Gold Nanoparticles Prepared with Phyllanthus emblica Fruit Extract and Bifidobacterium animalis subsp. lactis Can Induce Apoptosis via Mitochondrial Impairment with Inhibition of Autophagy in the Human Gastric Carcinoma Cell Line AGS
Abstract
:1. Introduction
2. Materials and Methods
2.1. Reagents
2.2. Biosynthesis of PEFE-AuNPs
2.3. Physicochemical Characterization of PEFE-AuNPs
2.4. Hemocompatibility Assay
2.5. Evaluation of Intracellular Uptake and Localization of PEFE-AuNPs
2.6. Cell Viability Assay
2.7. Colony Formation Analysis by Trypan Blue Staining
2.8. Hoechst 33,258 and Propidium Iodide (PI) Staining
2.9. Reactive Oxygen Species (ROS) and Mito-SOX Quantification
2.10. Cytosolic-Associated Protein Light Chain 3 (LC3) Immunofluorescence Staining
2.11. Quantitative Real-Time PCR (qRT-PCR)
2.12. Quantitative Real-Time PCR (qRT-PCR)
2.13. Statistical Analysis
3. Results and Discussion
3.1. Biosynthesis and Physiochemical Characterization of PEFE-AuNPs
3.2. Cytotoxic Effect of PEFE-AuNPs against Normal and Cancer Cells
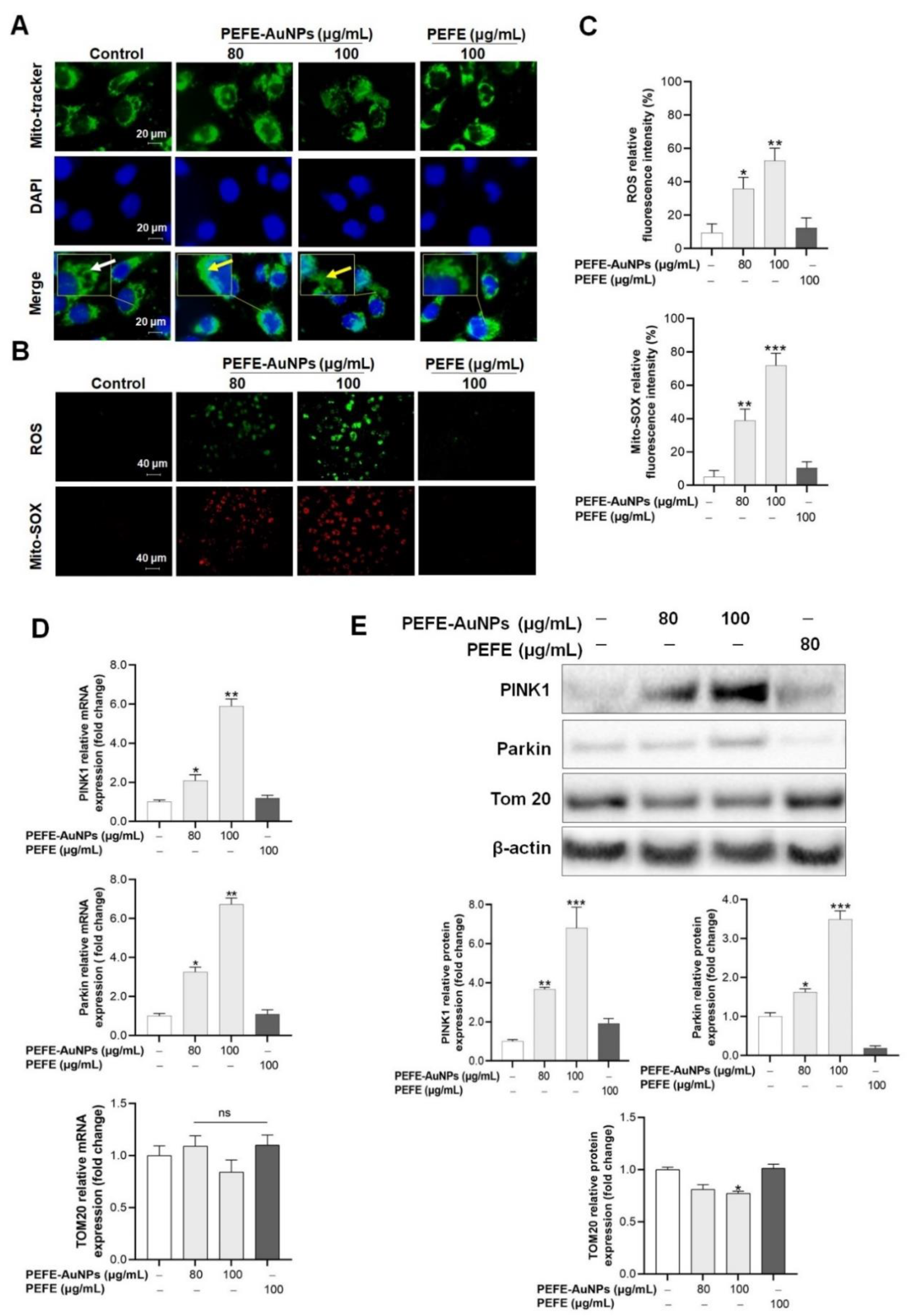
3.3. Mitochondrial Damage Is Induced by PEFE-AuNPs in AGS Cells
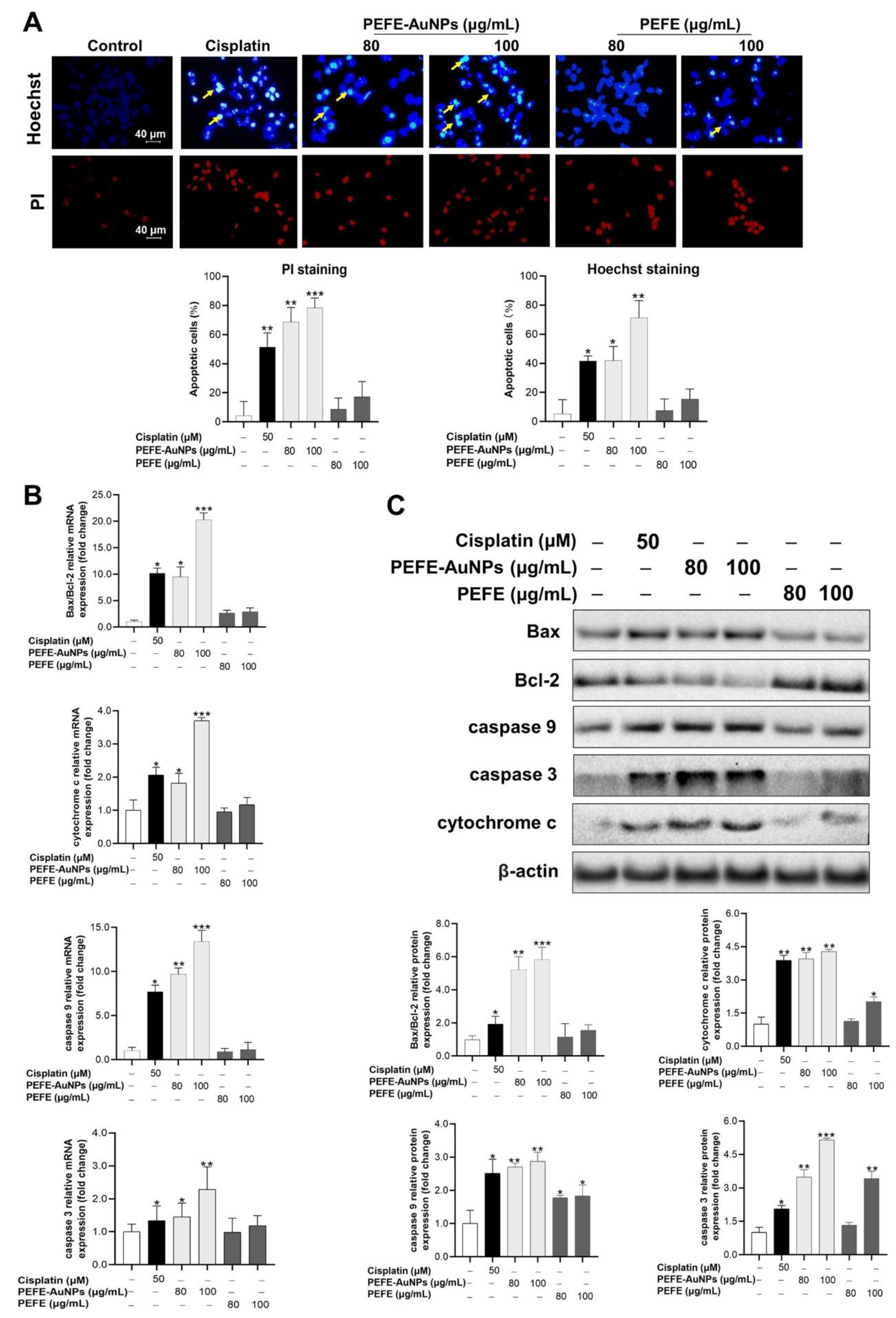
3.4. Apoptosis Is Induced by PEFE-AuNPs in AGS Cells
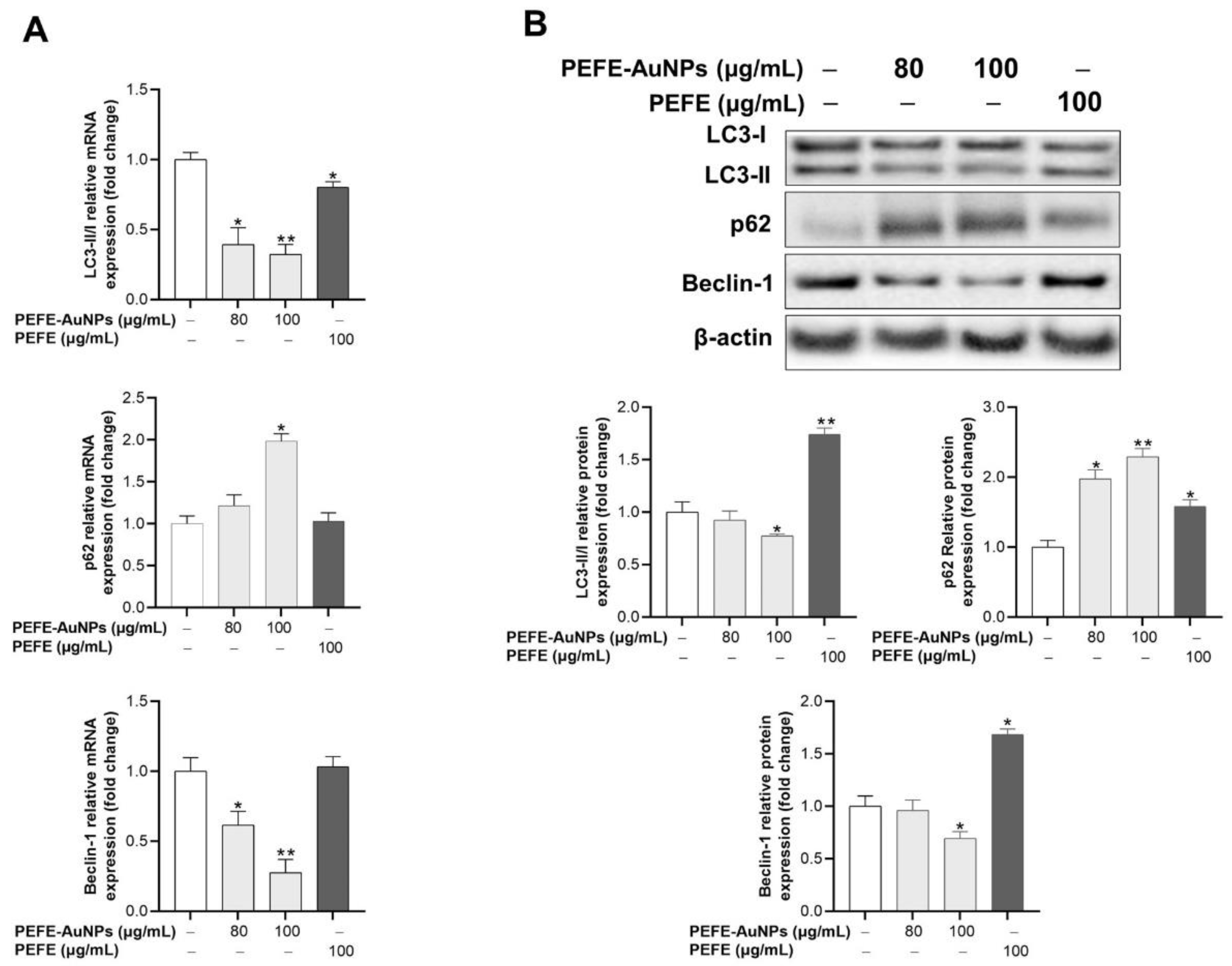
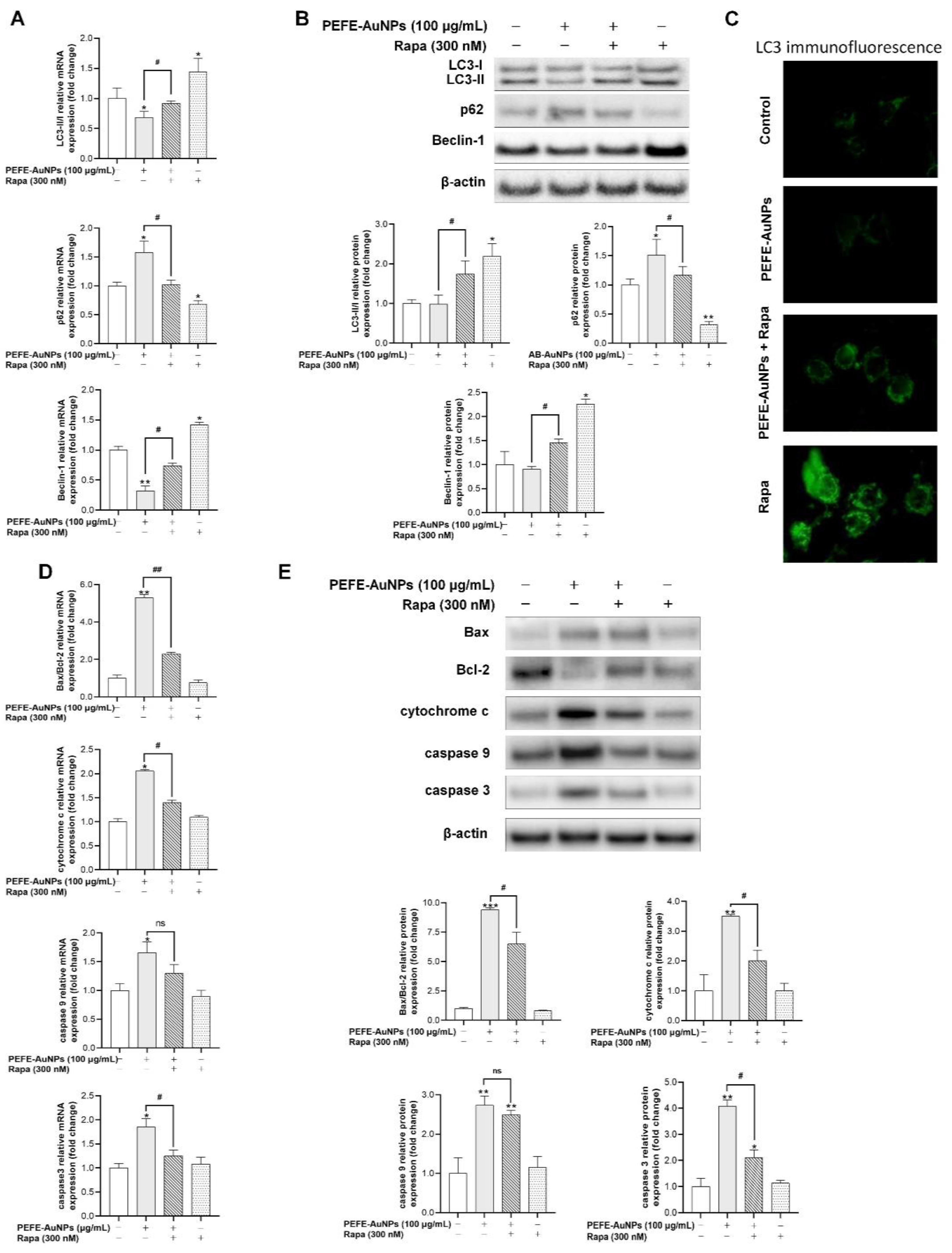
3.5. PEFE-AuNPs Promote Apoptosis by Inhibiting Autophagy in AGS Cells
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sitarz, R.; Skierucha, M.; Mielko, J.; Offerhaus, J.; Maciejewski, R.; Polkowski, W. Gastric cancer: Epidemiology, prevention, classification, and treatment. Cancer Manag. Res. 2018, 10, 239–248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jemal, A.; Bray, F.; Center, M.M.; Ferlay, J.; Ward, E.; Forman, D. Global cancer statistics. CA Cancer J. Clin. 2011, 61, 69–90. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schulte, N.; Ebert, M.P.; Härtel, N. Gastric Cancer: New Drugs—New Strategies. Gastrointest. Tumors 2015, 1, 180–194. [Google Scholar] [CrossRef] [PubMed]
- Haghighat, P.; Bekaii-Saab, T. An update on biochemotherapy of advanced gastric and gastroesophageal adenocarcinoma. J. Natl. Compr. Cancer Netw. 2008, 6, 895–900. [Google Scholar] [CrossRef]
- Dang, Y.; Liu, T.; Yan, J.; Reinhardt, J.D.; Yin, C.; Ye, F.; Zhang, G. Gastric cancer proliferation and invasion is reduced by macrocalyxin C via activation of the miR-212-3p/Sox6 Pathway. Cell. Signal. 2020, 66, 109430. [Google Scholar] [CrossRef]
- Herizchi, R.; Abbasi, E.; Milani, M.; Akbarzadeh, A. Current methods for synthesis of gold nanoparticles. Artif. Cells Nanomed. Biotechnol. 2016, 44, 596–602. [Google Scholar] [CrossRef]
- Salapa, J.; Bushman, A.; Lowe, K.; Irudayaraj, J. Nano drug delivery systems in upper gastrointestinal cancer therapy. Nano Converg. 2020, 7, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Patra, J.K.; Das, G.; Fraceto, L.F.; Campos, E.V.R.; del Pilar Rodriguez-Torres, M.; Acosta-Torres, L.S.; Diaz-Torres, L.A.; Grillo, R.; Swamy, M.K.; Sharma, S.; et al. Nano based drug delivery systems: Recent developments and future prospects. J. Nanobiotechnol. 2018, 16, 71. [Google Scholar] [CrossRef] [Green Version]
- Sekhon, B.S. Nanotechnology in agri-food production: An overview. Nanotechnol. Sci. Appl. 2014, 7, 31–53. [Google Scholar] [CrossRef] [Green Version]
- Dykman, L.; Khlebtsov, N. Gold nanoparticles in biomedical applications: Recent advances and perspectives. Chem. Soc. Rev. 2011, 41, 2256–2282. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Chaves, C.; Soto-Alvaredo, J.; Montes-Bayon, M.; Bettmer, J.; Llopis, J.; Sanchez-Gonzalez, C. Gold nanoparticles: Distribution, bioaccumulation and toxicity. In vitro and in vivo studies. Nanomed. Nanotechnol. Biol. Med. 2018, 14, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, P.M.; Vig, K.; Dennis, V.A.; Singh, S.R. Functionalized Gold Nanoparticles and Their Biomedical Applications. Nanomaterials 2011, 1, 31–63. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, S.; Ahmad, M.; Swami, B.L.; Ikram, S. A review on plants extract mediated synthesis of silver nanoparticles for antimicrobial applications: A green expertise. J. Adv. Res. 2016, 7, 17–28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahmed, S.; Ikram, S. Biosynthesis of gold nanoparticles: A green approach. J. Photochem. Photobiol. B Biol. 2016, 161, 141–153. [Google Scholar] [CrossRef]
- Durazzo, A.; Nazhand, A.; Lucarini, M.; Atanasov, A.G.; Souto, E.B.; Novellino, E.; Capasso, R.; Santini, A. An Updated Overview on Nanonutraceuticals: Focus on Nanoprebiotics and Nanoprobiotics. Int. J. Mol. Sci. 2020, 21, 2285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ali, J.; Ali, N.; Wang, L.; Waseem, H.; Pan, G. Revisiting the mechanistic pathways for bacterial mediated synthesis of noble metal nanoparticles. J. Microbiol. Methods 2019, 159, 18–25. [Google Scholar] [CrossRef] [Green Version]
- Davis, M.E.; Chen, Z.; Shin, D.M. Nanoparticle therapeutics: An emerging treatment modality for cancer. Nat. Rev. Drug Discov. 2008, 7, 771–782. [Google Scholar] [CrossRef]
- Gaire, B.P.; Subedi, L. Phytochemistry, pharmacology and medicinal properties of Phyllanthus emblica Linn. Chin. J. Integr. Med. 2014, 2014, 828. [Google Scholar] [CrossRef]
- Khopde, S.M.; Priyadarsini, K.I.; Mohan, H.; Gawandi, V.B.; Satav, J.G.; Yakhmi, J.V.; Banavaliker, M.M.; Biyani, M.K.; Mittal, J.P. Characterizing the antioxidant activity of amla (Phyllanthus emblica) extract. Curr. Aging Sci. 2001, 81, 185–190. [Google Scholar]
- Tahir, I.; Khan, M.R.; Shah, N.A.; Aftab, M. Evaluation of phytochemicals, antioxidant activity and amelioration of pulmonary fibrosis with Phyllanthus emblica leaves. BMC Complement. Altern. Med. 2016, 16, 406. [Google Scholar] [CrossRef] [Green Version]
- Zhao, T.; Sun, Q.; Marques, M.; Witcher, M. Anticancer Properties of Phyllanthus emblica (Indian Gooseberry). Oxid. Med. Cell. Longev. 2015, 2015, 950890. [Google Scholar] [CrossRef] [Green Version]
- Ihantola-Vormisto, A.; Summanen, J.; Kankaanranta, H.; Vuorela, H.; Asmawi, Z.; Moilanen, E. Anti-Inflammatory Activity of Extracts from Leaves of Phyllanthus emblica. Planta Med. 1997, 63, 518–524. [Google Scholar] [CrossRef] [PubMed]
- Dhale, D.; Mogle, U. Phytochemical screening and antibacterial activity of Phyllanthus emblica (L.). Sci. Res. Rep. 2011, 1, 138–142. [Google Scholar]
- Balusamy, S.R.; Veerappan, K.; Ranjan, A.; Kim, Y.-J.; Chellappan, D.K.; Dua, K.; Lee, J.; Perumalsamy, H. Phyllanthus emblica fruit extract attenuates lipid metabolism in 3T3-L1 adipocytes via activating apoptosis mediated cell death. Phytomedicine 2020, 66, 153129. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, P.; Vijayakumar, S.; Kothandaraman, S.; Palani, M. Anti-diabetic activity of quercetin extracted from Phyllanthus emblica L. fruit: In silico and in vivo approaches. J. Pharm. Anal. 2018, 8, 109–118. [Google Scholar] [CrossRef]
- Pramyothin, P.; Samosorn, P.; Poungshompoo, S.; Chaichantipyuth, C. The protective effects of Phyllanthus emblica Linn. extract on ethanol induced rat hepatic injury. J. Ethnopharmacol. 2006, 107, 361–364. [Google Scholar] [CrossRef]
- Markus, J.; Mathiyalagan, R.; Kim, Y.-J.; Abbai, R.; Singh, P.; Ahn, S.; Perez, Z.E.J.; Hurh, J.; Yang, D.C. Intracellular synthesis of gold nanoparticles with antioxidant activity by probiotic Lactobacillus kimchicus DCY51 T isolated from Korean kimchi. Enzym. Microb. Technol. 2016, 95, 85–93. [Google Scholar] [CrossRef]
- Kim, T.H.; Kim, W.J.; Park, S.Y.; Kim, H.; Chung, D.K. In Vitro Anti-Wrinkle and Skin-Moisturizing Effects of Evening Primrose (Oenothera biennis) Sprout and Identification of Its Active Components. Processes 2021, 9, 145. [Google Scholar] [CrossRef]
- Rao, K.J.; Paria, S. Aegle marmelos Leaf Extract and Plant Surfactants Mediated Green Synthesis of Au and Ag Nanoparticles by Optimizing Process Parameters Using Taguchi Method. ACS Sustain. Chem. Eng. 2015, 3, 483–491. [Google Scholar] [CrossRef]
- Gan, P.P.; Ng, S.H.; Huang, Y.; Li, S.F.Y. Green synthesis of gold nanoparticles using palm oil mill effluent (POME): A low-cost and eco-friendly viable approach. Bioresour. Technol. 2012, 113, 132–135. [Google Scholar] [CrossRef]
- Shameli, K.; Bin Ahmad, M.; Shabanzadeh, P.; Al-Mulla, E.A.J.; Zamanian, A.; Abdollahi, Y.; Jazayeri, S.D.; Eili, M.; Jalilian, F.A.; Haroun, R.Z. Effect of Curcuma longa tuber powder extract on size of silver nanoparticles prepared by green method. Res. Chem. Intermed. 2014, 40, 1313–1325. [Google Scholar] [CrossRef]
- Das, R.K.; Gogoi, N.; Bora, U. Green synthesis of gold nanoparticles using Nyctanthes arbortristis flower extract. Bioprocess Biosyst. Eng. 2011, 34, 615–619. [Google Scholar] [CrossRef] [PubMed]
- Bogireddy, N.K.R.; Anand, K.K.H.; Mandal, B.K. Gold nanoparticles—Synthesis by Sterculia acuminata extract and its catalytic efficiency in alleviating different organic dyes. J. Mol. Liq. 2015, 211, 868–875. [Google Scholar] [CrossRef]
- Liu, Y.; Kim, S.; Kim, Y.-J.; Perumalsamy, H.; Lee, S.; Hwang, E.; Yi, T.-H. Green synthesis of gold nanoparticles using Euphrasia officinalis leaf extract to inhibit lipopolysaccharide-induced inflammation through NF-κB and JAK/STAT pathways in RAW 264.7 macrophages. Int. J. Nanomed. 2019, 14, 2945–2959. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abbai, R.; Ramya Mathiyalagan, J.M.; Kim, Y.J.; Wang, C.; Singh, P.; Ahn, S.; Farh, M.E.A.; Yang, D.C. Green synthesis of multifunctional silver and gold nanoparticles from the oriental herbal adaptogen: Siberian ginseng. Int. J. Nanomed. 2016, 11, 3131–3143. [Google Scholar] [CrossRef] [Green Version]
- Colacio, E.; Cuesta, R.; Gutiérrez-Zorrilla, J.M.; Luque, A.; Román, P.; Giraldi, T.; Taylor, M.R. Gold(I)−Purine Interactions: Synthesis and Characterization of Cyclic and Open Chain Polynuclear Gold(I) Complexes Containing Xanthine Derivatives and Bis(phosphine) as Bridging Ligands. Crystal Structures of [Au2(μ-HX)(μ-dmpe)]·3H2O and [Au2(μ-TT)(μ-dmpe)]·H2O (H3X = Xanthine; H2TT = 8-Mercaptotheophylline). Inorg. Chem. 1996, 35, 4232–4238. [Google Scholar] [CrossRef]
- Lubran, M.M. Hematologic side effects of drugs. Ann. Clin. Lab. Sci. 1989, 19, 114–121. [Google Scholar]
- Osterberg, R.E.; See, N.A. Toxicity of Excipients—A Food and Drug Administration Perspective. Int. J. Toxicol. 2003, 22, 377–380. [Google Scholar] [CrossRef]
- Choudhury, A.J.; Gogoi, D.; Kandimalla, R.; Kalita, S.; Chaudhari, Y.B.; Khan, M.R.; Kotoky, J.; Chutia, J. Penicillin impregnation on oxygen plasma surface functionalized chitosan/ Antheraea assama silk fibroin: Studies of antibacterial activity and antithrombogenic property. Mater. Sci. Eng. C 2016, 60, 475–484. [Google Scholar] [CrossRef] [PubMed]
- Singhal, J.P.; Ray, A.R. Synthesis of blood compatible polyamide block copolymers. Biomaterials 2002, 23, 1139–1145. [Google Scholar] [CrossRef]
- Wu, S.; Zhou, F.; Zhang, Z.; Xing, D. Mitochondrial oxidative stress causes mitochondrial fragmentation via differential modulation of mitochondrial fission-fusion proteins. FEBS J. 2011, 278, 941–954. [Google Scholar] [CrossRef] [PubMed]
- Kudryavtseva, A.V.; Krasnov, G.S.; Dmitriev, A.A.; Alekseev, B.Y.; Kardymon, O.L.; Sadritdinova, A.F.; Fedorova, M.S.; Pokrovsky, A.V.; Melnikova, N.V.; Kaprin, A.D.; et al. Mitochondrial dysfunction and oxidative stress in aging and cancer. Oncotarget 2016, 7, 44879–44905. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liou, G.Y.; Storz, P. Reactive oxygen species in cancer. Free Radic. Res. 2010, 44, 479–496. [Google Scholar] [CrossRef] [Green Version]
- McWilliams, T.G.; Muqit, M.M. PINK1 and Parkin: Emerging themes in mitochondrial homeostasis. Curr. Opin. Cell Biol. 2017, 45, 83–91. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Okatsu, K.; Oka, T.; Iguchi, M.; Imamura, K.; Kosako, H.; Tani, N.; Kimura, M.; Go, E.; Koyano, F.; Funayama, M.; et al. PINK1 autophosphorylation upon membrane potential dissipation is essential for Parkin recruitment to damaged mitochondria. Nat. Commun. 2012, 3, 1016. [Google Scholar] [CrossRef] [Green Version]
- Rao, S.; Schmidt, O.; Harbauer, A.B.; Schönfisch, B.; Guiard, B.; Pfanner, N.; Meisinger, C. Biogenesis of the preprotein translocase of the outer mitochondrial membrane: Protein kinase A phosphorylates the precursor of Tom40 and impairs its import. Mol. Biol. Cell 2012, 23, 1618–1627. [Google Scholar] [CrossRef]
- Lowe, S.W.; Lin, A.W. Apoptosis in cancer. Carcinogenesis 2000, 21, 485–495. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frenzel, A.; Grespi, F.; Chmelewskij, W.; Villunger, A. Bcl2 family proteins in carcinogenesis and the treatment of cancer. Apoptosis 2009, 14, 584–596. [Google Scholar] [CrossRef] [Green Version]
- Adams, J.M.; Cory, S. The Bcl-2 apoptotic switch in cancer development and therapy. Oncogene 2007, 26, 1324–1337. [Google Scholar] [CrossRef] [Green Version]
- Mariño, G.; Niso-Santano, M.; Baehrecke, E.H.; Kroemer, G. Self-consumption: The interplay of autophagy and apoptosis. Nat. Rev. Mol. Cell Biol. 2014, 15, 81–94. [Google Scholar] [CrossRef] [Green Version]
- Tanida, I.; Ueno, T.; Kominami, E. LC3 and autophagy. Methods Mol. Biol. 2008, 445, 77–88. [Google Scholar] [CrossRef] [PubMed]
- Park, J.M.; Huang, S.; Wu, T.-T.; Foster, N.R.; Sinicrope, F.A. Prognostic impact of Beclin 1, p62/sequestosome 1 and LC3 protein expression in colon carcinomas from patients receiving 5-fluorouracil as adjuvant chemotherapy. Cancer Biol. Ther. 2013, 14, 100–107. [Google Scholar] [CrossRef] [Green Version]
- Yoshii, S.R.; Mizushima, N. Monitoring and Measuring Autophagy. Int. J. Mol. Sci. 2017, 18, 1865. [Google Scholar] [CrossRef]
- Bjørkøy, G.; Lamark, T.; Brech, A.; Outzen, H.; Perander, M.; Øvervatn, A.; Stenmark, H.; Johansen, T. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J. Cell Biol. 2005, 171, 603–614. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pankiv, S.; Clausen, T.H.; Lamark, T.; Brech, A.; Bruun, J.-A.; Outzen, H.; Øvervatn, A.; Bjørkøy, G.; Johansen, T. p62/SQSTM1 Binds Directly to Atg8/LC3 to Facilitate Degradation of Ubiquitinated Protein Aggregates by Autophagy. J. Biol. Chem. 2007, 282, 24131–24145. [Google Scholar] [CrossRef] [Green Version]
- Kang, R.; Zeh, H.J.; Lotze, M.T.; Tang, D. The Beclin 1 network regulates autophagy and apoptosis. Cell Death Differ. 2011, 18, 571–580. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, R.; Xu, X.; Puja, A.M.; Perumalsamy, H.; Balusamy, S.R.; Kim, H.; Kim, Y.-J. Gold Nanoparticles Prepared with Phyllanthus emblica Fruit Extract and Bifidobacterium animalis subsp. lactis Can Induce Apoptosis via Mitochondrial Impairment with Inhibition of Autophagy in the Human Gastric Carcinoma Cell Line AGS. Nanomaterials 2021, 11, 1260. https://doi.org/10.3390/nano11051260
Wang R, Xu X, Puja AM, Perumalsamy H, Balusamy SR, Kim H, Kim Y-J. Gold Nanoparticles Prepared with Phyllanthus emblica Fruit Extract and Bifidobacterium animalis subsp. lactis Can Induce Apoptosis via Mitochondrial Impairment with Inhibition of Autophagy in the Human Gastric Carcinoma Cell Line AGS. Nanomaterials. 2021; 11(5):1260. https://doi.org/10.3390/nano11051260
Chicago/Turabian StyleWang, Rongbo, Xingyue Xu, Aditi Mitra Puja, Haribalan Perumalsamy, Sri Renukadevi Balusamy, Hoon Kim, and Yeon-Ju Kim. 2021. "Gold Nanoparticles Prepared with Phyllanthus emblica Fruit Extract and Bifidobacterium animalis subsp. lactis Can Induce Apoptosis via Mitochondrial Impairment with Inhibition of Autophagy in the Human Gastric Carcinoma Cell Line AGS" Nanomaterials 11, no. 5: 1260. https://doi.org/10.3390/nano11051260






