The Effect of Stereocomplexation and Crystallinity on the Degradation of Polylactide Nanoparticles
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Preparation of SC
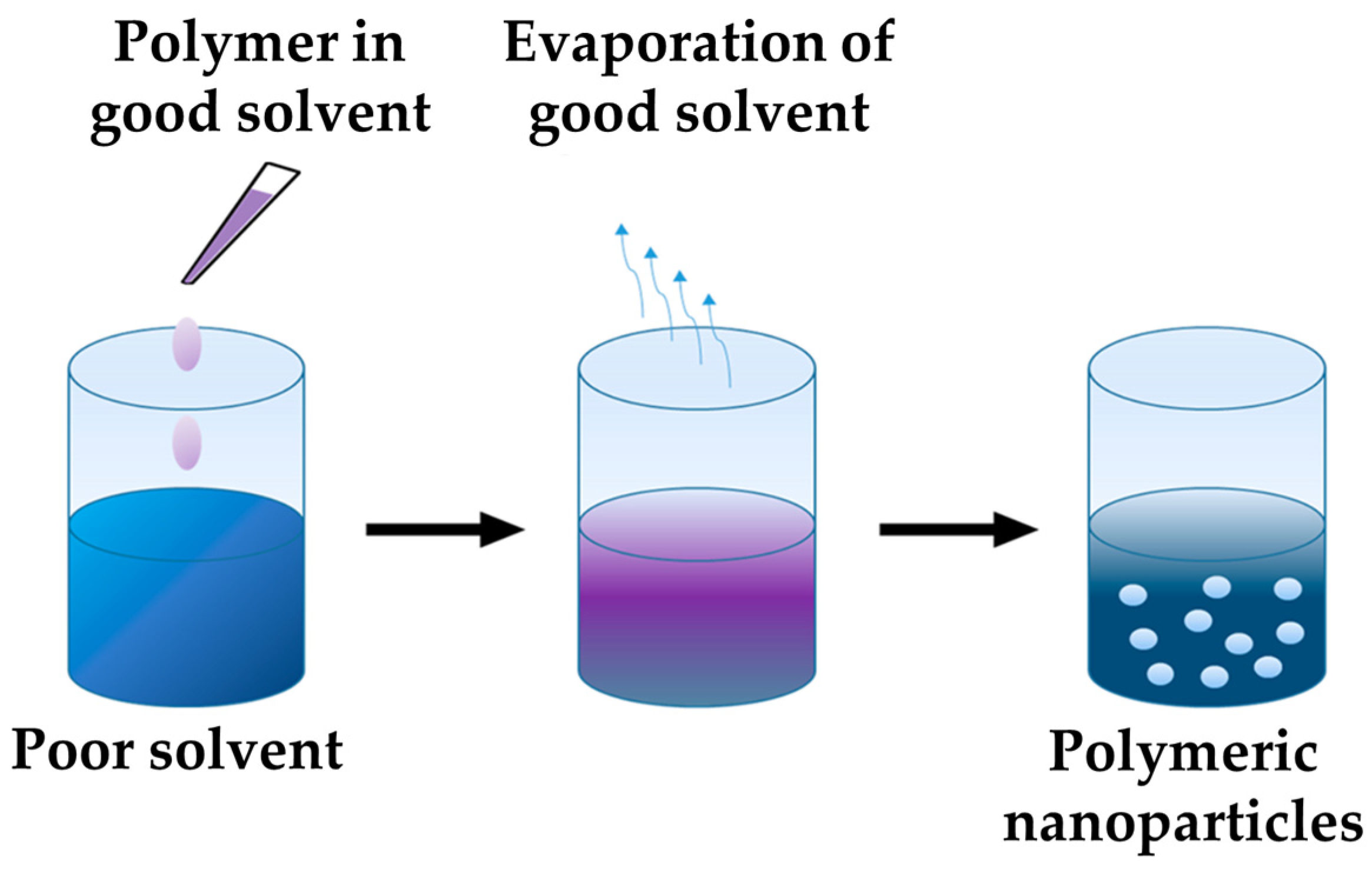
2.3. Preparation of PNPs via a Nanoprecipitation Process
2.4. Immobilization of PNPs by Surface Functionalization
2.5. Degradation Evaluation
2.6. Dynamic Light Scattering (DLS) Measurement
2.7. Atomic Force Microscopy (AFM) Measurement
2.8. Quartz Crystal Microbalance with Dissipation (QCM-D)
2.9. Statistical Analysis
3. Results and Discussions
3.1. Particle Sizes
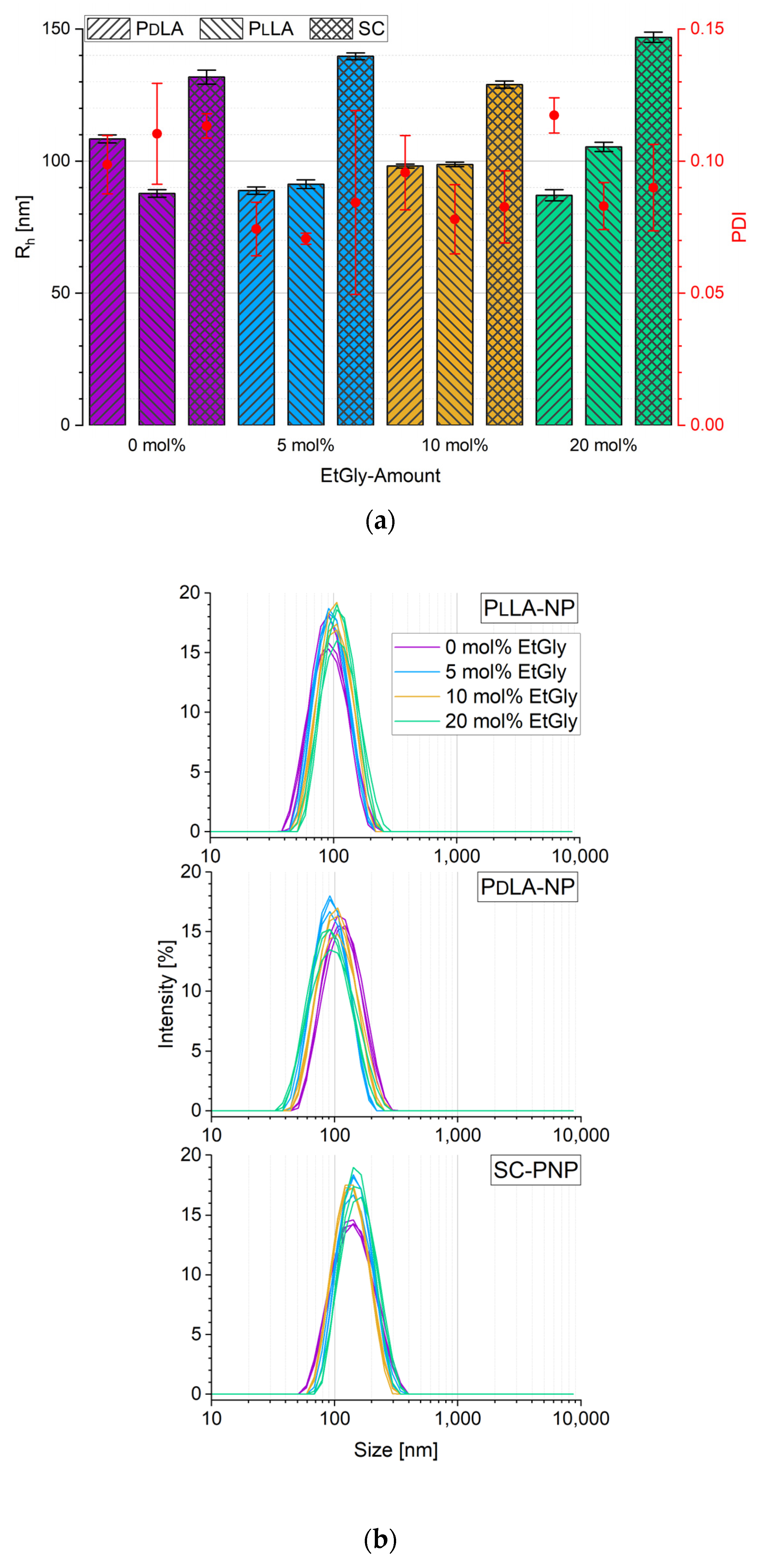
3.1.1. Particle Sizes Measured Using DLS
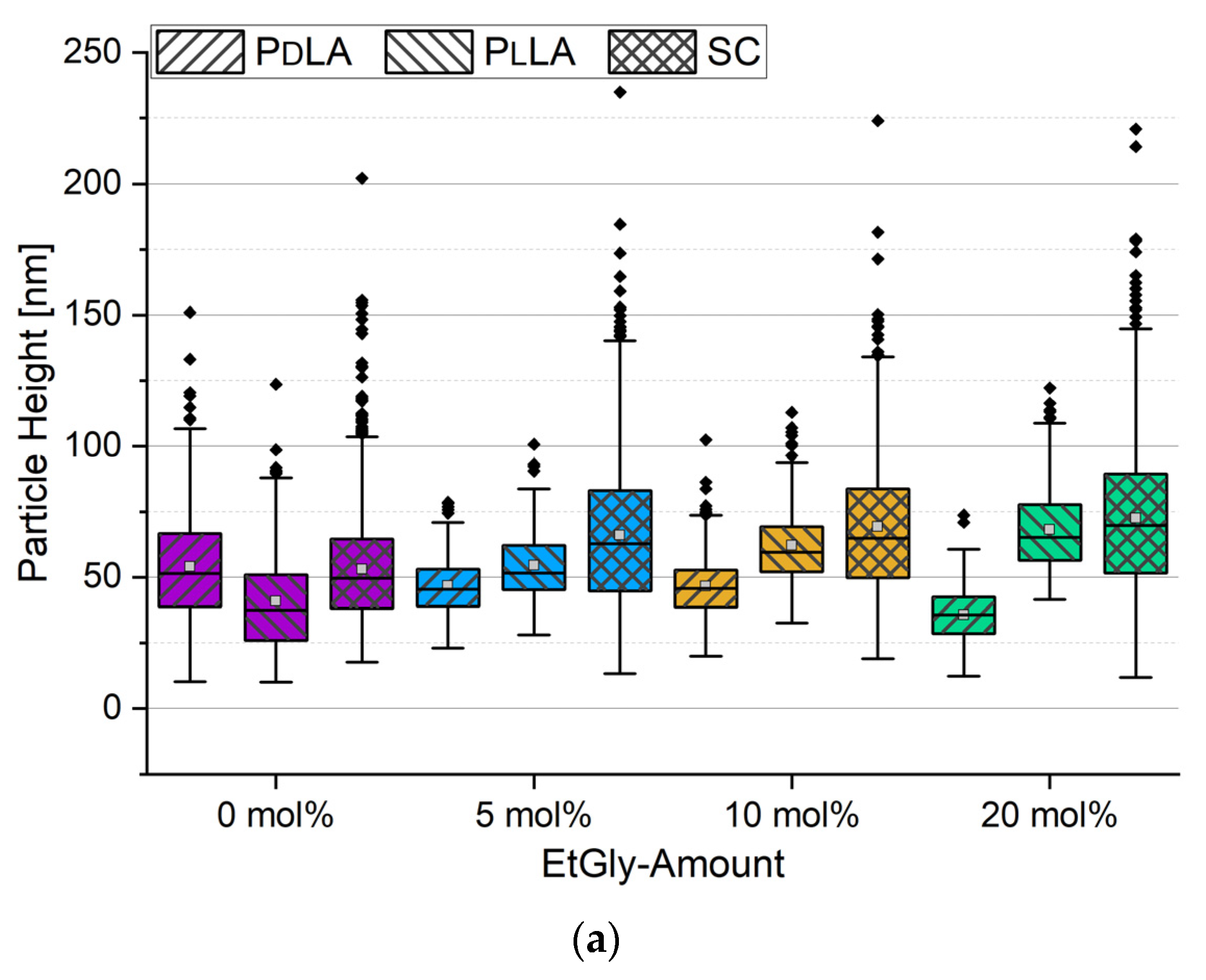
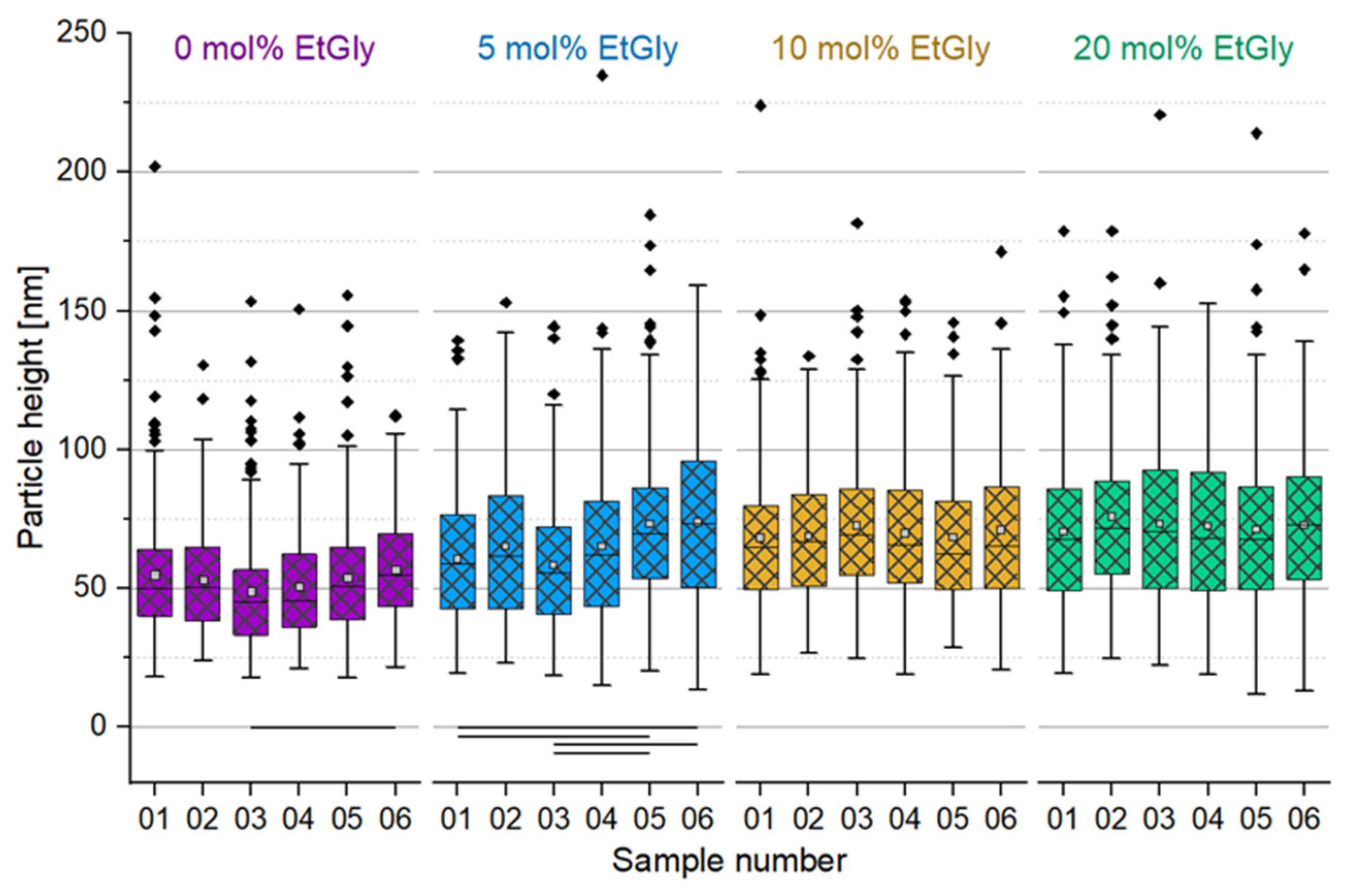
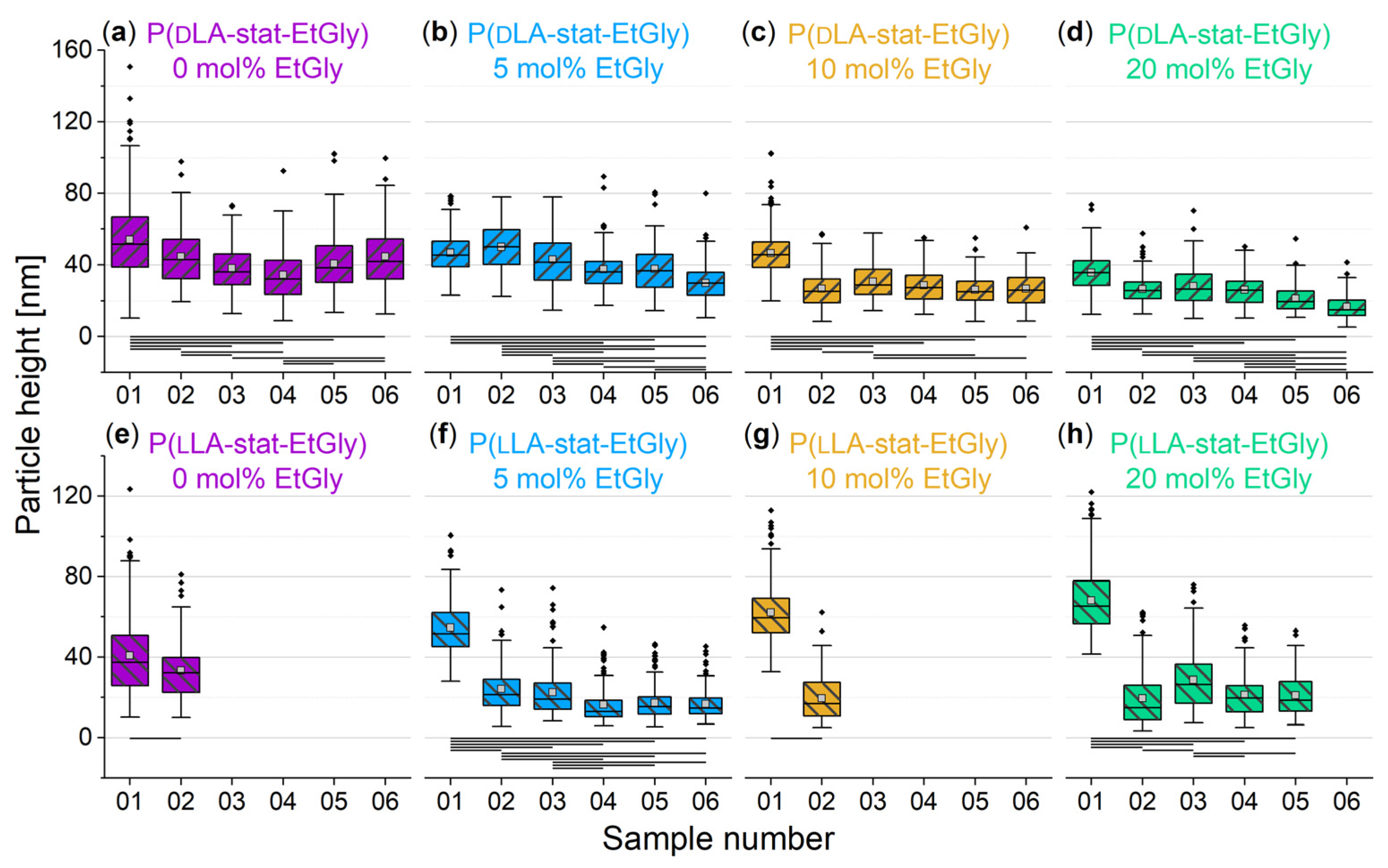
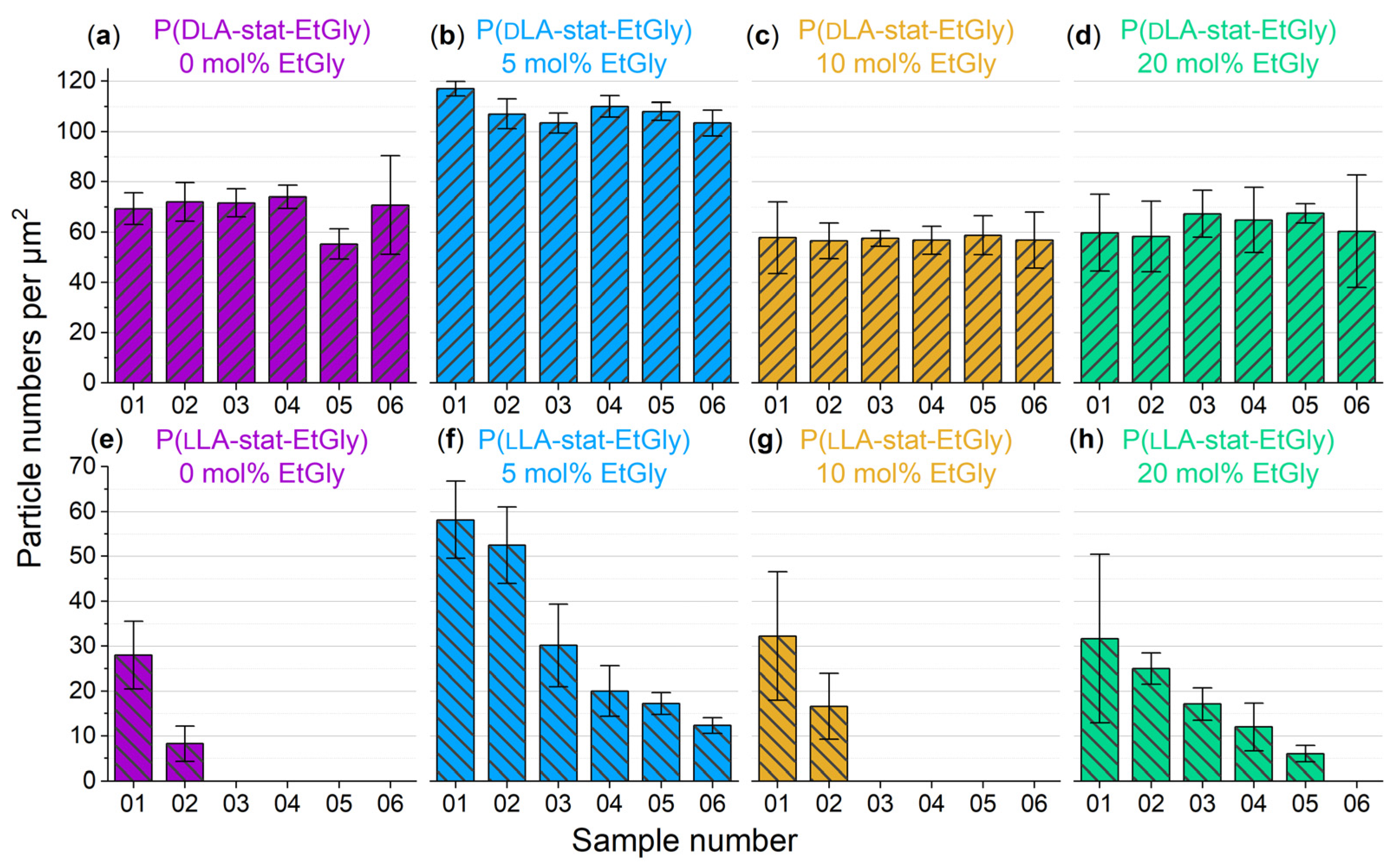
3.1.2. Particle Sizes Measured Using AFM
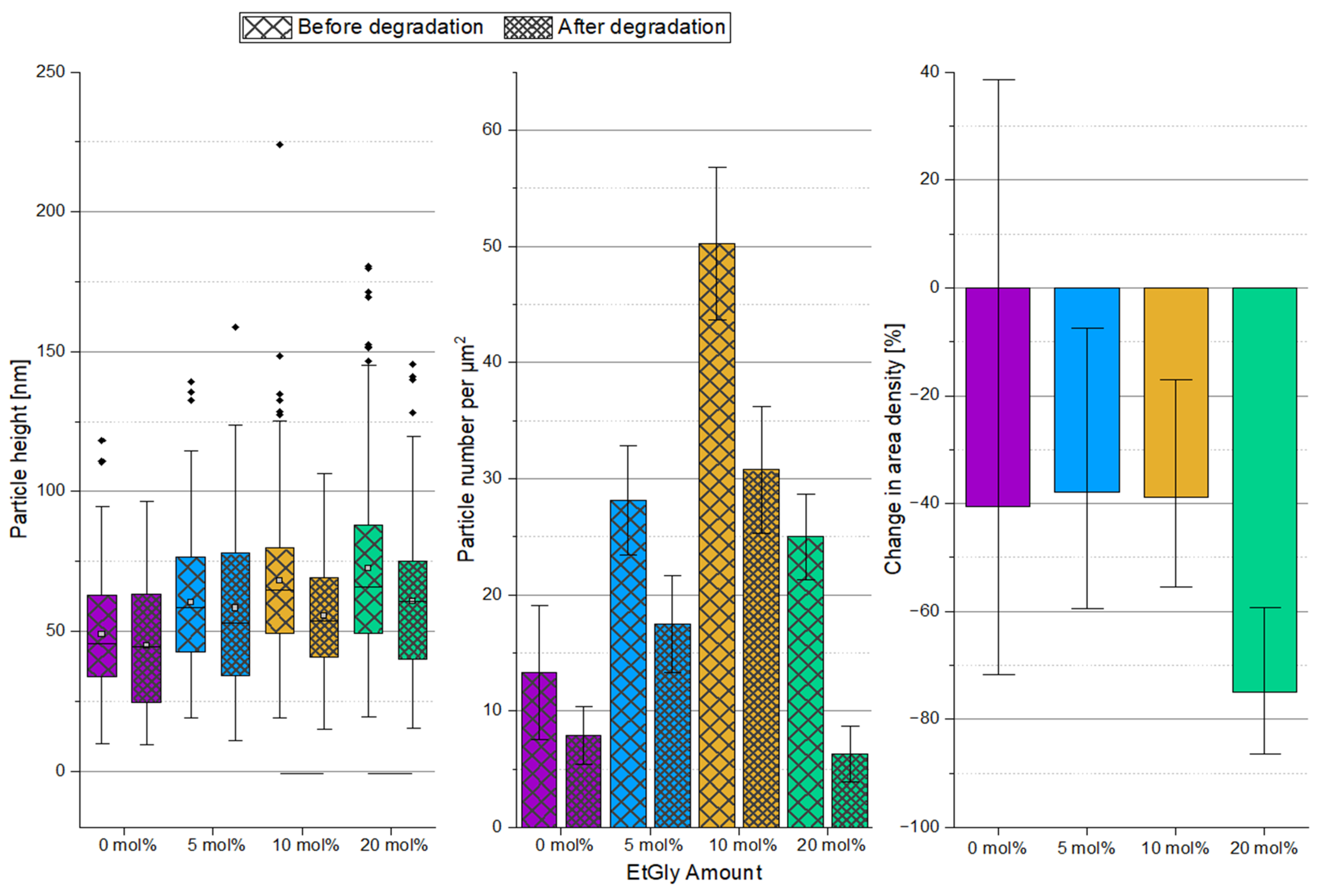
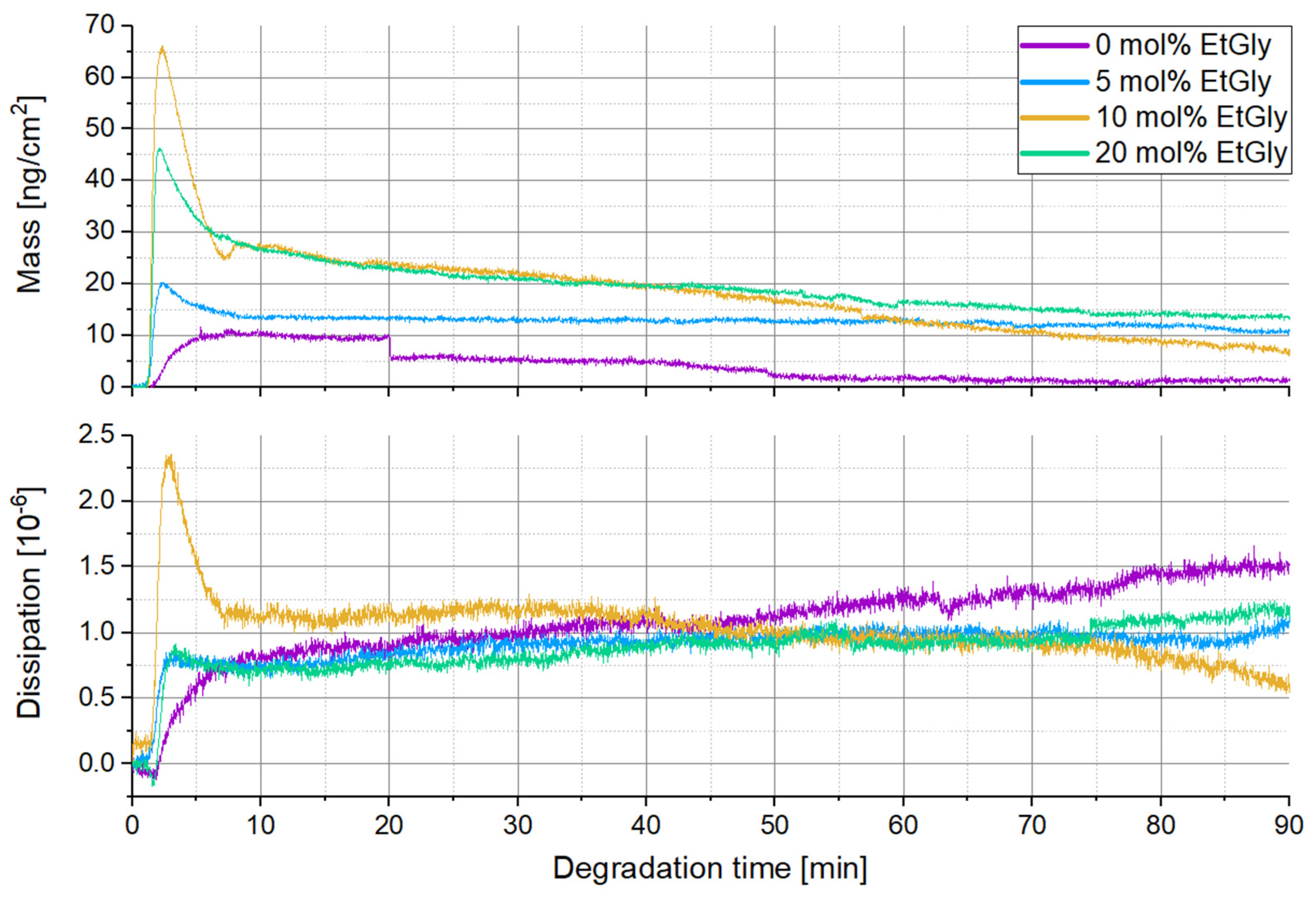
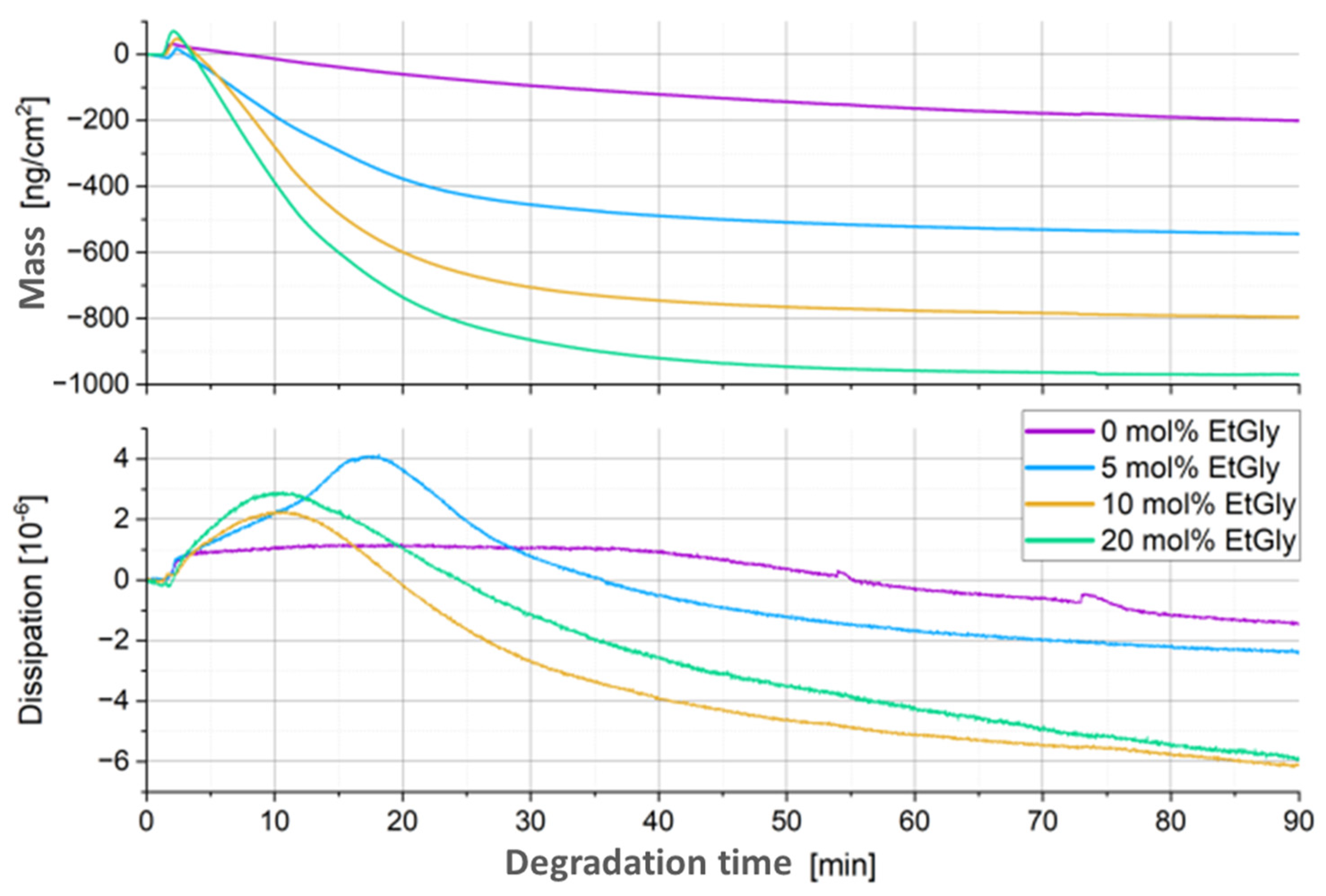
3.1.3. Degradation Evaluation of SC-PNPs
3.1.4. Degradation Evaluation of PNPs without Stereocomplexation
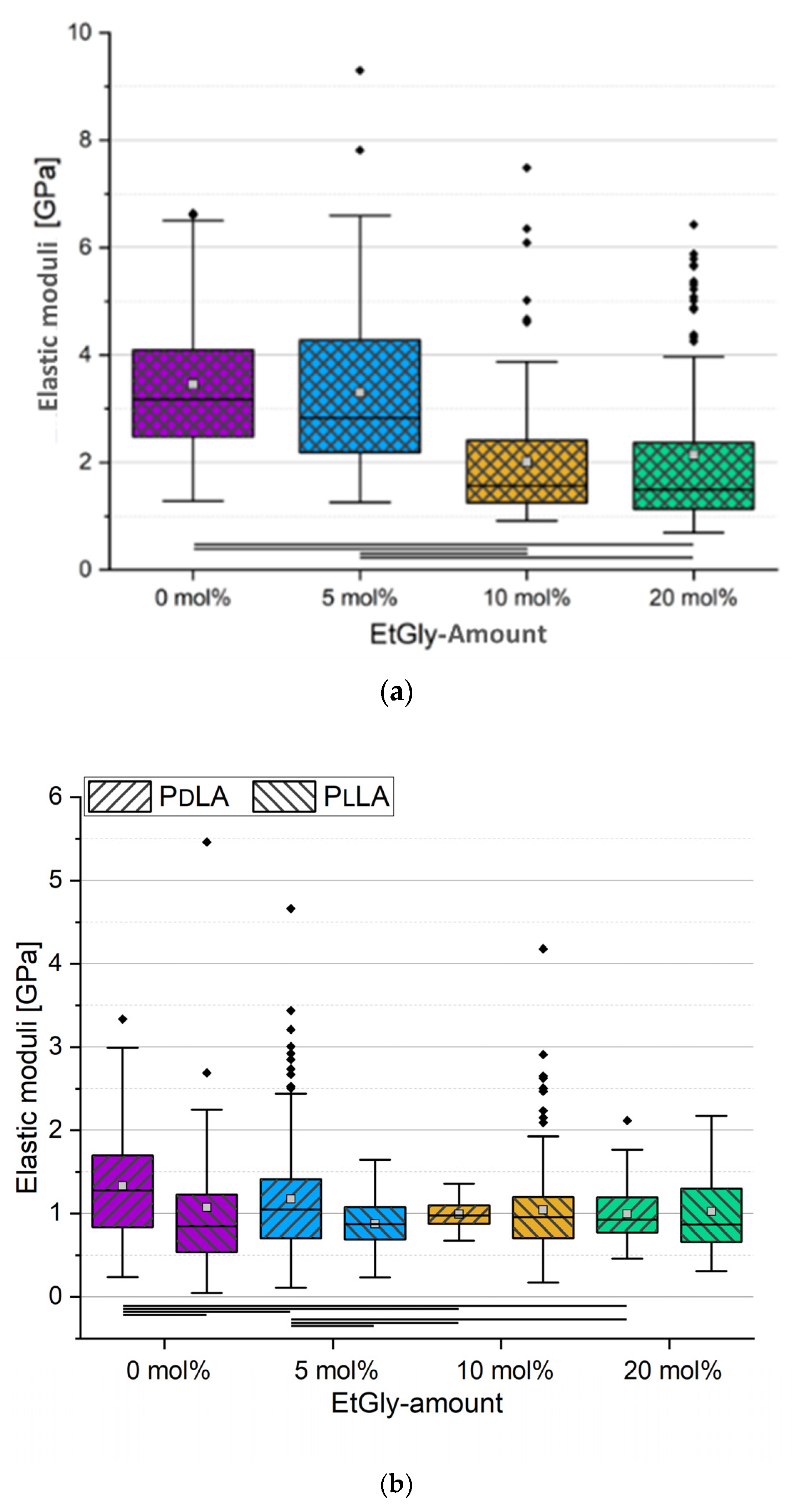
3.2. Influence of the SC, the Crystallinity, and the EtGly Amount on the Properties
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Park, K. Controlled drug delivery systems: Past forward and future back. J. Control. Release 2014, 190, 3–8. [Google Scholar] [CrossRef]
- de Faria, C.M.G.; Bissoli, M.; Vago, R.; Spinelli, A.E.; Amendola, V. Cytotoxicity of PEG-Coated Gold and Gold–Iron Alloy Nanoparticles: ROS or Ferroptosis? Nanomaterials 2023, 13, 3044. [Google Scholar] [CrossRef]
- Langer, R. Drug delivery and targeting. Nature 1998, 392 (Suppl. S6679), 5–10. [Google Scholar]
- Uhrich, K.E.; Cannizzaro, S.M.; Langer, R.S.; Shakesheff, K.M. Polymeric systems for controlled drug release. Chem. Rev. 1999, 99, 3181–3198. [Google Scholar] [CrossRef]
- Wu, Y.; Angelova, A. Recent Uses of Lipid Nanoparticles, Cell-Penetrating and Bioactive Peptides for the Development of Brain-Targeted Nanomedicines against Neurodegenerative Disorders. Nanomaterials 2023, 13, 3004. [Google Scholar] [CrossRef]
- Dong, P.; Rakesh, K.; Manukumar, H.; Mohammed, Y.H.E.; Karthik, C.; Sumathi, S.; Mallu, P.; Qin, H.-L. Innovative nano-carriers in anticancer drug delivery-a comprehensive review. Bioorg. Chem. 2019, 85, 325–336. [Google Scholar] [CrossRef]
- Prajapati, S.K.; Jain, A.; Jain, A.; Jain, S. Biodegradable polymers and constructs: A novel approach in drug delivery. Eur. Polym. J. 2019, 120, 109191. [Google Scholar] [CrossRef]
- Richert, A.; Dąbrowska, G. Enzymatic degradation and biofilm formation during biodegradation of polylactide and polycaprolactone polymers in various environments. Int. J. Biol. Macromol. 2021, 176, 226–232. [Google Scholar] [CrossRef]
- Li, S.; Girard, A.; Garreau, H.; Vert, M. Enzymatic degradation of polylactide stereocopolymers with predominant d-lactyl contents. Polym. Degrad. Stab. 2001, 71, 61–67. [Google Scholar] [CrossRef]
- Göpferich, A. Mechanisms of polymer degradation and erosion. Biomaterials 1996, 17, 103–114. [Google Scholar] [CrossRef]
- Lin, C.-C.; Anseth, K.S. Chapter ii.4.3—The biodegradation of biodegradable polymeric biomaterials. In Biomaterials Science, 3rd ed.; Ratner, B.D., Hoffman, A.S., Schoen, F.J., Lemons, J.E., Eds.; Academic Press: Cambridge, MA, USA, 2013; pp. 716–728. [Google Scholar]
- Pillai, O.; Panchagnula, R. Polymers in drug delivery. Curr. Opin. Chem. Biol. 2001, 5, 447–451. [Google Scholar] [CrossRef]
- Herzog, K.; Müller, R.-J.; Deckwer, W.-D. Mechanism and kinetics of the enzymatic hydrolysis of polyester nanoparticles by lipases. Polym. Degrad. Stab. 2006, 91, 2486–2498. [Google Scholar] [CrossRef]
- Azevedo, H.S.; Reis, R.L. Understanding the Enzymatic Degradation of Biodegradable Polymers and Strategies to Control Their Degradation Rate; CRC Press LLC: Boca Raton, FL, USA, 2005. [Google Scholar]
- Karavelidis, V.; Giliopoulos, D.; Karavas, E.; Bikiaris, D. Nanoencapsulation of a water soluble drug in biocompatible polyesters. effect of polyesters melting point and glass transition temperature on drug release behavior. Eur. J. Pharm. Sci. 2010, 41, 636–643. [Google Scholar] [CrossRef]
- Karavelidis, V.; Karavas, E.; Giliopoulos, D.; Papadimitriou, S.; Bikiaris, D. Evaluating the effects of crystallinity in new biocompatible polyester nanocarriers on drug release behavior. Int. J. Nanomed. 2011, 6, 3021. [Google Scholar]
- He, S.; Cong, Y.; Zhou, D.; Li, J.; Xie, Z.; Chen, X.; Jing, X.; Huang, Y. Dextran–platinum (IV) conjugate as reduction-responsive carrier for triggered drug release. J. Mater. Chem. B 2015, 3, 8203–8211. [Google Scholar] [CrossRef]
- Scheuer, K.; Bandelli, D.; Helbing, C.; Weber, C.; Alex, J.; Max, J.B.; Hocken, A.; Stranik, O.; Seiler, L.; Gladigau, F.; et al. Self-assembly of copolyesters into stereocomplex crystallites tunes the properties of polyester nanoparticles. Macromolecules 2020, 53, 8340–8351. [Google Scholar] [CrossRef]
- Bandelli, D.; Helbing, C.; Weber, C.; Seifert, M.; Muljajew, I.; Jandt, K.D.; Schubert, U.S. Maintaining the hydrophilic–hydrophobic balance of polyesters with adjustable crystallinity for tailor-made nanoparticles. Macromolecules 2018, 51, 5567–5576. [Google Scholar] [CrossRef]
- Bandelli, D.; Alex, J.; Helbing, C.; Ueberschaar, N.; Görls, H.; Bellstedt, P.; Weber, C.; Jandt, K.D.; Schubert, U.S. Poly (3-ethylglycolide): A well-defined polyester matching the hydrophilic hydrophobic balance of pla. Polym. Chem. 2019, 10, 5440–5451. [Google Scholar] [CrossRef]
- Tsuji, H. Poly (lactide) stereocomplexes: Formation, structure, properties, degradation, and applications. Macromol. Biosci. 2005, 5, 569–597. [Google Scholar] [CrossRef]
- Tsuji, H.; Fukui, I. Enhanced thermal stability of poly (lactide) s in the melt by enantiomeric polymer blending. Polymer 2003, 44, 2891–2896. [Google Scholar] [CrossRef]
- Wojtczak, E.; Biedroń, T.; Bednarek, M. Hydrolytic stability of polylactide stereocomplex microparticles containing metal ions. Polym. Bull. 2019, 76, 1135–1149. [Google Scholar] [CrossRef]
- Slager, J.; Domb, A.J. Biopolymer stereocomplexes. Adv. Drug Deliv. Rev. 2003, 55, 549–583. [Google Scholar] [CrossRef]
- Fessi, H.; Puisieux, F.; Devissaguet, J.P.; Ammoury, N.; Benita, S. Nano capsule formation by interfacial polymer deposition following solvent displacement. Int. J. Pharm. 1989, 55, R1–R4. [Google Scholar] [CrossRef]
- Yamashita, K.; Kikkawa, Y.; Kurokawa, K.; Doi, Y. Enzymatic degradation of poly (l-lactide) film by proteinase k: Quartz crystal microbalance and atomic force microscopy study. Biomacromolecules 2005, 6, 850–857. [Google Scholar] [CrossRef]
- Wang, Y.; Li, P.; Tran, T.T.-D.; Zhang, J.; Kong, L. Manufacturing techniques and surface engineering of polymer-based nanoparticles for targeted drug delivery to cancer. Nanomaterials 2016, 6, 26. [Google Scholar] [CrossRef]
- Tsuji, H.; Ikada, Y. Stereocomplex formation between enantiomeric poly(lactic acid)s. XI. Mechanical properties and morphology of solution-cast films. Polymer 1999, 40, 6699–6708. [Google Scholar] [CrossRef]
- Reeve, M.S.; McCarthy, S.P.; Downey, M.J.; Gross, R.A. Polylactide stereochemistry: Effect on enzymic degradability. Macromolecules 1994, 27, 825–831. [Google Scholar] [CrossRef]
- Utsel, S.; Bruce, C.; Pettersson, T.; Fogelström, L.; Carlmark, A.; Malmström, E.; Wågberg, L. Physical tuning of cellulose-polymer interactions utilizing cationic block copolymers based on pcl and quaternized pdmaema. ACS Appl. Mater. Interfaces 2012, 4, 6796–6807. [Google Scholar] [CrossRef]
- Schönherr, H.; Vancso, G.J. Scanning Force Microscopy of Polymers; Springer: Berlin/Heidelberg, Germany, 2010; Volume 27. [Google Scholar]
- Hoo, C.M.; Starostin, N.; West, P.; Mecartney, M.L. A comparison of atomic force microscopy (afm) and dynamic light scattering (dls) methods to characterize nanoparticle size distributions. J. Nanoparticle Res. 2008, 10, 89–96. [Google Scholar] [CrossRef]
- Li, S. Hydrolytic degradation characteristics of aliphatic polyesters derived from lactic and glycolic acids. J. Biomed. Mater. Res. Off. J. Soc. Biomater. Jpn. Soc. Biomater. Aust. Soc. Biomater. 1999, 48, 342–353. [Google Scholar] [CrossRef]
- Tonda-Turo, C.; Carmagnola, I.; Ciardelli, G. Quartz crystal microbalance with dissipation monitoring: A powerful method to predict the in vivo behavior of bioengineered surfaces. Front. Bioeng. Biotechnol. 2018, 6, 158. [Google Scholar] [CrossRef]
- Tsuji, H.; Miyauchi, S. Poly (l-lactide): 7. enzymatic hydrolysis of free and restricted amorphous regions in poly (l-lactide) films with different crystallinities and a fixed crystalline thickness. Polymer 2001, 42, 4463–4467. [Google Scholar] [CrossRef]
- Andersson, S.R.; Hakkarainen, M.; Inkinen, S.; Södergård, A.; Albertsson, A.-C. Customizing the hydrolytic degradation rate of stereocomplex pla through different PdLA architectures. Biomacromolecules 2012, 13, 1212–1222. [Google Scholar] [CrossRef]
- Betzel, C.; Pal, G.P.; Saenger, W. Three-dimensional structure of proteinase k at 0.15-nm resolution. Eur. J. Biochem. 1988, 178, 155–171. [Google Scholar] [CrossRef]
- Tsuji, H. Poly (lactic acid) stereocomplexes: A decade of progress. Adv. Drug Deliv. Rev. 2016, 107, 97–135. [Google Scholar] [CrossRef]
- Cappella, B.; Silbernagl, D. Nanomechanical properties of polymer thin films measured by force–distance curves. Thin Solid Film. 2008, 516, 1952–1960. [Google Scholar] [CrossRef]
- Cappella, B.; Dietler, G. Force-distance curves by atomic force microscopy. Surf. Sci. Rep. 1999, 34, 1–104. [Google Scholar] [CrossRef]
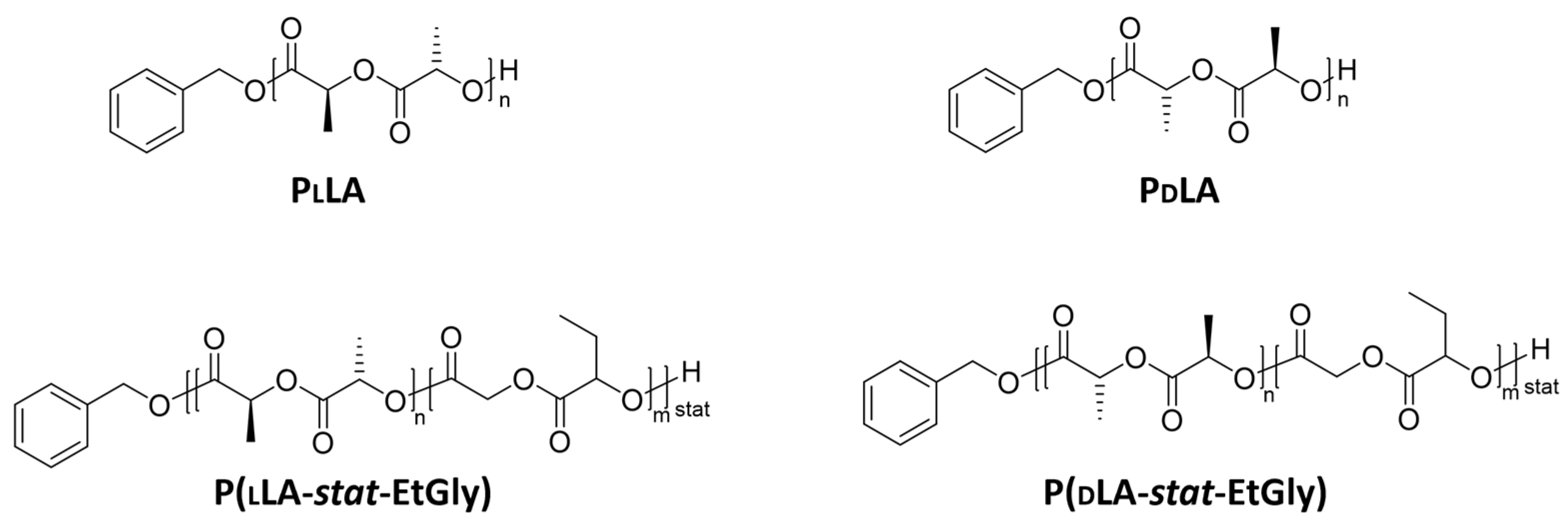
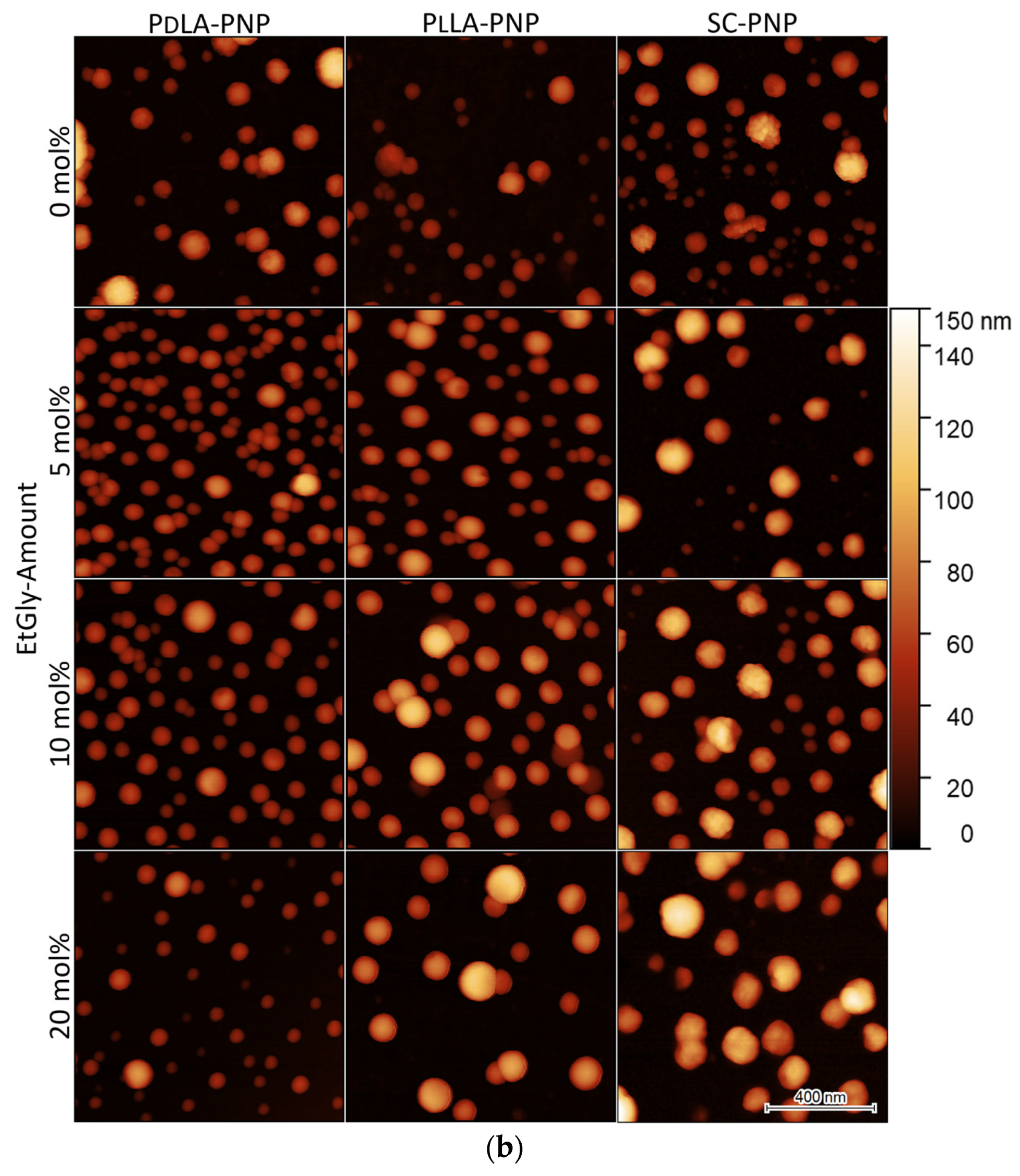



| Polymer | mol% LA/EtGly | Mn [kg mol−1] SEC3 | Đ SEC3 |
|---|---|---|---|
| PlLA | 100/0 | 16 | 1.06 |
| PdLA | 100/0 | 19 | 1.06 |
| P(lLA-stat-EtGly) | 95/05 | 16 | 1.11 |
| P(lLA-stat-EtGly) | 90/10 | 13 | 1.20 |
| P(lLA-stat-EtGly) | 80/20 | 12 | 1.23 |
| P(dLA-stat-EtGly) | 95/05 | 19 | 1.10 |
| P(dLA-stat-EtGly) | 90/10 | 18 | 1.21 |
| P(dLA-stat-EtGly) | 80/20 | 15 | 1.28 |
| Sample number | 01 | 02 | 03 | 04 | 05 | 06 |
| Degradation time [h] | 0.0 | 1.5 | 3.0 | 4.5 | 6.0 | 24.0 |
| EtGly Amount | |||
|---|---|---|---|
| At the Beginning | At the End | ||
| 0 mol% | 11.6 | −0.37 | −0.12 |
| 5 mol% | 20.3 | −1.33 | −0.21 |
| 10 mol% | 66.1 | −8.55 | −0.34 |
| 20 mol% | 46.3 | −3.44 | −0.18 |
| EtGly Amount | |||
|---|---|---|---|
| At the Beginning | At the End | ||
| 0 mol% | 30.9 | −5.76 | −1.14 |
| 5 mol% | 16.7 | −26.21 | −0.66 |
| 10 mol% | 47.1 | −39.17 | −0.46 |
| 20 mol% | 70.5 | −57.80 | −0.10 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yin, C.; Hemstedt, J.; Scheuer, K.; Struczyńska, M.; Weber, C.; Schubert, U.S.; Bossert, J.; Jandt, K.D. The Effect of Stereocomplexation and Crystallinity on the Degradation of Polylactide Nanoparticles. Nanomaterials 2024, 14, 440. https://doi.org/10.3390/nano14050440
Yin C, Hemstedt J, Scheuer K, Struczyńska M, Weber C, Schubert US, Bossert J, Jandt KD. The Effect of Stereocomplexation and Crystallinity on the Degradation of Polylactide Nanoparticles. Nanomaterials. 2024; 14(5):440. https://doi.org/10.3390/nano14050440
Chicago/Turabian StyleYin, Chuan, Jenny Hemstedt, Karl Scheuer, Maja Struczyńska, Christine Weber, Ulrich S. Schubert, Jörg Bossert, and Klaus D. Jandt. 2024. "The Effect of Stereocomplexation and Crystallinity on the Degradation of Polylactide Nanoparticles" Nanomaterials 14, no. 5: 440. https://doi.org/10.3390/nano14050440





