carbon nanodots: Sensitive analysis of transglutaminase 2. ACS Appl. Mater. Interfaces 2021, 13, 36919–36925. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.; Ma, X.; Sun, T.; Liu, L.; Hao, Y. Electrochemical detection of kinase by converting homogeneous analysis into heterogeneous assay through avidin-biotin interaction. Talanta 2021, 234, 122649–122654. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Sun, T.; Liu, L.; Xia, N.; Zhao, Y.; Yi, X. Surface plasmon resonance biosensor for the detection of miRNAs by combining the advantages of homogeneous reaction and heterogeneous detection. Talanta 2021, 234, 122622–122628. [Google Scholar] [CrossRef] [PubMed]
- La, M.; Wu, D.; Gao, Y.; Xia, N.; Niu, Y.; Liu, L.; Yi, X. Competitive impedimetric aptasensors for detection of small molecule pollutants by the signal amplification of self-assembled biotin-phenylalanine nanoparticle networks. Electrochem. Commun. 2020, 118, 106791–106796. [Google Scholar] [CrossRef]
- Xia, N.; Huang, Y.; Cui, Z.; Liu, S.; Deng, D.; Liu, L.; Wang, J. Impedimetric biosensor for assay of caspase-3 activity and evaluation of cell apoptosis using self-assembled biotin-phenylalanine network as signal enhancer. Sens. Actuators B Chem. 2020, 320, 128436–128442. [Google Scholar] [CrossRef]
- La, M.; Zhang, Y.; Gao, Y.; Li, M.; Liu, L.; Chang, Y. Impedimetric detection of microRNAs by the signal amplification of streptavidin induced in situ formation of biotin phenylalanine nanoparticle networks. J. Electrochem. Soc. 2020, 167, 117505–117512. [Google Scholar] [CrossRef]
- Hu, Q.; Wang, Q.; Kong, J.; Li, L.; Zhang, X. Electrochemically mediated in situ growth of electroactive polymers for highly sensitive detection of double-stranded DNA without sequence-preference. Biosens. Bioelectron. 2018, 101, 1–6. [Google Scholar] [CrossRef]
- Hu, Q.; Wang, Q.; Jiang, C.; Zhang, J.; Kong, J.; Zhang, X. Electrochemically mediated polymerization for highly sensitive detection of protein kinase activity. Biosens. Bioelectron. 2018, 110, 52–57. [Google Scholar] [CrossRef] [PubMed]
- Hu, Q.; Wang, Q.; Sun, G.; Kong, J.; Zhang, X. Electrochemically mediated surface-initiated de novo growth of polymers for amplified electrochemical detection of DNA. Anal. Chem. 2017, 89, 9253–9259. [Google Scholar] [CrossRef] [PubMed]
- Hu, Q.; Han, D.; Gan, S.; Bao, Y.; Niu, L. Surface-initiated-reversible-addition-fragmentation-chain-transfer polymerization for electrochemical DNA biosensing. Anal. Chem. 2018, 90, 12207–12213. [Google Scholar] [CrossRef]
- Hu, Q.; Kong, J.; Han, D.; Zhang, Y.; Bao, Y.; Zhang, X.; Niu, L. Electrochemically controlled RAFT polymerization for highly sensitive electrochemical biosensing of protein kinase activity. Anal. Chem. 2019, 91, 1936–1943. [Google Scholar] [CrossRef] [PubMed]
- Hu, Q.; Su, L.; Chen, Z.; Huang, Y.; Qin, D.; Niu, L. Coenzyme-mediated electro-RAFT polymerization for amplified electrochemical interrogation of trypsin activity. Anal. Chem. 2021, 93, 9602–9608. [Google Scholar] [CrossRef]
- Hu, Q.; Bao, Y.; Gan, S.; Zhang, Y.; Han, D.; Niu, L. Amplified electrochemical biosensing of thrombin activity by RAFT polymerization. Anal. Chem. 2020, 92, 3470–3476. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Hao, L.; Yang, F.; Liu, Y.; Yang, H.; Yan, S. Ultrasensitive electrochemical detection of CYFRA 21-1 via in-situ initiated ROP signal amplification strategy. Anal. Chim. Acta 2021, 1180, 338889–338895. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Zhang, B.; Yuan, L.; Liu, L. A signal amplification strategy based on peptide self-assembly for the identification of amyloid-β oligomer. Sens. Actuators B Chem. 2021, 335, 129697–129707. [Google Scholar] [CrossRef]
- Xia, N.; Sun, Z.; Ding, F.; Wang, Y.; Sun, W.; Liu, L. Protease biosensor by conversion of a homogeneous assay into a surface-tethered electrochemical analysis based on streptavidin-biotin interactions. ACS Sens. 2021, 6, 1166–1173. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Deng, D.; Wu, D.; Hou, W.; Wang, L.; Li, N.; Sun, Z. Duplex-specific nuclease-based electrochemical biosensor for the detection of microRNAs by conversion of homogeneous assay into surface-tethered electrochemical analysis. Anal. Chim. Acta 2021, 1149, 338199–338206. [Google Scholar] [CrossRef]
- Liu, L.; Wu, D.; Zhen, S.; Lu, K.; Yi, X.; Sun, Z. Electrochemical detection of telomerase in cancer cells based on the in-situ formation of streptavidin-biotin-DNA-biotin networks for signal amplification. Sens. Actuators B Chem. 2021, 334, 129659–129665. [Google Scholar] [CrossRef]



| Interaction | Nanomaterials | Target | Detection Range | Detection Limit | Ref. |
|---|---|---|---|---|---|
| DNA hybridization | DNA-AuNPs | lysozyme | 1 pM–1 nM | 0.32 pM | [36] |
| DNA-AgNPs | PDGF and thrombin | 5 pg/mL–1000 ng/mL | 1.6 pg/mL | [37] | |
| DNA-AuNPs | PKA | 0.03–40 U/mL | 0.03 U/mL | [38] | |
| DNA-AuNPs | miR-141 | 0.1 fM–10 nM | 25.1 aM | [39] | |
| DNA-CeO2 | VEGF | 1 fg/mL–0.1 ng/mL | 0.27 fg/mL | [41] | |
| Thi-modified DNA-Fe3O4 NPs and Fc-CHO-modified DNA-Fe3O4 NPs | miR-141 and miR-21 | 1 fM–1 nM | 0.44 fM for miR-141 and 0.46 fM for miR-21 | [42] | |
| DNA-AgNPs | miR-17-5p | 100 aM–100 pM | 2 aM | [43] | |
| DNA-based electrostatic interaction | Ag@Au CSNPs | Hg2+ | 10 pM–2.5 nM | 3.6 pM | [44] |
| CTAB-capped AgNPs | PSA | 0.1 pg/mL–75 ng/mL | 0.033 pg/mL | [45] | |
| AuNPs | DNA | 15 pM–1.0 nM | 2.6 pM | [46] | |
| DNA-based in situ metallization | AgNCs | DNA | 0.2 fM–1 pM | 0.16 fM | [48] |
| AgNCs | miR-199a | 1.0 fM–0.1 nM | 0.64 fM | [49] | |
| AgNCs | methyltransferase | 0.02–10 U/mL | 0.0073 U/mL | [50] | |
| AgNPs | Pb2+ | 1 pM–100 nM | 0.24 pM | [51] | |
| AgNPs | Type b3a2 | 10 fM–10 nM | 0.56 fM | [52] | |
| AgNCs | HAT | 0.5–100 nM | 0.49 nM | [53] | |
| CuNPs | folate receptor | 0.01–100 ng/mL | 3 pg/mL | [54] | |
| CuNCs | miR-21 | 10 pM–0.1 fM | 10 aM | [55] | |
| Metal ion–ligand coordination | thymine-modified AgNPs | Hg2+ | 50 pM–50 nM | 5 pM | [28] |
| MBA-modified AgNPs | Cu2+ | 0.1–100 nM | 0.08 nM | [56] | |
| MPA-modified AgNPs | Cr3+ | 200–5000 ppb | 278 ppb | [57] | |
| Thymine-modified UCNPs | Hg2+ | 10 pM–100 nM | 0.4 pM | [58] | |
| Au@Ag NPs | D-tryptophan | 5 pM–1 nM | 1.21 pM | [59] | |
| L-cysteine-modified AuNPs | lipopolysaccharide | 1.0–10 pg/mL | 0.033 pg/mL | [60] | |
| Metal–thiol and boronate ester interactions | citrate-capped AgNPs | tyrosinase | 0.001–0.5 mU/mL | 0.1 mU/mL | [61] |
| citrate-capped AgNPs | thrombin | 0.025–5 ng/mL | 0.02 ng/mL | [61] | |
| citrate-capped AgNPs | H2O2 | 1 nM–0.6 μM | Not reported | [62] | |
| citrate-capped AgNPs | miR-21 | 0.1–50 fM | 20 aM | [67] | |
| citrate-capped AgNPs | tyrosine kinase | 0.1–25 ng/mL | 0.1 ng/mL | [68] | |
| citrate-capped AgNPs | PSA | 0.5–200 pg/mL | 0.2 pg/mL | [69] | |
| citrate-capped AgNPs | wild-type p53 | 0.1–100 pM | 0.1 pM | [70] | |
| Peptide-induced assembly | citrate-capped AuNPs | PKA | 0.01–1 U/mL | 20 mU/mL | [71] |
| citrate-capped AuNPs | DPP-IV | 0.001–0.5 mU/mL | 0.55 µU/mL | [72] | |
| citrate-capped AgNPs | AβO | 0.01–200 nM | 6 pM | [73] | |
| citrate-capped AgNPs | AβO | 20 pM–100 nM | 8 pM | [78] | |
| citrate-capped AuNPs | hCG | 0.001–0.2 IU/mL | 0.6 mIU/mL | [79] | |
| citrate-capped AgNPs | hCG | 0.001–0.2 IU/mL | 0.4 mIU/mL | [80] | |
| Carbon nanodots | transglutaminase 2 | 1 pg/mL–50 ng/mL | 0.25 pg/mL | [81] | |
| SA–biotin interaction | biotin-FNPs | aflatoxin B1 | 0.05–3 pg/mL | Not reported | [84] |
| biotin-FNPs | caspase-3 | 1–125 pg/mL | 1 pg/mL | [85] | |
| biotin-FNPs | miR-21 | 0.1–250 fM | 0.1 fM | [86] | |
| In situ assembly of small molecules and biomolecules | Fc derivate | DNA | 1.0 fM–1.0 nM | 0.47 fM | [87] |
| Fc derivate | PKA | 0–140 mU/mL | 1.63 mU/mL | [88] | |
| Fc derivate | DNA | 0.1 fM–0.1 nM | 0.072 fM | [89] | |
| Fc derivate | DNA | 10 aM–10 pM | 3.2 aM | [90] | |
| Fc derivate | PKA | 0–140 mU/mL | 1.02 mU/mL | [91] | |
| Fc derivate | trypsin | 25–175 μU/mL | 18.2 μU/mL | [92] | |
| Fc derivate | thrombin | 10–250 μU/mL | 2.7 μU/mL | [93] | |
| Fc derivate | CYFRA 21-1 | 1 pg/mL–1 μg/mL | 9.08 fg/mL | [94] | |
| Fc-labeled peptide | AβO | 0.005–5 μM | 0.6 nM | [95] | |
| (SA–biotin–peptide–biotin)n | caspase-3 | 0–50 pg/mL | 0.2 pg/mL | [96] | |
| (SA–biotin–DNA–biotin)n | miR-21 | 0.01–2.5 fM | 10 aM | [97] | |
| (SA–biotin–DNA–biotin)n | telomerase | 20–5000 cells/mL | 20 cells/mL | [98] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chang, Y.; Xia, N.; Huang, Y.; Sun, Z.; Liu, L. In Situ Assembly of Nanomaterials and Molecules for the Signal Enhancement of Electrochemical Biosensors. Nanomaterials 2021, 11, 3307. https://doi.org/10.3390/nano11123307
Chang Y, Xia N, Huang Y, Sun Z, Liu L. In Situ Assembly of Nanomaterials and Molecules for the Signal Enhancement of Electrochemical Biosensors. Nanomaterials. 2021; 11(12):3307. https://doi.org/10.3390/nano11123307
Chicago/Turabian StyleChang, Yong, Ning Xia, Yaliang Huang, Zhifang Sun, and Lin Liu. 2021. "In Situ Assembly of Nanomaterials and Molecules for the Signal Enhancement of Electrochemical Biosensors" Nanomaterials 11, no. 12: 3307. https://doi.org/10.3390/nano11123307






