A High-Quality CdSe/CdS/ZnS Quantum-Dot-Based FRET Aptasensor for the Simultaneous Detection of Two Different Alzheimer’s Disease Core Biomarkers
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals and Reagents
2.2. Instruments and Characterization
2.3. Preparation of Water-Soluble Dual-Color CdSe/CdS/ZnS Core/Shell/Shell QDs
2.4. Fabrication of the Aptamer-Functionalized QDs
2.5. Synthesis of Au NRs-CTAB
2.6. Synthesis of Au NRs@PDA
2.7. Formation of Soluble AβO
2.8. Simultaneous Detection of AβO and Tau Protein Using the FRET Aptasensor
3. Results
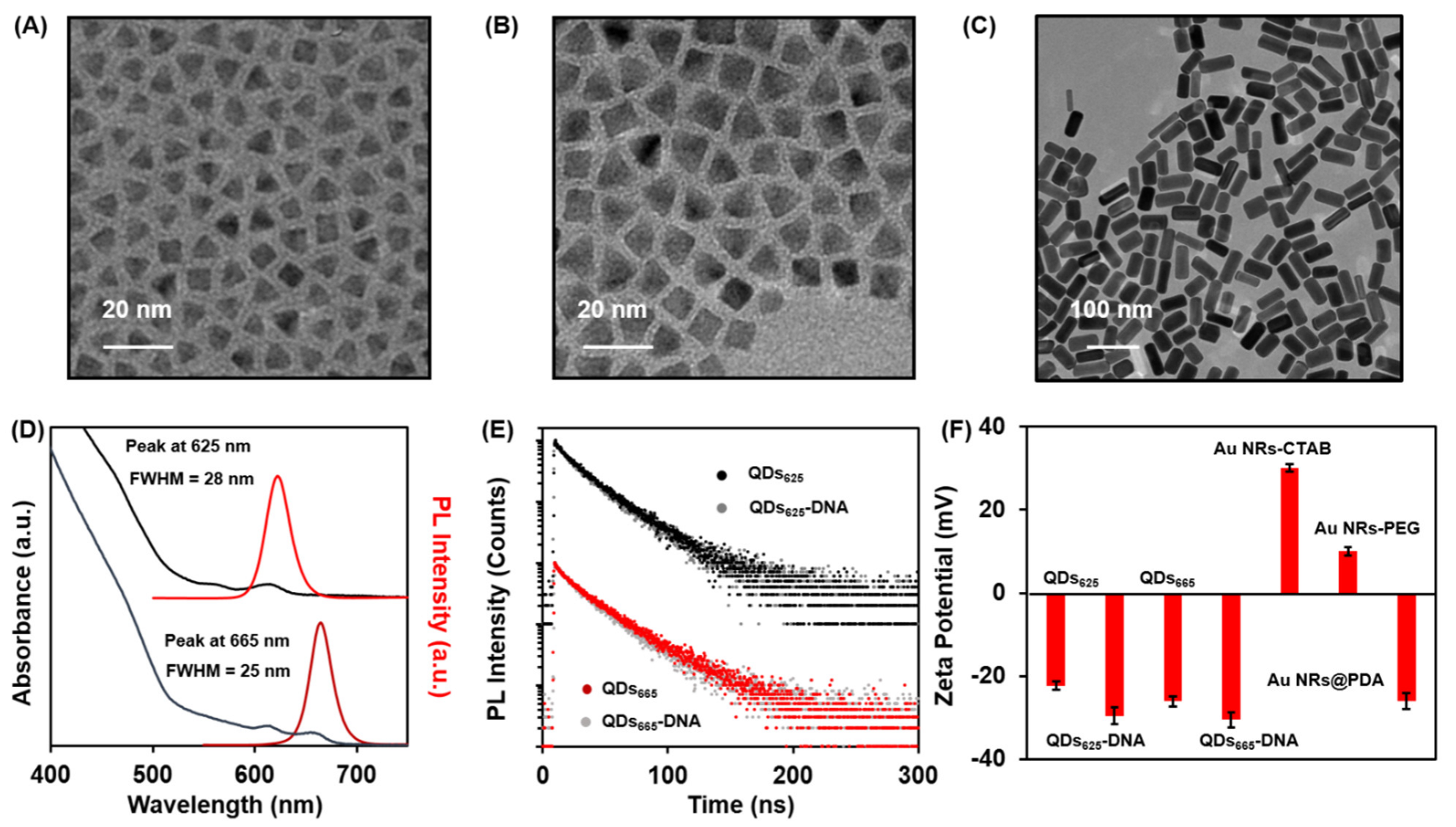
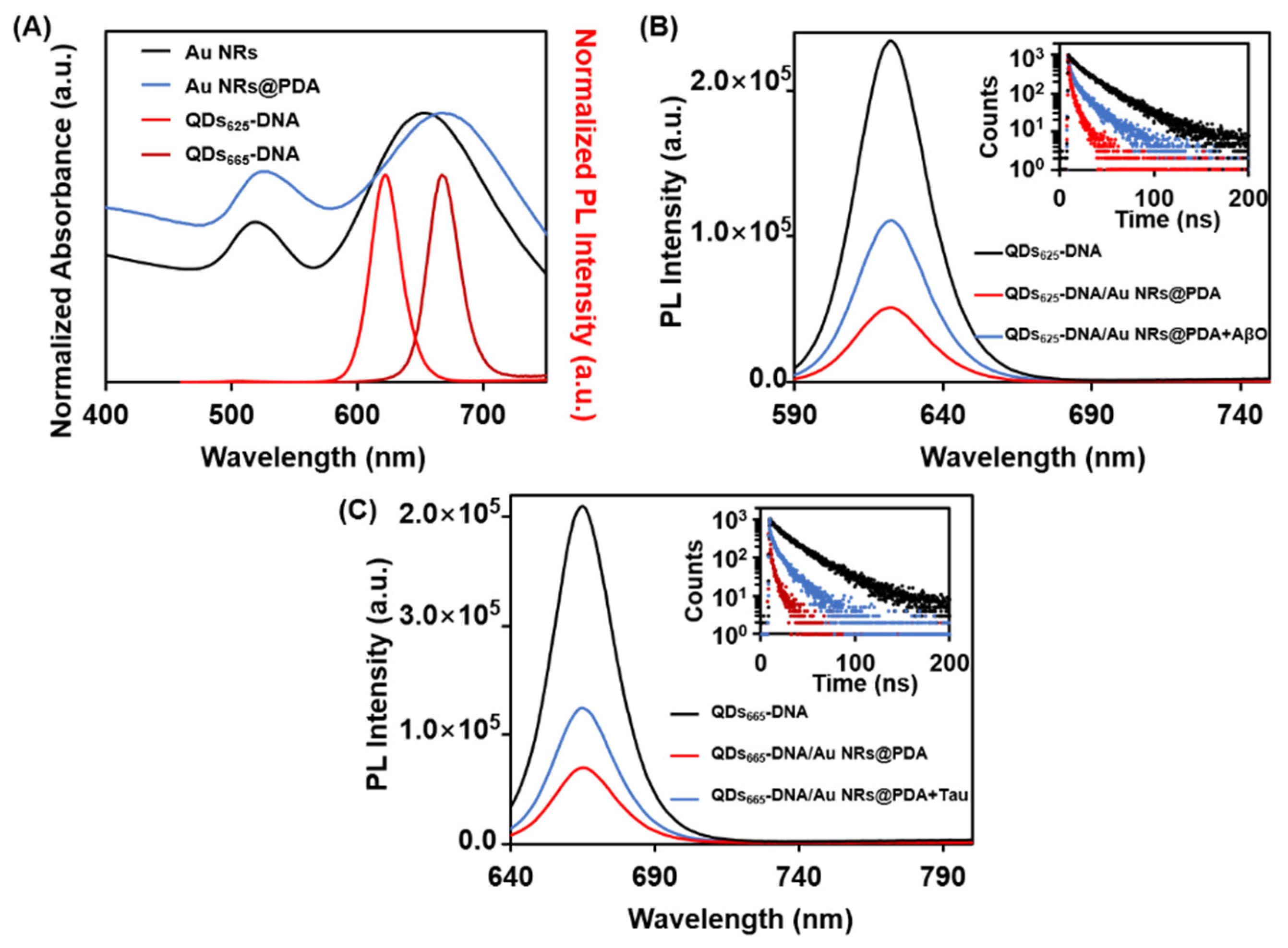
3.1. Characterization of the QDs, QDs-DNA Conjugates, and Au NRs
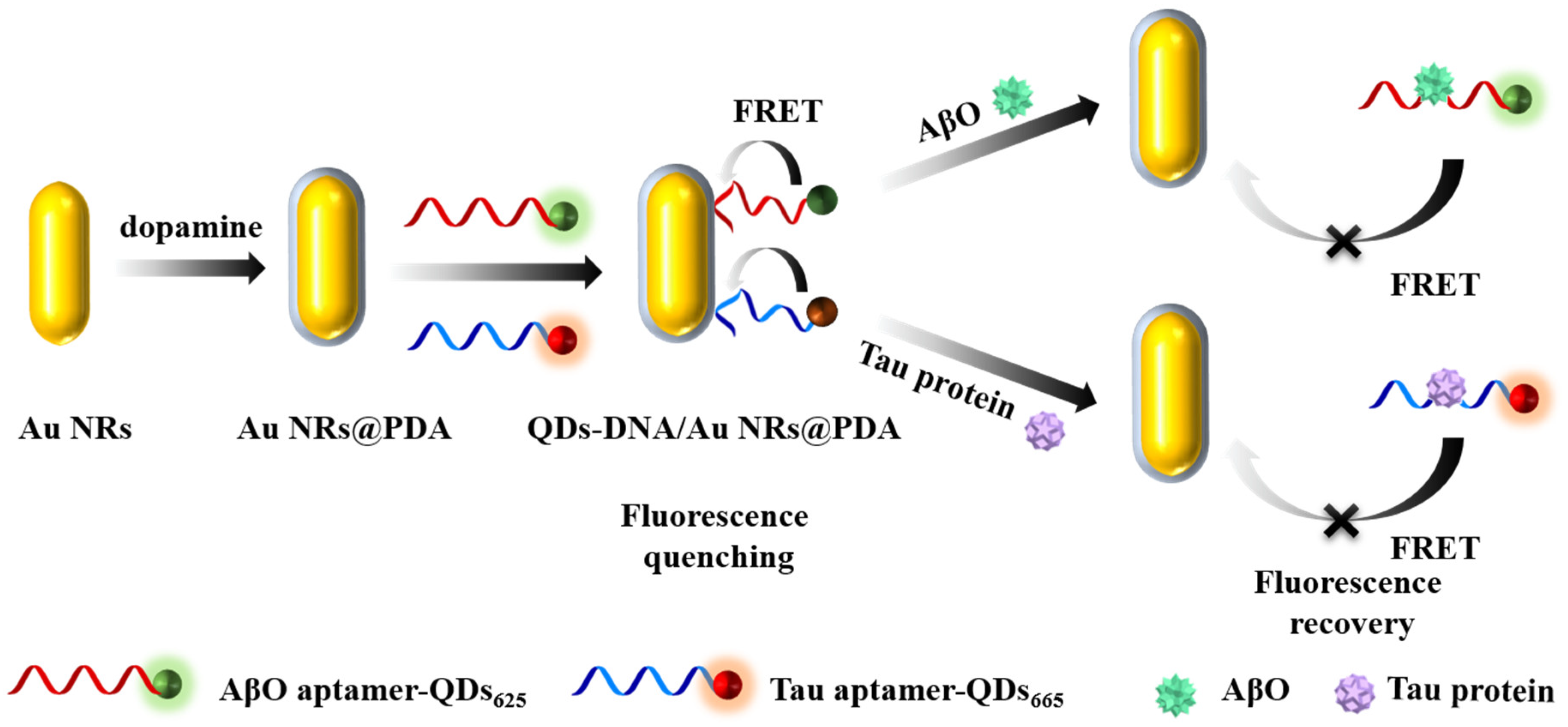
3.2. Feasibility of the Proposed Aptasensor
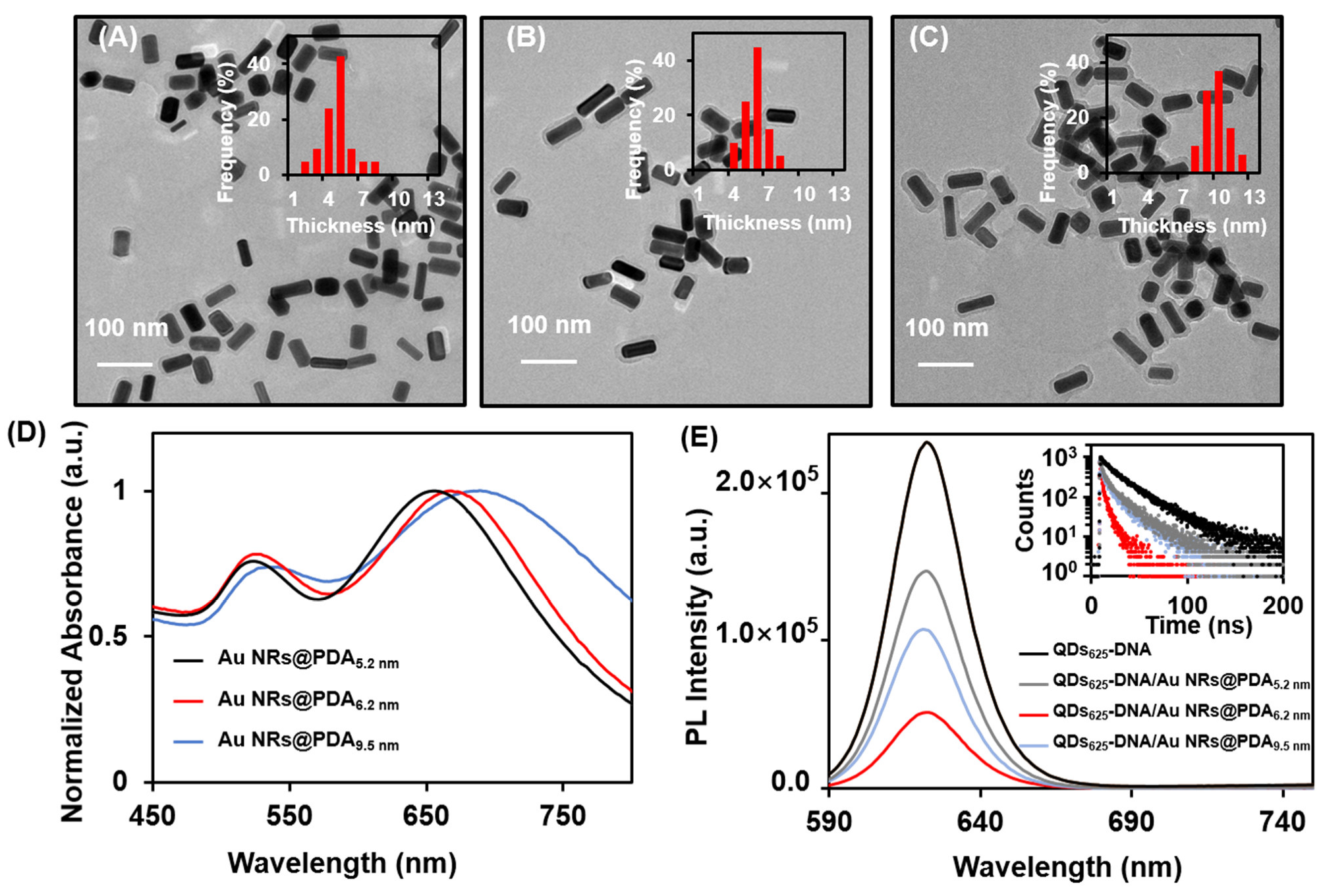
3.3. Optimization of the Experimental Conditions
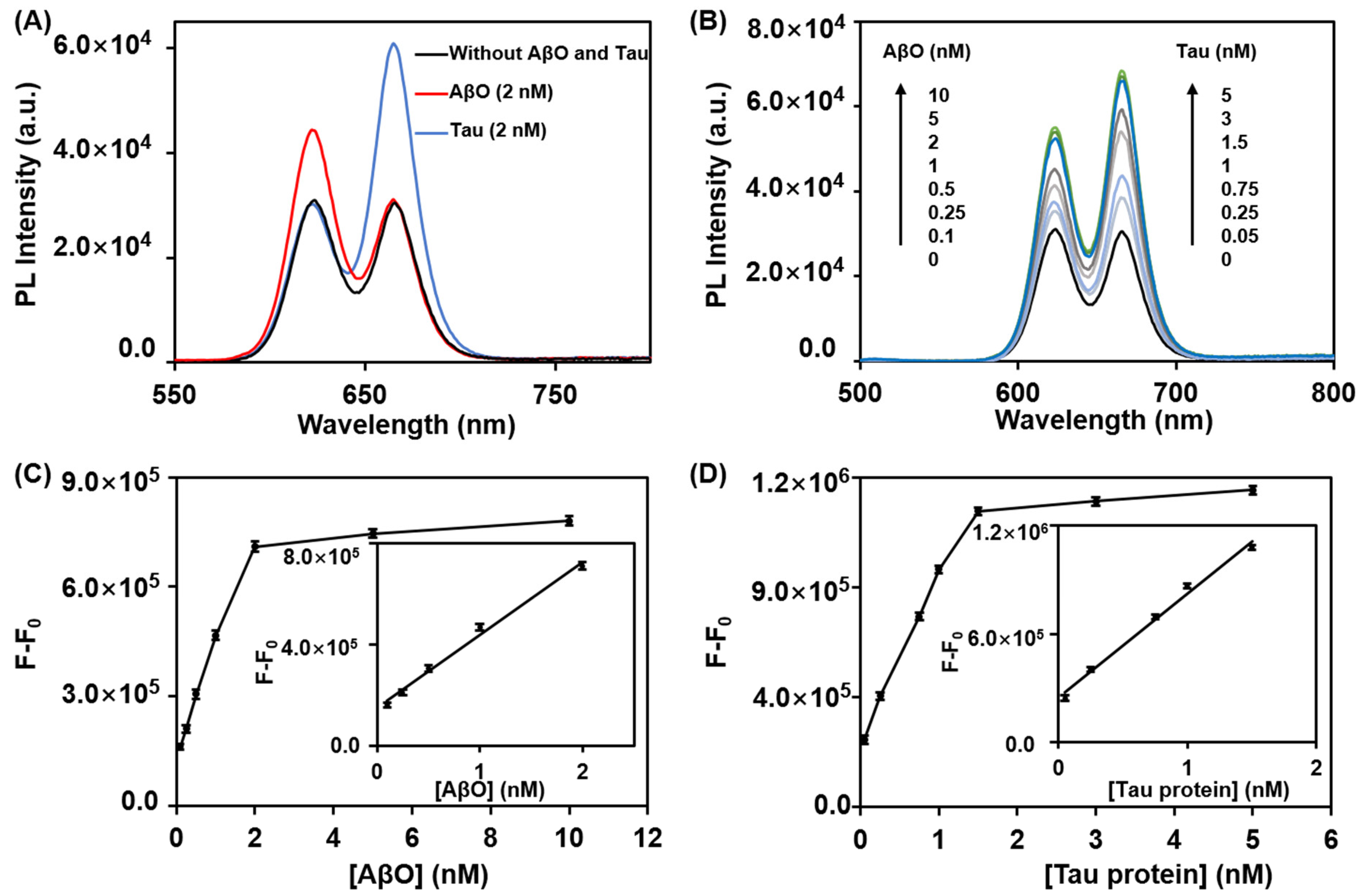
3.4. Cross-Reactivity Analysis and Simultaneous Detection of the AβO and Tau Protein
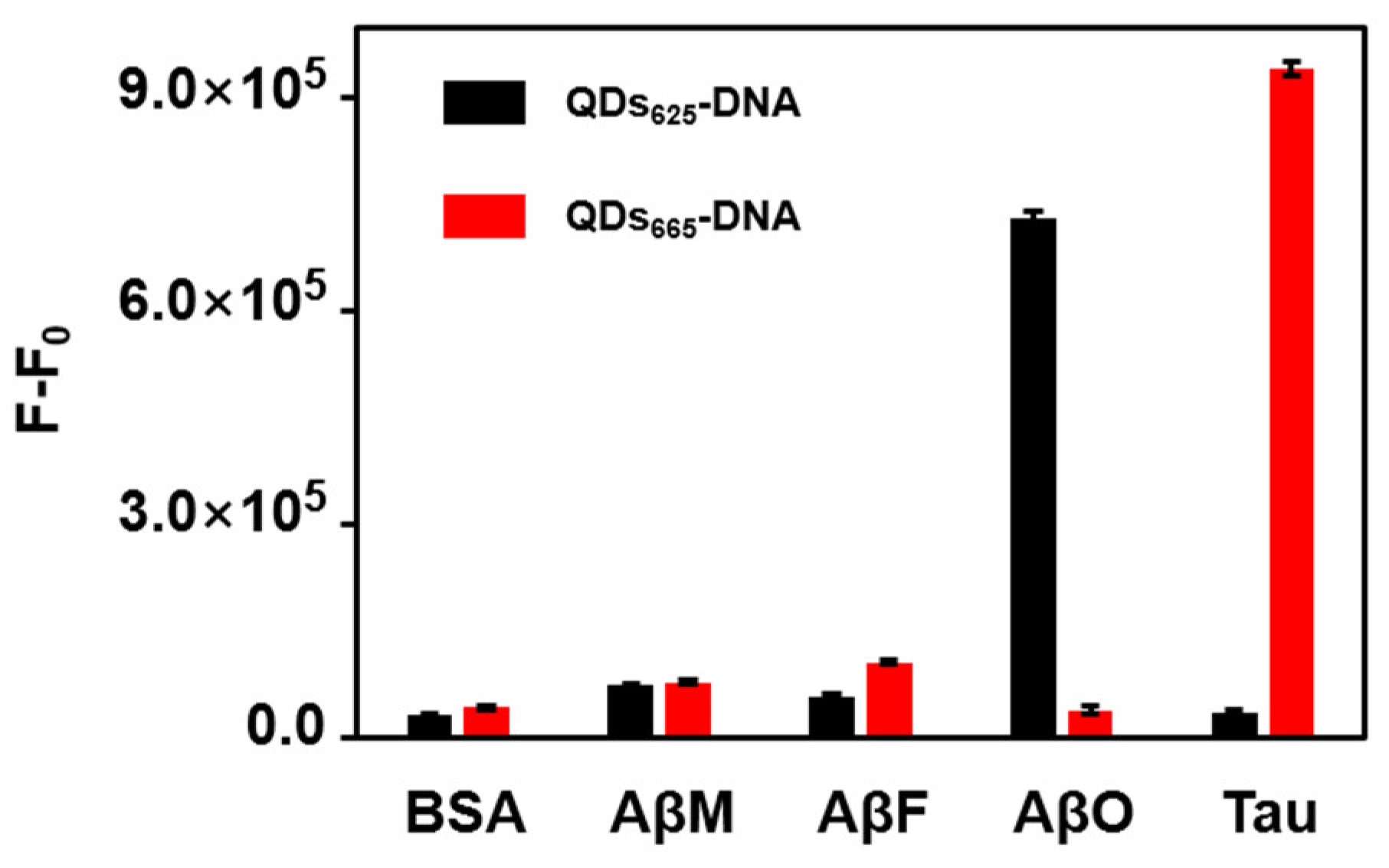
3.5. Selectivity of the FRET Aptasensor
3.6. Real Sample Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Knopman, D.S.; Amieva, H.; Petersen, R.C.; Chételat, G.; Holtzman, D.M.; Hyman, B.T.; Nixon, R.A.; Jones, D.T. Alzheimer disease. Nat. Rev. Dis. Primers 2021, 7, 33. [Google Scholar] [CrossRef] [PubMed]
- Masters, C.L.; Bateman, R.; Blennow, K.; Rowe, C.C.; Sperling, R.A.; Cummings, J.L. Alzheimer’s disease. Nat. Rev. Dis. Primers 2015, 1, 15056. [Google Scholar] [CrossRef] [PubMed]
- Panza, F.; Lozupone, M.; Logroscino, G.; Imbimbo, B.P. A critical appraisal of amyloid-β-targeting therapies for Alzheimer disease. Nat. Rev. Neurol. 2019, 15, 73–88. [Google Scholar] [CrossRef] [PubMed]
- Congdon, E.E.; Sigurdsson, E.M. Tau-targeting therapies for Alzheimer disease. Nat. Rev. Neurol. 2018, 14, 399–415. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.M.; Chen, G.F.; Gong, L.; Ge, K.Z.; Pan, W.Z.; Li, N.; Machuki, J.O.; Yu, Y.Y.; Geng, D.Q.; Dong, H.F.; et al. DNAzyme-powered three-dimensional DNA walker nanoprobe for detection amyloid β-peptide oligomer in living cells and In Vivo. Anal. Chem. 2020, 92, 9247–9256. [Google Scholar] [CrossRef] [PubMed]
- Ngoc Le, H.T.; Park, J.; Chinnadayyala, S.R.; Cho, S. Sensitive electrochemical detection of amyloid beta peptide in human serum using an interdigitated chain-shaped electrode. Biosens. Bioelectron. 2019, 144, 111694. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.C.; Xu, T.L.; Zhu, Q.L.; Zhang, X.J. Integrated individually electrochemical array for simultaneously detecting multiple Alzheimer’s biomarkers. Biosens. Bioelectron. 2020, 162, 112253. [Google Scholar] [CrossRef]
- Qin, H.X.; Gao, X.; Yang, X.Y.; Cao, W.; Liu, S.F. A label-free and signal-on electrochemiluminescence strategy for sensitive amyloid-beta assay. Biosens. Bioelectron. 2019, 141, 111438. [Google Scholar] [CrossRef]
- Wang, Y.F.; Zhang, Y.; Sha, H.; Xiong, X.; Jia, N. Design and biosensing of a ratiometric electrochemiluminescence resonance energy transfer aptasensor between a g-C3N4 nanosheet and Ru@MOF for amyloid-β protein. ACS Appl. Mater. Interfaces 2019, 11, 36299–36306. [Google Scholar] [CrossRef]
- Zhang, J.L.; Zhang, X.C.; Gao, Y.; Yan, J.Y.; Song, W.B. Integrating CuO/g-C3N4 p-n heterojunctioned photocathode with MoS2 QDs@Cu NWs multifunctional signal amplifier for ultrasensitive detection of AβO. Biosens. Bioelectron. 2021, 176, 112945. [Google Scholar] [CrossRef]
- Gao, N.; Zhang, Y.F.; Gao, K.K.; Xie, J.D.; Liu, L.; Li, Y.Y.; Qiu, L.P.; Wei, Q.; Ma, H.M.; Pang, X.H. Ultrasensitive label-free photoelectrochemical immunosensor for the detection of amyloid β-protein based on Zn:SnO2/SnS2-Au nanocomposites. Sens. Actuators B Chem. 2020, 308, 127576. [Google Scholar] [CrossRef]
- Deng, C.Y.; Liu, H.; Zhang, M.M.; Deng, H.H.; Lei, C.Y.; Shen, L.; Jiao, B.; Tu, Q.Y.; Jin, Y.; Xiang, L.; et al. Light-up nonthiolated aptasensor for low-mass, soluble amyloid-β40 oligomers at high salt concentrations. Anal. Chem. 2018, 90, 1710–1717. [Google Scholar] [CrossRef] [PubMed]
- Hu, T.; Lu, S.S.; Chen, C.X.; Sun, J.; Yang, X.R. Colorimetric sandwich immunosensor for Aβ(1-42) based on dual anti-body-modified gold nanoparticles. Sens. Actuators B Chem. 2017, 243, 792–799. [Google Scholar] [CrossRef]
- Zhang, X.; Liu, S.; Song, X.L.; Wang, H.W.; Wang, J.F.; Wang, Y.; Huang, J.D.; Yu, J.H. Robust and universal SERS sensing platform for multiplexed detection of Alzheimer’s disease core biomarkers using PAapt-AuNPs conjugates. ACS Sens. 2019, 4, 2140–2149. [Google Scholar] [CrossRef]
- Xia, N.; Liu, L.; Harrington, M.G.; Wang, J.X.; Zhou, F.M. Resonance detection of Aβ(1-40) and Aβ(1-42) peptides in cerebrospinal fluids with signal amplification by streptavidin conjugated to an N-terminus-specific antibody. Anal. Chem. 2010, 82, 10151–10157. [Google Scholar] [CrossRef] [Green Version]
- Palmqvist, S.; Tideman, P.; Cullen, N.; Zetterberg, H.; Blennow, K.; The Alzheimer’s Disease Neuroimaging Initiative; Dage, J.L.; Stomrud, E.; Janelidze, S.; Mattsson-Carlgren, N.; et al. Prediction of future Alzheimer’s disease dementia using plasma phos-pho-tau combined with other accessible measures. Nat. Med. 2021, 27, 1034–1042. [Google Scholar] [CrossRef]
- Kim, K.; Kim, M.J.; Kim, D.W.; Kim, S.Y.; Park, S.; Park, C.B. Clinically accurate diagnosis of Alzheimer’s disease via multi-plexed sensing of core biomarkers in human plasma. Nat. Commun. 2020, 11, 119. [Google Scholar] [CrossRef] [Green Version]
- Fang, W.K.; Liu, L.; Zhang, L.L.; Liu, D.; Liu, Y.; Tang, H.W. Detection of amyloid β oligomers by a fluorescence ratio strategy based on optically trapped highly doped upconversion nanoparticles-SiO2@metal−organic framework microspheres. Anal. Chem. 2021, 93, 12447–12455. [Google Scholar] [CrossRef]
- Zhang, X.J.; Hu, Y.; Yang, X.T.; Tang, Y.Y.; Han, S.Y.; Kang, A.; Deng, H.S.; Chi, Y.M.; Zhu, D.; Lu, Y. FÖrster resonance energy transfer (FRET)-based biosensors for biological applications. Biosens. Bioelectron. 2019, 138, 111314. [Google Scholar] [CrossRef]
- Quan, K.; Yi, C.P.; Yang, X.H.; He, X.X.; Huang, J.; Wang, K.M. FRET-based nucleic acid probes: Basic designs and applications in bioimaging. Trends Anal. Chem. 2020, 124, 115784. [Google Scholar] [CrossRef]
- Sheng, A.Z.; Wang, P.; Yang, J.Y.; Tang, L.F.; Chen, F.; Zhang, J. MXene coupled with CRISPR-Cas12a for analysis of endotoxin and bacteria. Anal. Chem. 2021, 93, 4676–4681. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.N.; Li, X.; Yang, Y.; Si, S.H.; Deng, C.Y.; Wu, H.Y. A simple aptasensor for Aβ40 oligomers based on tunable mismatched base pairs of dsDNA and graphene oxide. Biosens. Bioelectron. 2020, 149, 111840. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.Y.; Zhao, J.W.; Yang, G.M.; He, Y.; Chen, S.H.; Yuan, R. Ultrasensitive detection of amyloid beta oligomers based on the “DD-A” FRET binary probes and quadrivalent cruciform DNA nanostructure-mediated cascaded amplifier. ACS Appl. Mater. Interfaces 2021, 13, 32013–32021. [Google Scholar] [CrossRef] [PubMed]
- Chern, M.; Kays, J.C.; Bhuckory, S.; Dennis, A.M. Sensing with photoluminescent semiconductor quantum dots. Methods Appl. Fluores. 2019, 7, 012005. [Google Scholar] [CrossRef]
- Medintz, I.; Uyeda, H.; Goldman, E.; Mattoussi, H. Quantum dot bioconjugates for imaging, labelling and sensing. Nat. Mater. 2005, 4, 435–446. [Google Scholar] [CrossRef]
- Chan, W.C.W.; Nie, S.M. Quantum dot bioconjugates for ultrasensitive nonisotopic detection. Science 1998, 281, 2016–2018. [Google Scholar] [CrossRef] [Green Version]
- Bruchez, M.J.; Moronne, M.; Gin, P.; Weiss, S.; Alivisatos, A.P. Semiconductor nanocrystals as fluorescent biological labels. Science 1998, 281, 2013. [Google Scholar] [CrossRef] [Green Version]
- Magaryan, K.A.; Karimullin, K.R.; Vasil’eva, I.A.; Naumov, A.V. Analysis of the temperature dependence of the exciton lu-minescence spectra of cadmium selenide quantum dots grown in a liquid crystal matrix. Opt. Spectrosc. 2019, 126, 41–43. [Google Scholar] [CrossRef]
- Karimullin, K.R.; Arzhanov, A.I.; Eremchev, I.Y.; Kulnitskiy, B.A.; Surovtsev, N.V.; Naumov, A.V. Combined photon-echo, luminescence and raman spectroscopies of layered ensembles of colloidal quantum dots. Laser Phys. 2019, 29, 124009. [Google Scholar] [CrossRef]
- Hou, X.Q.; Kang, J.; Qin, H.Y.; Chen, X.W.; Ma, J.L.; Zhou, J.H.; Chen, L.P.; Wang, L.J.; Wang, L.W.; Peng, X.G. Engineering auger recombination in colloidal quantum dots via dielectric screening. Nat. Commun. 2019, 10, 1750. [Google Scholar] [CrossRef]
- Rosen, S.; Schwartz, O.; Oron, D. Transient fluorescence of the off state in blinking CdSe/CdS/ZnS semiconductor nanocrystals is not governed by auger recombination. Phys. Rev. Lett. 2010, 104, 157404. [Google Scholar] [CrossRef] [PubMed]
- Wegner, K.D.; Lanh, P.T.; Jennings, T.; Oh, E.; Jain, V.; Fairclough, S.M.; Smith, J.M.; Giovanelli, E.; Lequeux, N.; Pons, T.; et al. Influence of luminescence quantum yield, surface coating, and functionalization of quantum dots on the sensitivity of time resolved FRET bioassays. ACS Appl. Mater. Interfaces 2013, 5, 2881–2892. [Google Scholar] [CrossRef] [PubMed]
- Chen, O.; Zhao, J.; Chauhan, V.P.; Cui, J.; Wong, C.; Harris, D.K.; Wei, H.; Han, H.S.; Fukumura, D.; Jain, R.K.; et al. Compact high-quality CdSe-CdS core-shell nanocrystals with narrow emission linewidths and suppressed blinking. Nat. Mater. 2013, 12, 445–451. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, J.H.; Zhu, M.Y.; Meng, R.Y.; Qin, H.Y.; Peng, X.G. Ideal CdSe/CdS core/shell nanocrystals enabled by entropic ligands and their core size-, shell thickness-, and ligand-dependent photoluminescence properties. J. Am. Chem. Soc. 2017, 139, 16556–16567. [Google Scholar] [CrossRef]
- Manner, V.W.; Koposov, A.Y.; Szymanski, P.; Klimov, V.I.; Sykora, M. Role of solvent-oxygen ion pairs in photooxidation of CdSe nanocrystal quantum dots. ACS Nano 2012, 6, 2371–2377. [Google Scholar] [CrossRef]
- Gupta, R.; Peveler, W.J.; Lix, K.; Algar, W.R. Comparison of semiconducting polymer dots and semiconductor quantum dots for smartphone-based fluorescence assays. Anal. Chem. 2019, 91, 10955–10960. [Google Scholar] [CrossRef]
- Goryacheva, O.A.; Guhrenz, C.; Schneider, K.; Beloglazova, N.V.; Goryacheva, I.Y.; Saeger, S.D.; Gaponik, N. Silanized luminescent quantum dots for the simultaneous multicolor lateral flow immunoassay of two mycotoxins. ACS Appl. Mater. Interfaces 2020, 12, 24575–24584. [Google Scholar] [CrossRef]
- Wang, W.L.; Guo, Y.; Tiede, C.; Chen, S.Y.; Kopytynski, M.; Kong, Y.F.; Kulak, A.; Tomlinson, D.; Chen, R.; McPherson, M.; et al. Ultraefficient cap-exchange protocol to compact biofunctional quantum dots for sensitive ratiometric biosensing and cell imaging. ACS Appl. Mater. Interfaces 2017, 9, 15232–15244. [Google Scholar] [CrossRef] [Green Version]
- Wu, Y.T.; Qiu, X.; Lindbo, S.; Susumu, K.; Medintz, I.L.; Hober, S.; Hildebrandt, N. Quantum dot–based FRET immunoassay for HER2 using ultrasmall affinity proteins. Small 2018, 14, 1802266. [Google Scholar] [CrossRef] [Green Version]
- Wang, W.L.; Kong, Y.F.; Jiang, J.; Tian, X.; Li, S.; Akshath, U.S.; Tiede, C.; Hondow, N.; Yu, A.C.; Guo, Y.; et al. Photon induced quantum yield regeneration of cap-exchanged CdSe/CdS quantum rods for ratiometric biosensing and cellular imaging. Nanoscale 2020, 12, 8647–8655. [Google Scholar] [CrossRef]
- Tong, P.; Zhao, W.W.; Zhang, L.; Xu, J.J.; Chen, H.Y. Double-probe signal enhancing strategy for toxin aptasensing based on rolling circle amplification. Biosens. Bioelectron. 2012, 33, 146–151. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.B.; Xu, Y.; Zhao, X.H.; Chang, Y.; Liu, Y.; Jiang, L.J.; Sharma, J.; Seo, D.K.; Yan, H. A facile one-step in situ functionalization of quantum dots with preserved photoluminescence for bioconjugation. J. Am. Chem. Soc. 2007, 129, 6380–6381. [Google Scholar] [CrossRef] [PubMed]
- Deka, S.; Quarta, A.; Lupo, M.G.; Falqui, A.; Boninelli, S.; Giannini, C.; Morello, G.; Giorgi, M.D.; Lanzani, G.; Spinella, C.; et al. CdSe/CdS/ZnS double shell nanorods with high photoluminescence efficiency and their exploitation as biolabeling probes. J. Am. Chem. Soc. 2009, 131, 2948–2958. [Google Scholar] [CrossRef] [PubMed]
- Hou, X.Q.; Qin, H.Y.; Peng, X.G. Enhancing dielectric screening for auger suppression in CdSe/CdS quantum dots by epitaxial growth of ZnS shell. Nano Lett. 2021, 21, 3871–3878. [Google Scholar] [CrossRef] [PubMed]
- Dabbousi, B.O.; Rodriguez-Viejo, J.; Mikulec, F.V.; Heine, J.R.; Mattoussi, H.; Ober, R.; Jensen, K.F.; Bawendi, M.G. (CdSe)ZnS core-shell quantum dots: Synthesis and characterization of a size series of highly luminescent nanocrystallites. J. Phys. Chem. B 1997, 101, 9463–9475. [Google Scholar] [CrossRef]
- Ye, X.C.; Jin, L.H.; Caglayan, H.; Chen, J.; Xing, G.Z.; Zheng, C.; Doan-Nguyen, V.; Kang, Y.J.; Engheta, N.; Kagan, C.R.; et al. Improved size-tunable synthesis of monodisperse gold nanorods through the use of aromatic additives. ACS Nano 2012, 6, 2804–2817. [Google Scholar] [CrossRef]
- Chen, H.Q.; Yuan, F.; Wang, S.Z.; Xu, J.; Zhang, Y.Y.; Wang, L. Aptamer-based sensing for thrombin in red region via fluorescence resonant energy transfer between NaYF4:Yb, Er upconversion nanoparticles and gold nanorods. Biosens. Bioelectron. 2013, 48, 19–25. [Google Scholar] [CrossRef]
- Choi, C.K.K.; Li, J.M.; Wei, K.C.; Xu, Y.J.; Ho, L.W.C.; Zhu, M.L.; To, K.K.W.; Choi, C.H.J.; Bian, L.M. A gold@polydopamine core–shell nanoprobe for long-term intracellular detection of microRNAs in differentiating stem cells. J. Am. Chem. Soc. 2015, 137, 7337–7346. [Google Scholar] [CrossRef]
- Ye, Q.; Zhou, F.; Liu, W.M. Bioinspired catecholic chemistry for surface modification. Chem. Soc. Rev. 2011, 40, 4244–4258. [Google Scholar] [CrossRef]
- Wang, J.; Gao, Y.H.; Liu, P.P.; Xu, S.H.; Luo, X.L. Core-shell multifunctional nanomaterial-based all-in-one nanoplatform for simultaneous multilayer imaging of dual types of tumor biomarkers and photothermal therapy. Anal. Chem. 2020, 92, 15169–15178. [Google Scholar] [CrossRef]
- Liu, L.; Chang, Y.; Yu, J.; Jiang, M.S.; Xia, N. Two-in-one polydopamine nanospheres for fluorescent determination of beta-amyloid oligomers and inhibition of beta-amyloid aggregation. Sens. Actuators B Chem. 2017, 251, 359–365. [Google Scholar] [CrossRef]
- Cao, Z.Y.; Shu, Y.F.; Qin, H.Y.; Su, B.; Peng, X.G. Quantum dots with highly efficient, stable, and multicolor electrochemiluminescence. ACS Cent. Sci. 2020, 6, 1129–1137. [Google Scholar] [CrossRef] [PubMed]
- Yu, W.W.; Qu, L.H.; Guo, W.Z.; Peng, X.G. Experimental determination of the extinction coefficient of CdTe, CdSe, and CdS nanocrystals. Chem. Mater. 2003, 15, 2854–2860. [Google Scholar] [CrossRef]
- Qiu, Y.; Liu, Y.; Wang, L.M.; Xu, L.G.; Bai, R.; Ji, Y.L.; Wu, X.C.; Zhao, Y.L.; Li, Y.F.; Chen, C.Y. Surface chemistry and aspect ratio mediated cellular uptake of Au nanorods. Biomaterials 2010, 31, 7606–7619. [Google Scholar] [CrossRef] [PubMed]
- Aldana, J.; Wang, Y.A.; Peng, X.G. Photochemical instability of CdSe nanocrystals coated by hydrophilic thiols. J. Am. Chem. Soc. 2001, 123, 8844–8850. [Google Scholar] [CrossRef]
- Hu, M.; He, Y.; Song, S.P.; Yan, J.; Lu, H.T.; Weng, L.X.; Wang, L.H.; Fan, C.H. DNA-bridged bioconjugation of fluorescent quantum dots for highly sensitive microfluidic protein chips. Chem. Commun. 2010, 46, 6126–6128. [Google Scholar] [CrossRef]
- Tian, F.; Lyu, J.; Shi, J.Y.; Yang, M. Graphene and graphene-like two-denominational materials based fluorescence resonance energy transfer (FRET) assays for biological applications. Biosens. Bioelectron. 2017, 89, 123–135. [Google Scholar] [CrossRef]
- Chen, W.; Gao, G.; Jin, Y.; Deng, C. A facile biosensor for Aβ40O based on fluorescence quenching of prussian blue nanoparticles. Talanta 2020, 216, 120930. [Google Scholar] [CrossRef]
- Jiang, L.; Chen, B.; Chen, B.; Li, X.; Liao, H.; Huang, H.; Guo, Z.; Zhang, W.; Wu, L. Detection of Aβ oligomers based on magnetic-field-assisted separation of aptamer-functionalized Fe3O4 magnetic nanoparticles and BaYF5:Yb,Er nanoparticles as upconver-sion fluorescence labels. Talanta 2017, 170, 350–357. [Google Scholar] [CrossRef]
- Chen, L.; Lin, J.; Yi, J.; Weng, Q.; Zhou, Y.; Han, Z.; Li, C.; Chen, J.; Zhang, Q. A tyrosinase-induced fluorescence immunoassay for detection of tau protein using dopamine-functionalized CuInS2/ZnS quantum dots. Anal. Bioanal. Chem. 2019, 411, 5277–5285. [Google Scholar] [CrossRef]
- Huang, A.; Zhang, L.; Li, W.; Ma, Z.; Shuo, S.; Yao, T. Controlled fluorescence quenching by antibody-conjugated graphene oxide to measure tau protein. R. Soc. Open Sci. 2018, 5, 171808. [Google Scholar] [CrossRef] [PubMed]
| Analytes | Spiked (pM) | Found (pM) | Recovery (%) | RSD (%) |
|---|---|---|---|---|
| 200 | 190 | 95 | 3.59 | |
| AβO | 500 | 510 | 102 | 4.16 |
| 1500 | 1470 | 98 | 2.35 | |
| 100 | 98 | 98 | 4.23 | |
| tau protein | 500 | 520 | 104 | 3.62 |
| 1000 | 1010 | 101 | 4.76 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lu, X.; Hou, X.; Tang, H.; Yi, X.; Wang, J. A High-Quality CdSe/CdS/ZnS Quantum-Dot-Based FRET Aptasensor for the Simultaneous Detection of Two Different Alzheimer’s Disease Core Biomarkers. Nanomaterials 2022, 12, 4031. https://doi.org/10.3390/nano12224031
Lu X, Hou X, Tang H, Yi X, Wang J. A High-Quality CdSe/CdS/ZnS Quantum-Dot-Based FRET Aptasensor for the Simultaneous Detection of Two Different Alzheimer’s Disease Core Biomarkers. Nanomaterials. 2022; 12(22):4031. https://doi.org/10.3390/nano12224031
Chicago/Turabian StyleLu, Xingchang, Xiaoqi Hou, Hailin Tang, Xinyao Yi, and Jianxiu Wang. 2022. "A High-Quality CdSe/CdS/ZnS Quantum-Dot-Based FRET Aptasensor for the Simultaneous Detection of Two Different Alzheimer’s Disease Core Biomarkers" Nanomaterials 12, no. 22: 4031. https://doi.org/10.3390/nano12224031





