A Visual Discrimination of Existing States of Virus Capsid Protein by a Giant Molybdate Cluster
Abstract
:1. Introduction
2. Results and Discussion
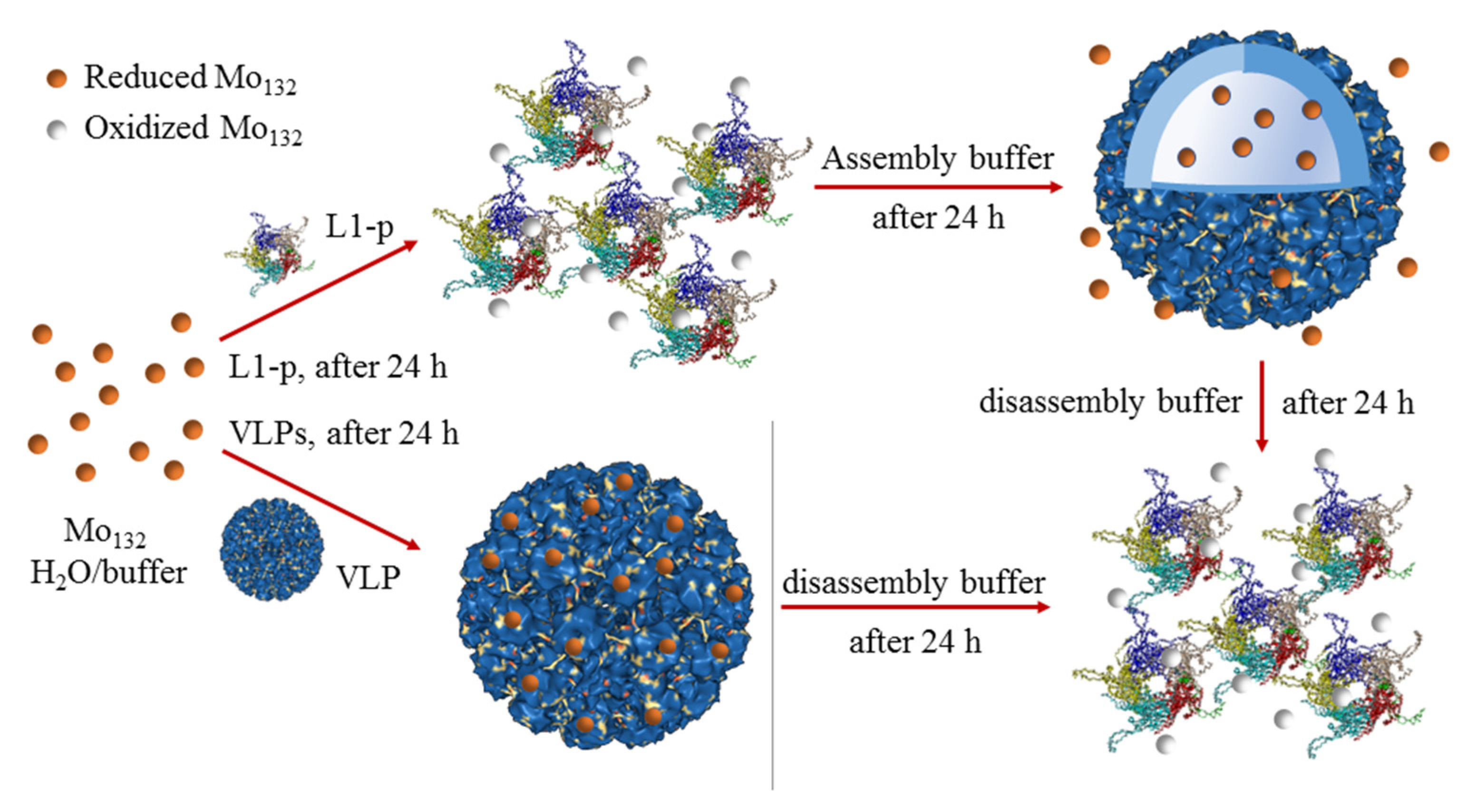
2.1. Hypochromic Response of [Mo132] to L1-p
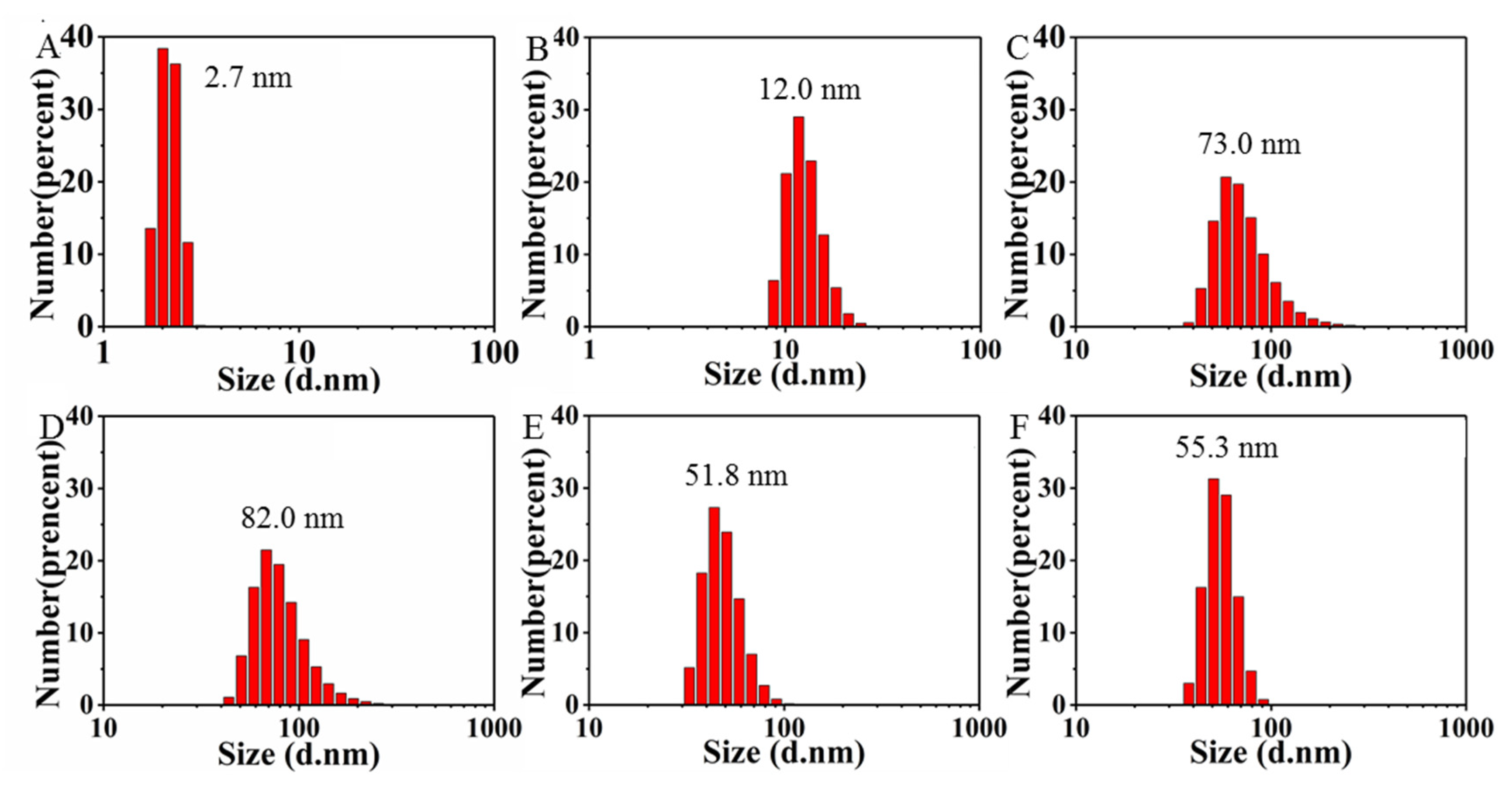
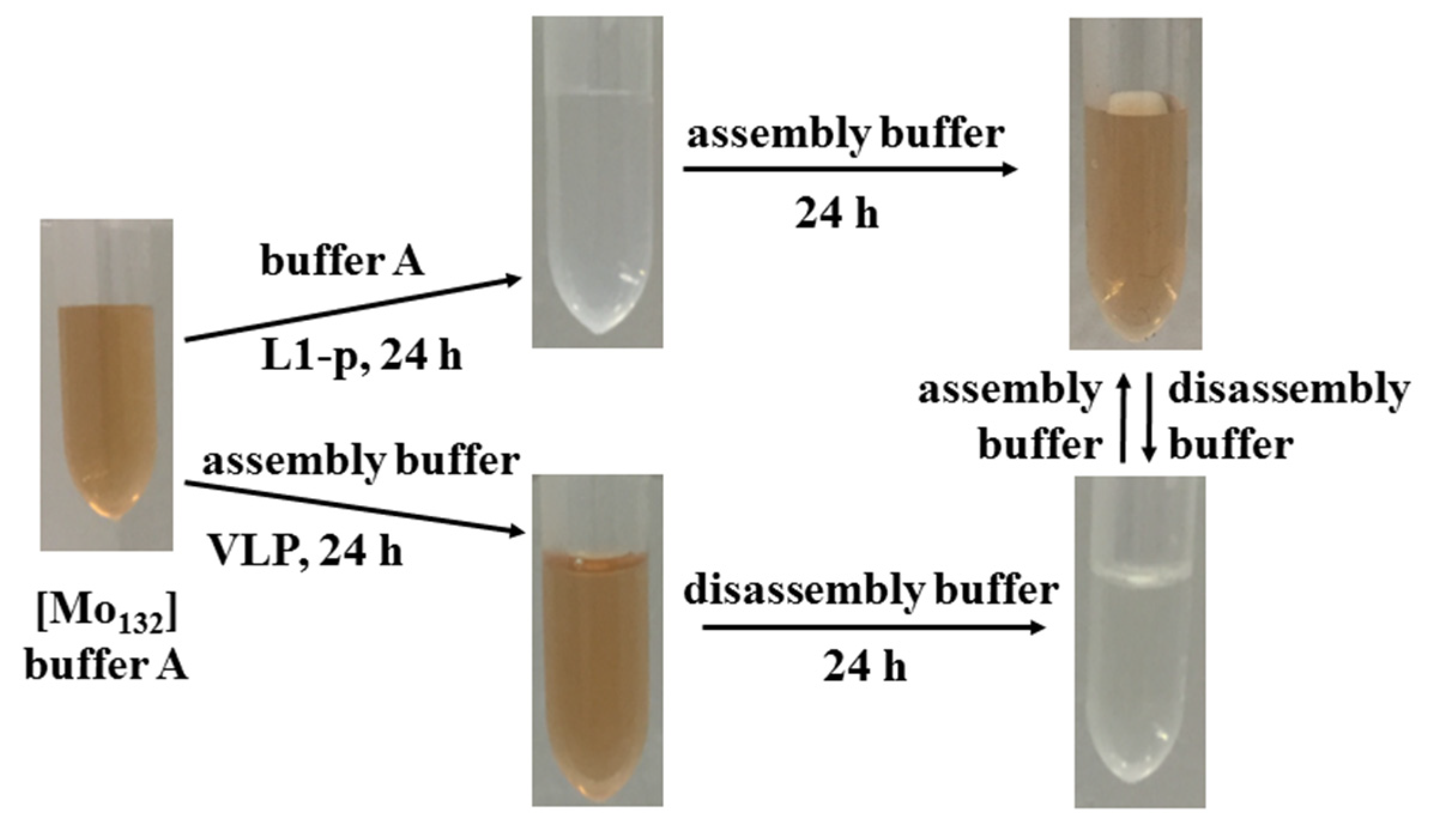
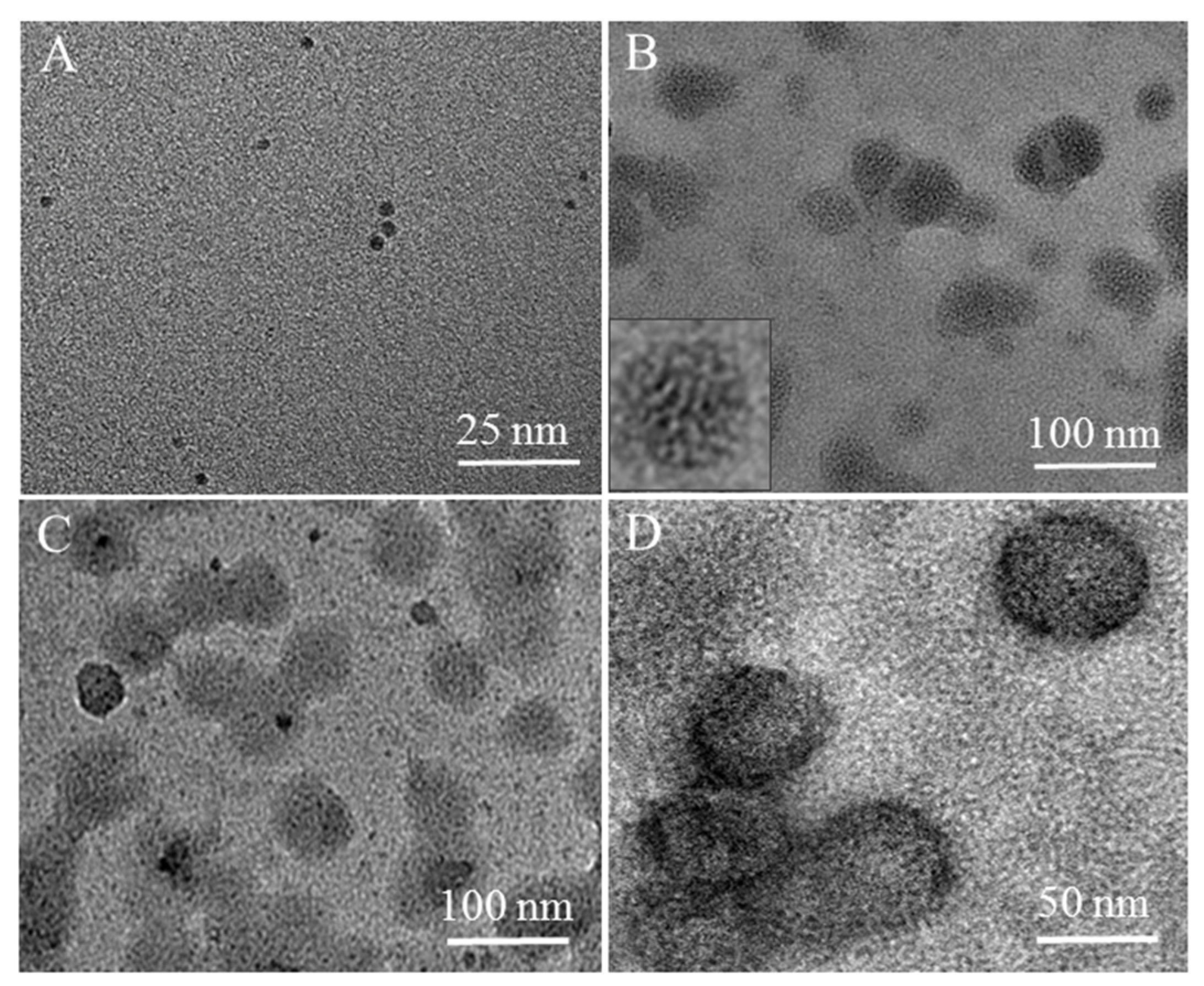
2.2. Visual Response of [Mo132] to the Isolated L1-p and VLPs
2.3. Prevention of [Mo132] Hypochromic by VLPs
2.4. Colorimetry Response Mechanism of Mo132 to L1-p and VLP
3. Conclusions and Perspectives
4. Experimental Section
4.1. Reagents and Materials
4.2. Expression, Purification of HPV 16 L1 Protein and Peptides
4.3. Preparation of (NH4)42[Mo132O372(CH3COO)30]∙72H2O
4.4. Assembly/Disassembly Monitoring of Mo132 and HPV 16 L1
4.5. Cesium Chloride Gradient Centrifugation
4.6. Instruments
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Frega, A.; Stentella, P.; Ioris, A.D.; Piazze, J.J.; Fambrini, M.; Marchionni, M.; Cosmi, E.V. Young women, cervical intraepithelial neoplasia and human papillomavirus: Risk factors for persistence and recurrence. Cancer Lett. 2003, 196, 127–134. [Google Scholar] [CrossRef]
- Colón-López, V.; Ortiz, A.P.; Palefsky, J. Burden of human papillomavirus infection and related comorbidities in men: Implications for research, disease prevention and health promotion among hispanic men. P. R. Health Sci. J. 2010, 29, 232–240. [Google Scholar]
- Dunjic, M.; Stanisic, S.; Krstic, D.; Stanisic, M.; Dunjic, M. Integrative approach to diagnosis of genital human papillomaviruses (HPV) infection of female. Acupunct. Electro-Ther. Res. 2014, 39, 229–239. [Google Scholar] [CrossRef] [PubMed]
- Pogoda, C.S.; Roden, R.B.S.; Garcea, R.L. Immunizing against anogenital cancer: HPV vaccines. PLoS Pathog. 2016, 12, e1005587. [Google Scholar] [CrossRef] [PubMed]
- Harper, D.M.; Franco, E.L.; Wheeler, C.; Ferris, D.G.; Jenkins, D.; Schuind, A.; Zahaf, T.; Innis, B.; Naud, P.; De Carvalho, N.S.; et al. GlaxoSmithKline HPV Vaccine Study Group. Efficacy of a Bivalent L1 Virus-Like Particle Vaccine in Prevention of Infection with Human Papillomavirus Types 16 and 18 in Young Women: A Randomised Controlled Trial. Lancet 2004, 364, 1757–1765. [Google Scholar] [CrossRef]
- Einstein, M.H. Acquired Immune Response to Oncogenic Human Papillomavirus Associated with Prophylactic Cervical Cancer Vaccines. Cancer Immunol. Immunother. 2008, 57, 443–451. [Google Scholar] [CrossRef]
- Lua, L.H.L.; Connors, N.K.; Sainsbury, F.; Chuan, Y.P.; Wibowo, N.; Middelberg, A.P.J. Bioengineering virus-like particles as vaccines. Biotechnol. Bioeng. 2014, 111, 425–440. [Google Scholar] [CrossRef]
- McCarthy, M.P.; White, W.I.; Palmer-Hill, F.; Koenig, S.; Suzich, J.A. Quantitative disassembly and reassembly of human papillomavirus type 11 virus-like particles in vitro. J. Virol. 1998, 72, 32–41. [Google Scholar] [CrossRef] [Green Version]
- Zdanowicz, M.; Chroboczek, J. Virus-like particles as drug delivery vectors. Acta Biochim. Pol. 2016, 63, 469–473. [Google Scholar] [CrossRef]
- Huber, B.; Schellenbacher, C.; Shafti-Keramat, S.; Jindra, C.; Christensen, N.; Kirnbauer, R. Chimeric L2-Based Virus-Like Particle (VLP) Vaccines Targeting Cutaneous Human Papillomaviruses (HPV). PLoS ONE 2017, 12, e0169533. [Google Scholar] [CrossRef]
- Mach, H.; Volkin, D.B.; Troutman, R.D.; Wang, B.; Luo, Z.; Jansen, K.U.; Shi, L. Disassembly and reassembly of yeast-derived recombinant human papillomavirus virus-like particles (HPV VLPs). J. Pharm. Sci. 2006, 95, 2195–2206. [Google Scholar] [CrossRef] [PubMed]
- Molina Sánchez, P. Polyoxometalate Self-Assembly: From Molecules to Hybrid Materials; University of Glasgow: Glasgow, UK, 2013. [Google Scholar]
- Wu, L.; Liang, J. Polyoxometalates and Their Complexes Toward Biological Application. In Supramolecular Chemistry of Biomimetic Systems; Li, J., Ed.; Springer Nature Singapore Pte Ltd.: Singapore, 2017; pp. 311–354. [Google Scholar]
- Bijelic, A.; Aureliano, M.; Rompel, A. Polyoxometalates as potential next-generation metallodrugs in the combat against cancer. Angew. Chem. Int. Ed. 2019, 58, 2980–2999. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, T.; Diemann, E.; Li, H.; Dress, A.W.M.; Müller, A. Self-assembly in aqueous solution of wheel-shaped Mo154 oxide clusters into vesicles. Nature 2003, 426, 59–62. [Google Scholar] [CrossRef] [PubMed]
- Müller, A.; Rehder, D. Molecular Metal Oxides in Protein Cages/Cavities. Coordination Chemistry in Protein Cages. In Coordination Chemistry in Protein Cages: Principles, Design, and Applications; Ueno, T., Watanabe, Y., Eds.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2013; pp. 25–42. [Google Scholar]
- Ren, H.; Shehzad, F.K.; Zhou, Y.; Zhang, L.; Iqbal, A.; Long, Y. Incorporation of Keplerate-type Mo–O based macroanions into Zn2Al-LDH results in the formation of all-inorganic composite films with remarkable third-order optical nonlinearity. Dalton Trans. 2018, 47, 6184–6188. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Chen, H.; Zhang, G.; Kong, X.; Yin, S.; Li, B.; Wu, L. An ultra-small thermosensitive nanocomposite with a Mo154-core as a comprehensive platform for NIR-triggered photothermal-chemotherapy. J. Mater. Chem. B 2018, 6, 241–248. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Li, W.; Li, H.; Wu, L. Ionic Complexes of Metal Oxide Clusters for Versatile Self-Assemblies. Acc. Chem. Res. 2017, 50, 1391–1399. [Google Scholar] [CrossRef]
- Li, J.; Chen, Z.; Zhou, M.; Jing, J.; Li, W.; Wang, Y.; Wu, L.; Wang, L.; Wang, Y.; Lee, M. Polyoxometalate-Driven Self-Assembly of Short Peptides into Multivalent Nanofibers with Enhanced Antibacterial Activity. Angew. Chem. Int. Ed. 2016, 55, 2592–2595. [Google Scholar] [CrossRef]
- Müller, A.; Serain, C. Soluble molybdenum blues-“des Pudels Kern”. Acc. Chem. Res. 2000, 33, 2–10. [Google Scholar] [CrossRef]
- Müller, A.; Krickemeyer, E.; Meyer, J.; Bögge, H.; Peters, F.; Plass, W.; Diemann, E.; Dillinger, S.; Nonnenbruch, F.; Randerath, M.; et al. [Mo154(NO)14O420(OH)28(H2O)70](25±5)−: A Water-Soluble Big Wheel with More than 700 Atoms and a Relative Molecular Mass of about 24000. Angew. Chem. Int. Ed. Engl. 1995, 34, 2122–2124. [Google Scholar] [CrossRef]
- Müller, A.; Krickemeyer, E.; Bögge, H.; Schmidtmann, M.; Peters, F. Organizational Forms of Matter: An Inorganic Super Fullerene and Keplerate Based on Molybdenum Oxide. Angew. Chem. Int. Ed. 1998, 37, 3359–3363. [Google Scholar] [CrossRef]
- Müller, A.; Sarkar, S.; Shah, S.Q.N.; Bögge, H.; Schmidtmann, M.; Kögerler, P.; Hauptfleisch, B.; Trautwein, A.X.; Schünemann, V. Archimedean Synthesis and Magic Numbers: “Sizing” Giant Molybdenum-Oxide-Based Molecular Spheres of the Keplerate Type. Angew. Chem. Int. Ed. 1999, 38, 3238–3241. [Google Scholar] [CrossRef]
- Guo, J.; Liu, W.; Dong, X.; Lei, H.; Li, X.M.; Sun, B.; Yin, Y.H. Expression and purification of human papillomavirus type 52 virus-like particles in Escherichia coli. Biotechnology 2019, 2, 127–132. [Google Scholar]
- Liu, T.; Langston, M.L.K.; Li, D.; Pigga, J.M.; Pichon, C.; Todea, A.M.; Müller, A. Self-recognition among different polyprotic macroions during assembly processes in dilute solution. Science 2011, 331, 1590–1592. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fu, D.-Y.; Zhang, S.; Qu, Z.; Yu, X.; Wu, Y.; Wu, L. Hybrid Assembly toward Enhanced Thermal Stability of Virus-like Particles and Antibacterial Activity of Polyoxometalates. ACS Appl. Mater. Interfaces 2018, 10, 6137–6145. [Google Scholar] [CrossRef]
- Jin, S.; Zheng, D.-D.; Sun, B.; Yu, X.; Zha, X.; Liu, Y.; Wu, S.; Wu, Y. Controlled Hybrid-Assembly of HPV16/18 L1 Bi VLPs in Vitro. ACS Appl. Mater. Interfaces. ACS Appl. Mater. Interfaces 2016, 8, 34244–34251. [Google Scholar] [CrossRef]
- Zheng, D.-D.; Fu, D.-Y.; Wu, Y.; Sun, Y.-L.; Tan, L.-L.; Zhou, T.; Ma, S.-Q.; Zha, X.; Yang, Y.-W. Efficient inhibition of human papillomavirus 16 L1 pentamer formation by a carboxylatopillarene and a p-sulfonatocalixarene. Chem. Commun. 2014, 50, 3201–3203. [Google Scholar] [CrossRef]
- Chen, X.S.; Garcea, R.L.; Goldberg, I.; Casini, G.; Harrison, S.C. Structure of Small Virus-like Particles Assembled from the L1 Protein of Human Papillomavirus 16. Mol. Cell 2000, 5, 557–567. [Google Scholar] [CrossRef]
- Zhang, T.; Li, H.-W.; Wu, Y.; Wang, Y.; Wu, L. Self-Assembly of an Europium-Containing Polyoxometalate and the Arginine/Lysine-Rich Peptides from Human Papillomavirus Capsid Protein L1 in Forming Luminescence-Enhanced Hybrid Nanospheres. J. Phys. Chem. C 2015, 119, 8321–8328. [Google Scholar] [CrossRef]
- Zhang, T.; Li, H.-W.; Wu, Y.; Wang, Y.; Wu, L. The Two-Step Assemblies of Basic-Amino-Acid-Rich Peptide with a Highly Charged Polyoxometalate. Chem.-Eur. J. 2015, 21, 9028–9033. [Google Scholar] [CrossRef]
- Bernardini, G.; Wedd, A.G.; Zhao, C.; Bond, A.M. Photochemical oxidation of water and reduction of polyoxometalate anions at interfaces of water with ionic liquids or diethylether. Proc. Natl. Acad. Sci. USA 2012, 109, 11552–11557. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Bond, A.M.; MacFarlane, D.R.; Forsyth, S.A.; Pringle, J.M.; Mariotti, A.W.A.; Glowinski, A.F.; Wedd, A.G. Voltammetric Studies on the Reduction of Polyoxometalate Anions in Ionic Liquids. Inorg. Chem. 2005, 44, 5123–5132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bijelic, A.; Rompel, A. The use of polyoxometalates in protein crystallography—An attempt to widen a well-known bottleneck. Coordin. Chem. Rev. 2015, 299, 22–38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bijelic, A.; Rompel, A. Ten Good Reasons for the Use of the Tellurium-Centered Anderson–Evans Polyoxotungstate in Protein Crystallography. Acc. Chem. Res. 2017, 50, 1441–1448. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cardone, G.; Moyer, A.L.; Cheng, N.; Thompson, C.D.; Dvoretzky, I.; Lowy, D.R.; Schiller, J.T.; Steven, A.C.; Buck, C.B.; Trus, B.L. Maturation of the Human Papillomavirus 16 Capsid. mBio 2014, 5, e01104–e01114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, M.; Cripe, T.P.; Estes, P.A.; Lyon, M.K.; Rose, R.C.; Garcea, R.L. Expression of the human papillomavirus type 11 capsid protein in escherichia coli: Characterization of protein domains involved in DNA binding and capsid assembly. J. Virol. 1997, 71, 2988–2995. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Newton, G.N.; Cameron, J.M.; Wales, D.J. Shining a light on the photo-sensitisation of organic-inorganic hybrid polyoxometalates. Dalton Trans. 2018, 47, 5120–5136. [Google Scholar]
- Shi, Z.; Zhou, Y.; Zhang, L.; Yang, D.; Mu, C.; Ren, H.; Shehzada, F.K.; Li, J. Fabrication and optical nonlinearities of composite films derived from the water-soluble Keplerate type polyoxometalate and chloroform-soluble porphyrin. Dalton Trans. 2015, 44, 4102–4107. [Google Scholar] [CrossRef]


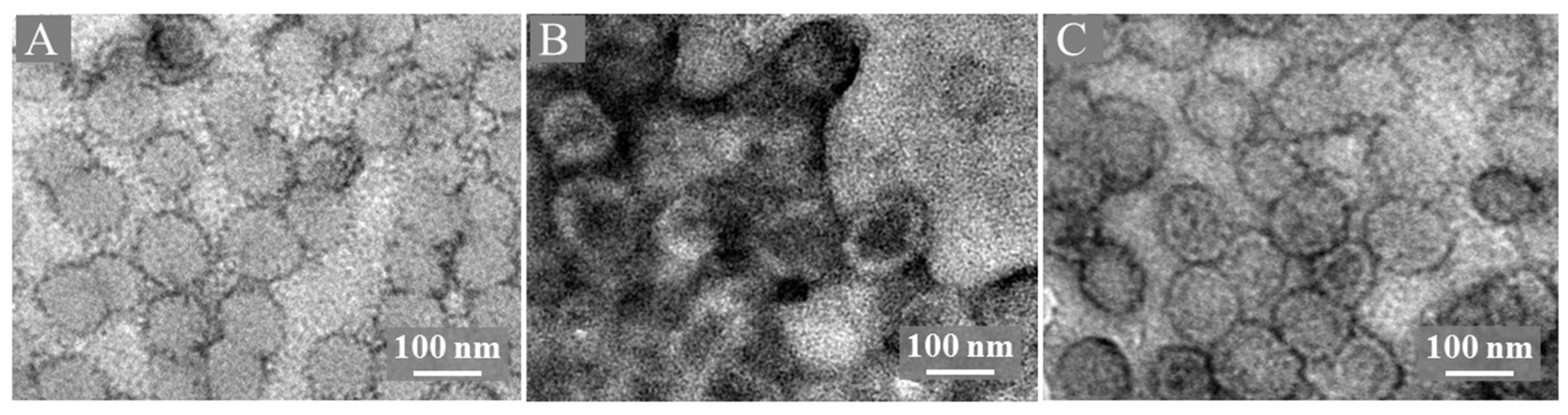
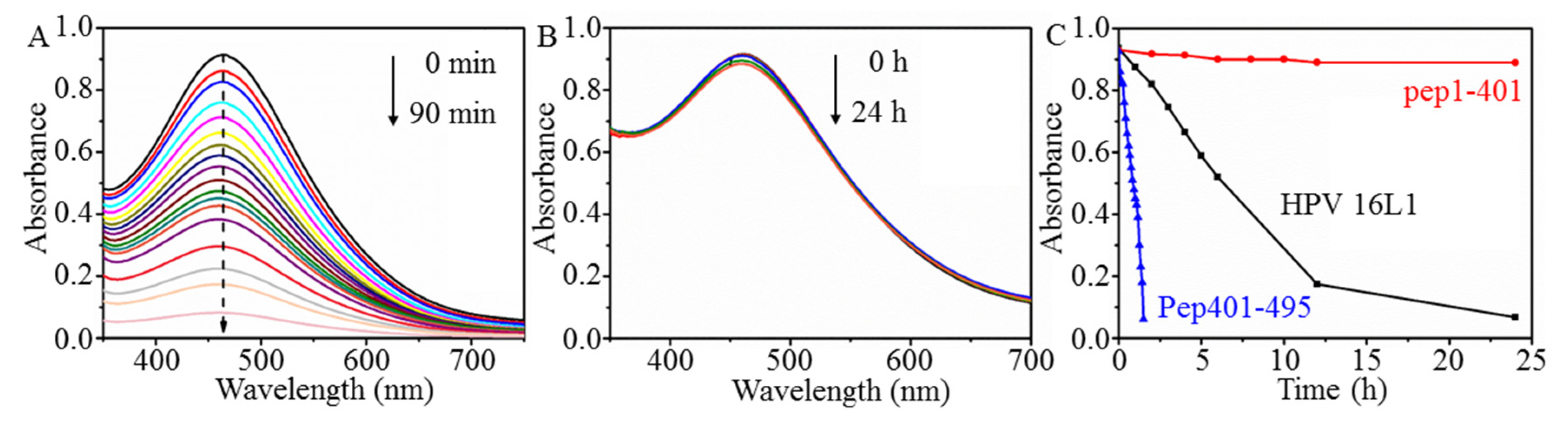
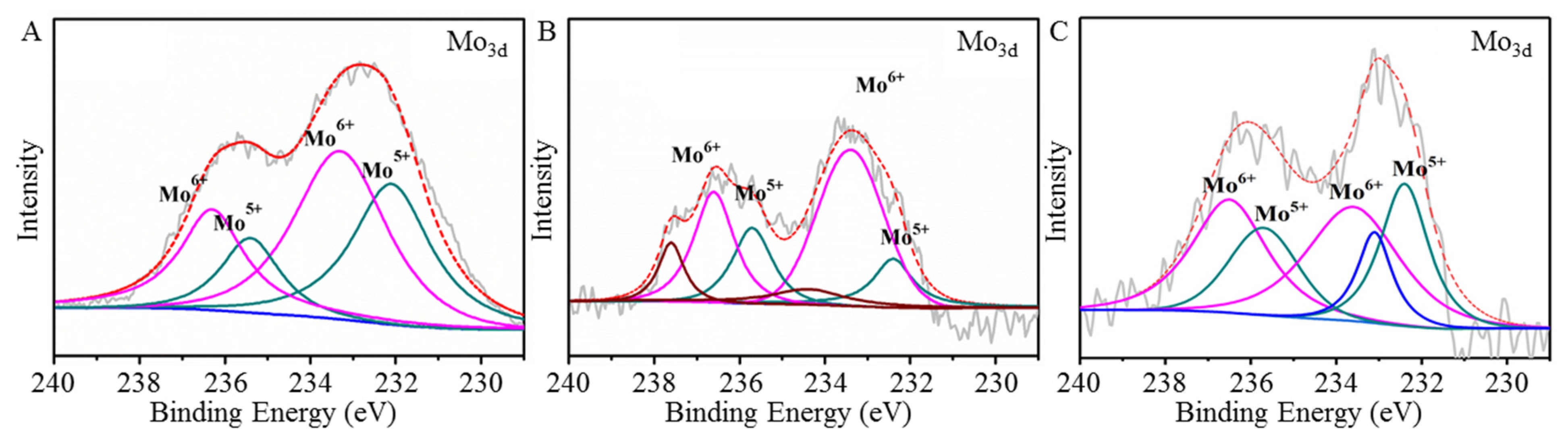
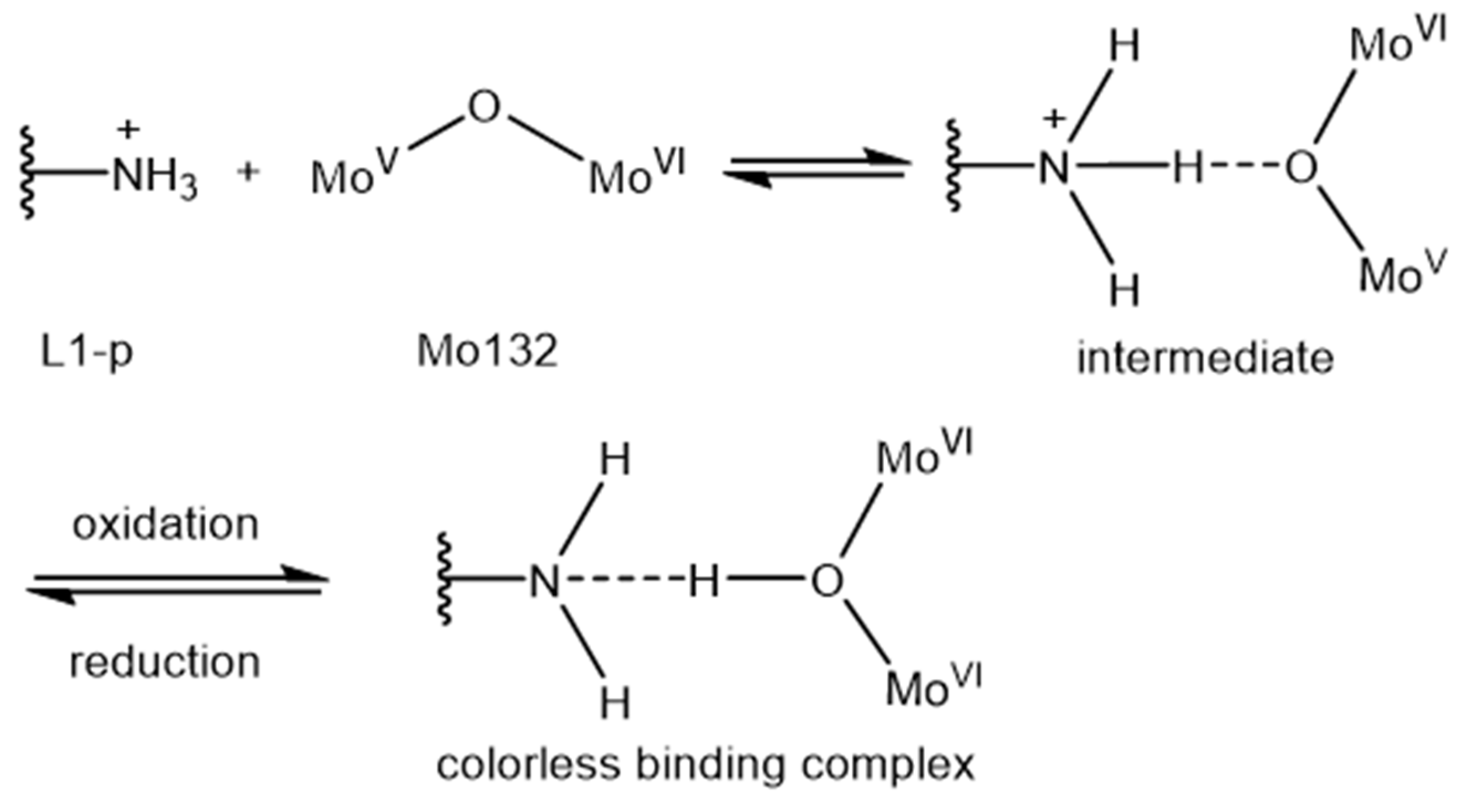
| Samples | [Mo132] | [Mo132] + L1-p | [Mo132] + VLPs |
|---|---|---|---|
| Mo(V) | 115,359.60 + 43,910.59 | 15,696.10 + 18,827.48 | 17,349.15 + 21,444.59 |
| Mo(VI) | 147,757.50 + 82,513.98 | 58,321.11 + 28,475.33 | 26,737.09 + 32,049.94 |
| Ratio of Mo(VI) to Mo(V) | 1.45:1 | 2.5:1 | 1.5:1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xue, Y.; Wei, M.; Fu, D.; Wu, Y.; Sun, B.; Yu, X.; Wu, L. A Visual Discrimination of Existing States of Virus Capsid Protein by a Giant Molybdate Cluster. Nanomaterials 2022, 12, 736. https://doi.org/10.3390/nano12050736
Xue Y, Wei M, Fu D, Wu Y, Sun B, Yu X, Wu L. A Visual Discrimination of Existing States of Virus Capsid Protein by a Giant Molybdate Cluster. Nanomaterials. 2022; 12(5):736. https://doi.org/10.3390/nano12050736
Chicago/Turabian StyleXue, Yarong, Mingfen Wei, Dingyi Fu, Yuqing Wu, Bo Sun, Xianghui Yu, and Lixin Wu. 2022. "A Visual Discrimination of Existing States of Virus Capsid Protein by a Giant Molybdate Cluster" Nanomaterials 12, no. 5: 736. https://doi.org/10.3390/nano12050736






