The Pd (II) Reduction Mechanisms in Bacillus megaterium Y-4 Revealed by Proteomic Analysis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Synthesis and Analysis of Microbial Pd (0) Nanoparticle
3. Results
3.1. Production of Pd-NPs by B. megaterium Y-4
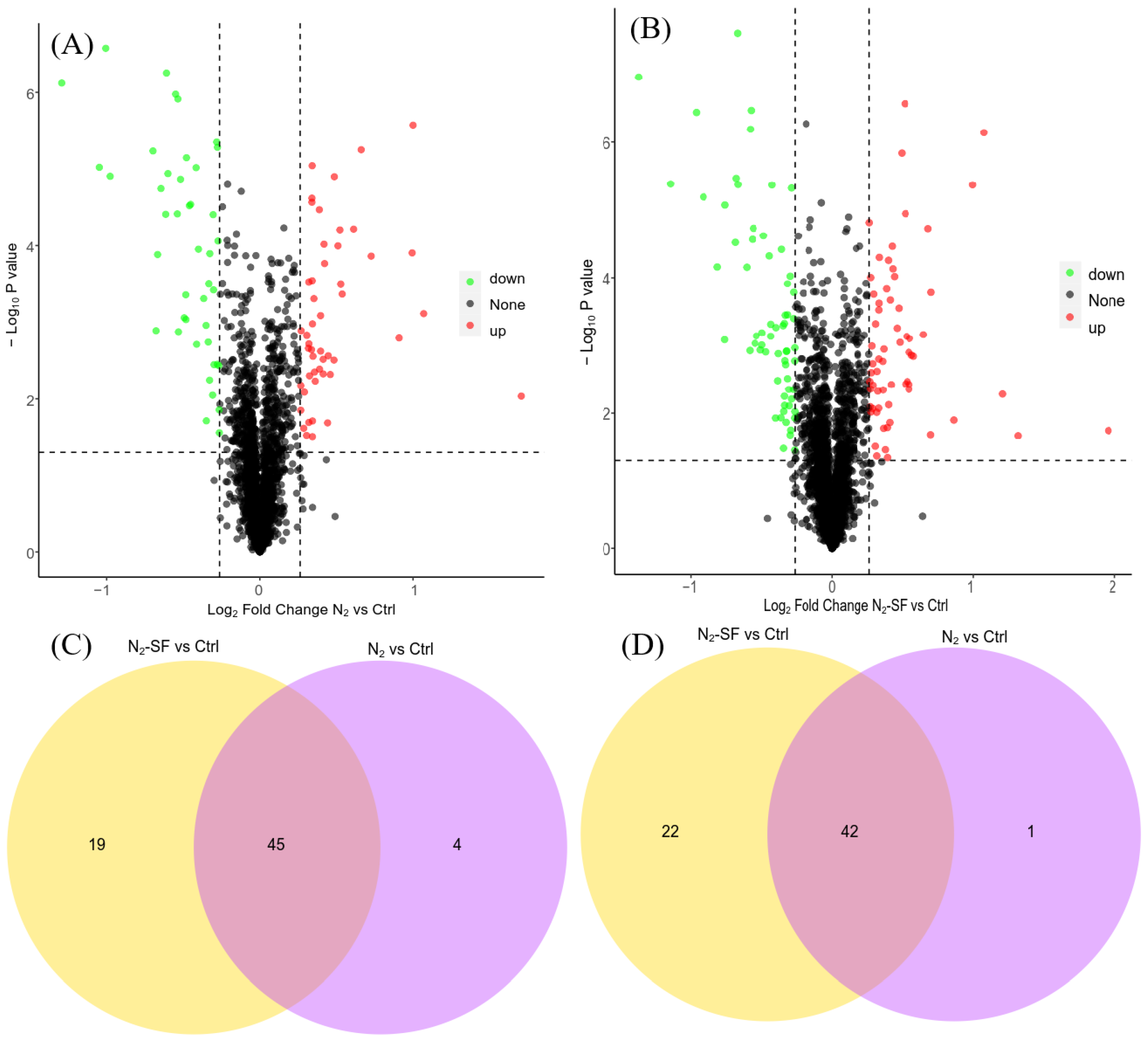
3.2. Protein Identification and Comparison during Palladium Reduction
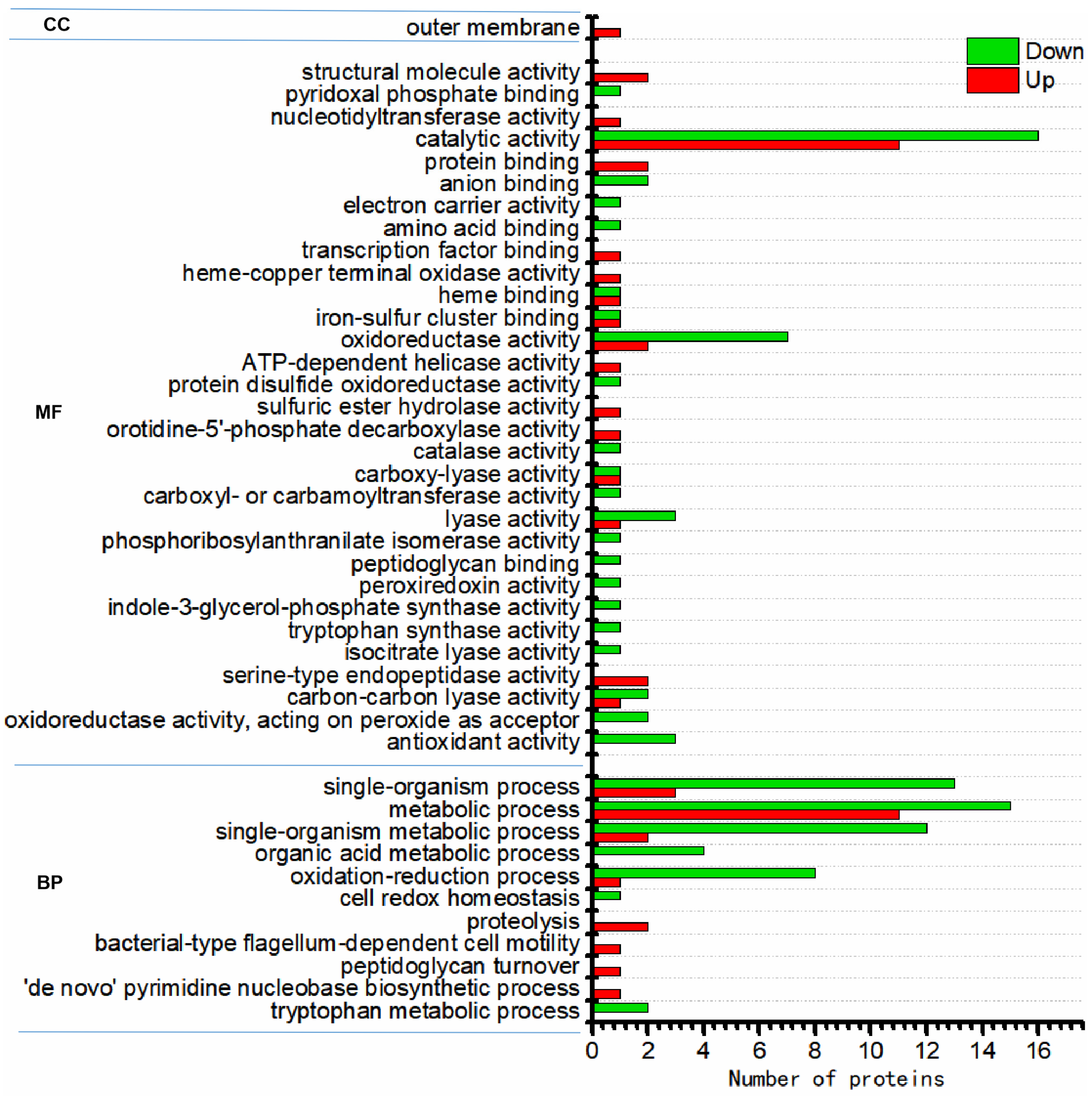
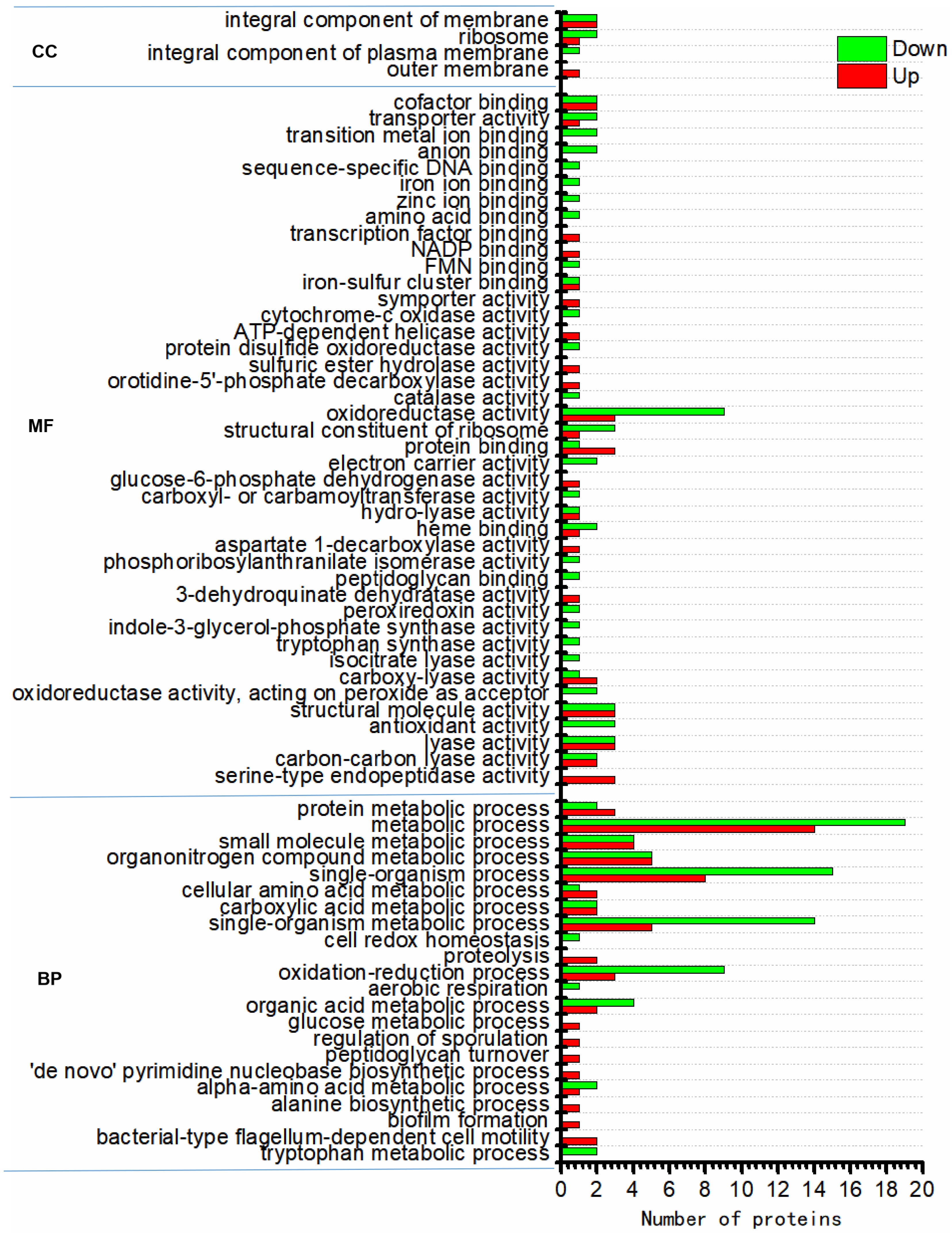
3.3. Functional Characterization of Differentially Expressed Proteins
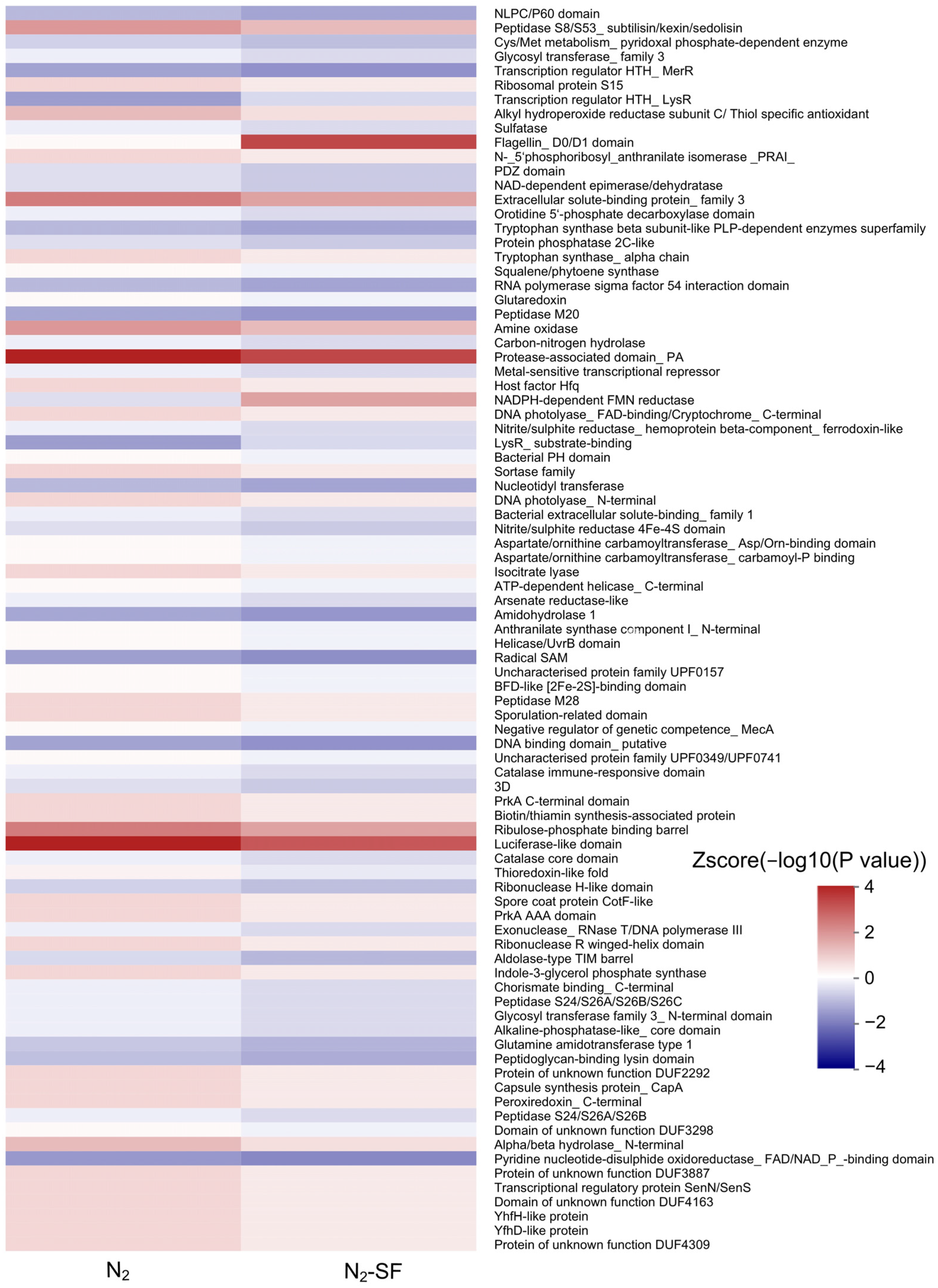
3.4. Functional Enrichment of Differentially Regulated Proteins
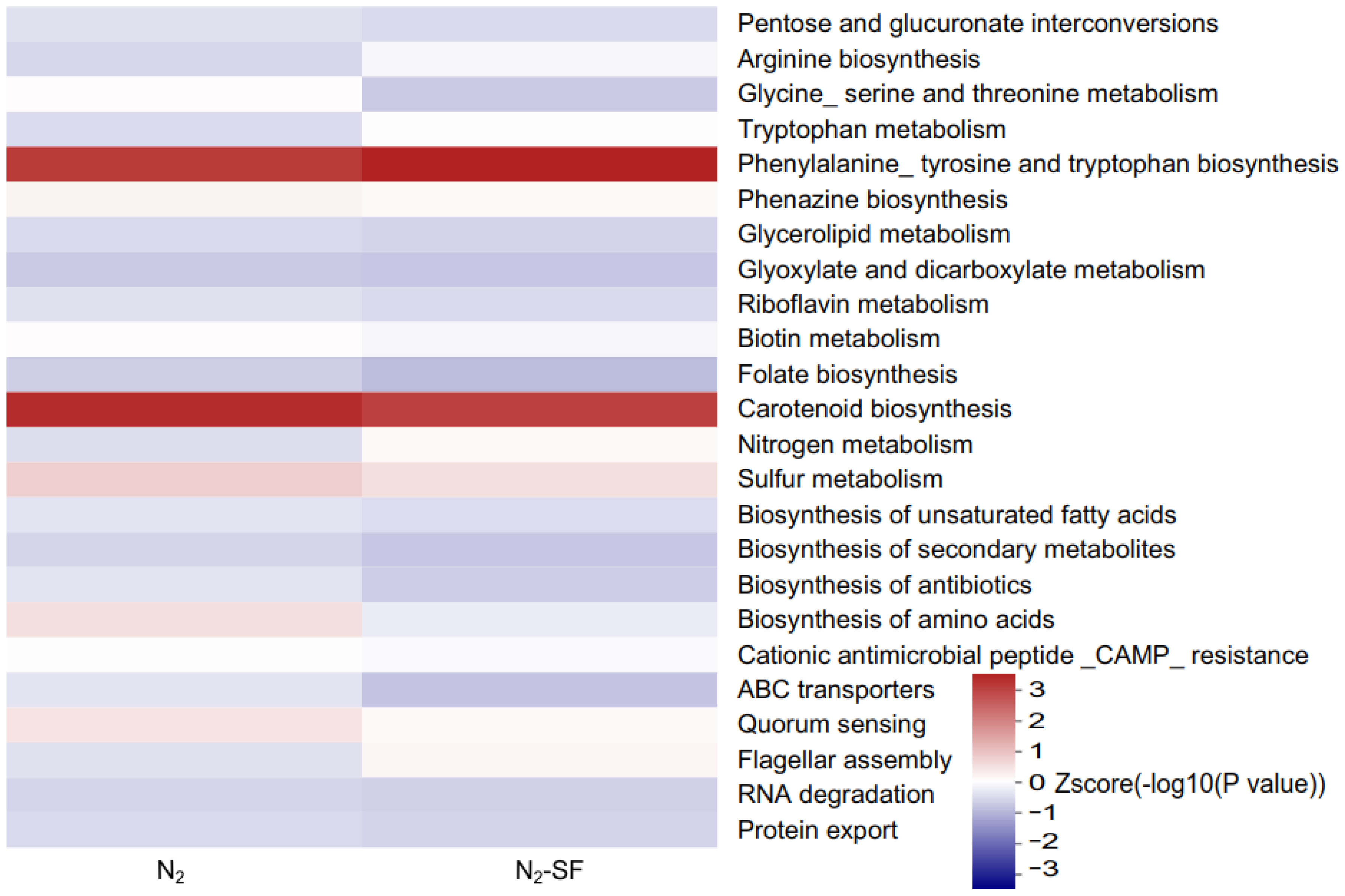
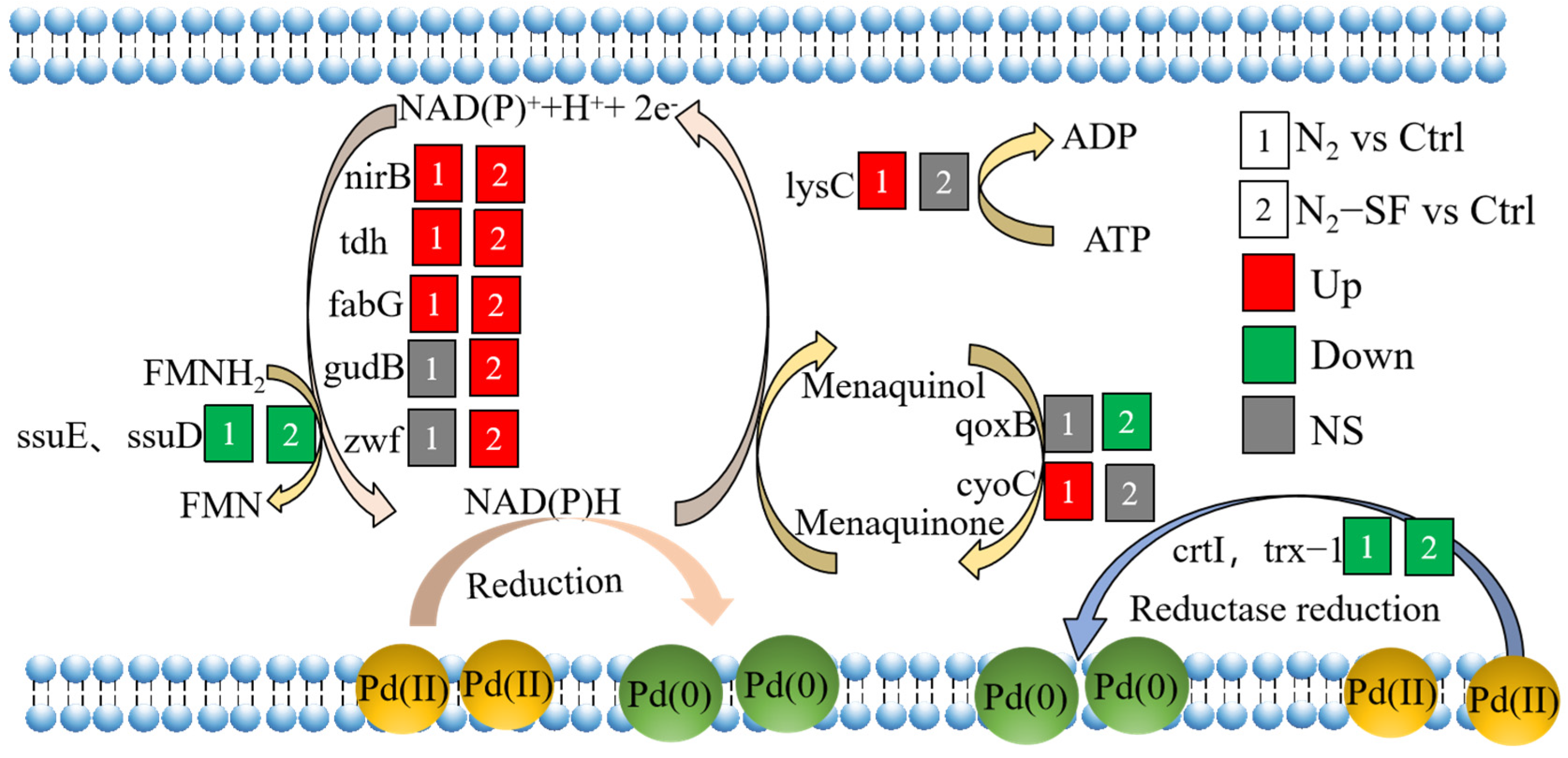
3.5. Metabolic Pathways Identified for Palladium Reduction
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Zhou, L.; Yang, J.; Ma, F.; Pi, S.; Tang, A.; Li, A. Recycling of Pd (0) catalysts by magnetic nanocomposites—Microbial extracellular polymeric substances@Fe3O4. J. Environ. Manag. 2021, 280, 111834. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Rajesh, N. Augmenting the adsorption of palladium from spent catalyst using a thiazole ligand tethered on an amine functionalized polymeric resin. Chem. Eng. J. 2016, 283, 999–1008. [Google Scholar] [CrossRef]
- Chen, Y.; Chen, Y. Difference in toxicity of Pd (II) and mechanism of action before and after reduction by Bacillus wiedmannii MSM. Environ. Sci. Pollut. R 2021, 29, 1824–1835. [Google Scholar] [CrossRef]
- Yates, M.D.; Cusick, R.D.; Logan, B.E. Extracellular palladium nanoparticle production using Geobacter sulfurreducens. ACS Sustain. Chem. Eng. 2013, 1, 1165–1171. [Google Scholar] [CrossRef]
- Tuo, Y.; Liu, G.; Zhou, J.; Wang, A.; Wang, J.; Jin, R.; Lv, H. Microbial formation of palladium nanoparticles by Geobacter sulfurreducens for chromate reduction. Bioresour. Technol. 2013, 133, 606–611. [Google Scholar] [CrossRef]
- De Windt, W.; Aelterman, P.; Verstraete, W. Bioreductive deposition of palladium (0) nanoparticles on Shewanella oneidensis with catalytic activity towards reductive dechlorination of polychlorinated biphenyls. Environ. Microbiol. 2005, 7, 314–325. [Google Scholar] [CrossRef]
- De Windt, W.; Boon, N.; Van den Bulcke, J.; Rubberecht, L.; Prata, F.; Mast, J.; Hennebel, T.; Verstraete, W. Biological control of the size and reactivity of catalytic Pd (0) produced by Shewanella oneidensis. Anton. Leeuw. Int. J. G 2006, 90, 377–389. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.; Zhu, N.; Kang, N.; Ha, C.; Shi, C.; Wu, P. Biorecovery mechanism of palladium as nanoparticles by Enterococcus faecalis: From biosorption to bioreduction. Chem. Eng. J. 2017, 328, 1051–1057. [Google Scholar] [CrossRef]
- You, L.; Pan, D.; Chen, N.; Lin, W.; Chen, Q.; Rensing, C.; Zhou, S. Extracellular electron transfer of Enterobacter cloacae SgZ-5T via bi-mediators for the biorecovery of palladium as nanorods. Environ. Int. 2019, 123, 1–9. [Google Scholar] [CrossRef]
- Asada, T.; Edamitsu, Y.; Wakashiba, K.; Nomura, T. Bioreduction of aqueous palladium ions by Shewanella algae under atmospheric conditions. Hydrometallurgy 2024, 224, 106244. [Google Scholar] [CrossRef]
- Bunge, M.; Sobjerg, L.S.; Rotaru, A.E.; Gauthier, D.; Lindhardt, A.T.; Hause, G.; Finster, K.; Kingshott, P.; Skrydstrup, T.; Meyer, R.L. Formation of palladium(0) nanoparticles at microbial surfaces. Biotechnol. Bioeng. 2010, 107, 206–215. [Google Scholar] [CrossRef]
- Chen, Y.; Hu, K.; Chen, Y. The effect of biotic and abiotic environmental factors on Pd (II) adsorption and reduction by Bacillus megaterium Y-4. Chemosphere 2019, 220, 1058–1066. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhao, Q.; Chen, B. Reduction and removal of Cr(VI) in water using biosynthesized palladium nanoparticles loaded Shewanella oneidensis MR-1. Sci. Total Environ. 2022, 805, 150336. [Google Scholar] [CrossRef] [PubMed]
- Schlüter, M.; Hentzel, T.; Suarez, C.; Koch, M.; Lorenz, W.G.; Böhm, L.; Düring, R.; Koinig, K.A.; Bunge, M. Synthesis of novel palladium(0) nanocatalysts by microorganisms from heavy-metal-influenced high-alpine sites for dehalogenation of polychlorinated dioxins. Chemosphere 2014, 117, 462–470. [Google Scholar] [CrossRef] [PubMed]
- Rybochkin, P.V.; Perchikov, R.N.; Ya Karlinskii, B.; Kamanina, O.A.; Arlyapov, V.A.; Kashin, A.S.; Ananikov, V.P. Aerobic bacteria-supported biohybrid palladium catalysts for efficient cross-coupling reactions. J. Catal. 2024, 429, 115238. [Google Scholar] [CrossRef]
- Wu, R.; Tian, X.; Xiao, Y.; Ulstrup, J.; Christensen, H.; Zhao, F.; Zhang, J. Selective electrocatalysis of biofuel molecular oxidation using palladium nanoparticles generated on Shewanella oneidensis MR-1. J. Mater. Chem. A 2018, 6, 10655–10662. [Google Scholar] [CrossRef]
- Yang, Z.; Hou, Y.; Zhang, B.; Cheng, H.; Yong, Y.; Liu, W.; Han, J.; Liu, S.; Wang, A. Insights into palladium nanoparticles produced by Shewanella oneidensis MR-1, Roles of NADH dehydrogenases and hydrogenases. Environ. Res. 2020, 191, 110196. [Google Scholar] [CrossRef]
- Chen, Y.; Chen, Y.; Wang, B.; Mao, C.; Liu, C.; Liu, R.; Yan, B. From trash to treasure: Microbial conversion of palladium contaminants into valuable Pd nanoparticles by Bacillus thuringiensis Y9. J. Clean. Prod. 2022, 366, 132880. [Google Scholar] [CrossRef]
- Matsumoto, T.; Kamino, M.; Yamada, R.; Konishi, Y.; Ogino, H. Identification of genes responsible for reducing palladium ion in Escherichia coli. J. Biotechnol. 2020, 324, 7–10. [Google Scholar] [CrossRef]
- Gang, H.; Xiao, C.; Xiao, Y.; Yan, W.; Bai, R.; Ding, R.; Yang, Z.; Zhao, F. Proteomic analysis of the reduction and resistance mechanisms of Shewanella oneidensis MR-1 under long-term hexavalent chromium stress. Environ. Int. 2019, 127, 94–102. [Google Scholar] [CrossRef]
- Perez-Riverol, Y.; Bai, J.; Bandla, C.; García-Seisdedos, D.; Hewapathirana, S.; Kamatchinathan, S.; Kundu, D.J.; Prakash, A.; Frericks-Zipper, A.; Eisenacher, M.; et al. The PRIDE database resources in 2022, a hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res. 2022, 50, D543–D552. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.; Park, T.J.; Lee, D.C.; Lee, S.Y. Recombinant Escherichia coli as a biofactory for various single- and multi-element nanomaterials. Proc. Natl. Acad. Sci. USA 2018, 115, 5944–5949. [Google Scholar] [CrossRef]
- Dundas, C.M.; Graham, A.J.; Romanovicz, D.K.; Keitz, B.K. Extracellular electron transfer by Shewanella oneidensis controls palladium nanoparticle phenotype. ACS Synth. Biol. 2018, 7, 2726–2736. [Google Scholar] [CrossRef]
- Li, L.; Yan, W.; Zhang, B.; Zhang, H.; Geng, R.; Sun, S.; Guan, X. Coupling of selenate reduction and pyrrhotite oxidation by indigenous microbial consortium in natural aquifer. Water Res. 2023, 238, 119987. [Google Scholar] [CrossRef] [PubMed]
- Pat-Espadas, A.M.; Razo-Flores, E.; Rangel-Mendez, J.R.; Cervantes, F.J. Direct and quinone-mediated palladium reduction by Geobacter sulfurreducens: Mechanisms and modeling. Environ. Sci. Technol. 2014, 48, 2910–2919. [Google Scholar] [CrossRef] [PubMed]
- Brutinel, E.D.; Gralnick, J.A. Shuttling happens: Soluble flavin mediators of extracellular electron transfer in Shewanella. Appl. Microbiol. Biot. 2012, 93, 41–48. [Google Scholar] [CrossRef]
- Ohtake, H.; Cervantes, C.; Silver, S. Decreased chromate uptake in Pseudomonas fluorescens carrying a chromate resistance plasmid. J. Bacteriol. 1987, 169, 3853–3856. [Google Scholar] [CrossRef]
- Bulut, H.; Moniot, S.; Licht, A.; Scheffel, F.; Gathmann, S.; Saenger, W.; Schneider, E. Crystal Structures of Two Solute Receptors for l-Cystine and l-Cysteine, Respectively, of the Human Pathogen Neisseria gonorrhoeae. J. Mol. Biol. 2012, 415, 560–572. [Google Scholar] [CrossRef]
- Qin, Q.L.; Li, Y.; Zhang, Y.J.; Zhou, Z.M.; Zhang, W.X.; Chen, X.L.; Zhang, X.Y.; Zhou, B.C.; Wang, L.; Zhang, Y.Z. Comparative genomics reveals a deep-sea sediment-adapted life style of Pseudoalteromonas sp. SM9913. ISME J. 2011, 5, 274–284. [Google Scholar] [CrossRef]
- Wang, L.; Huang, L.; Su, Y.; Qin, Y.; Kong, W.; Ma, Y.; Xu, X.; Lin, M.; Zheng, J.; Yan, Q. Involvement of the flagellar assembly pathway in Vibrio alginolyticus adhesion under environmental stresses. Front Cell. Infect Microbiol. 2015, 5, 59. [Google Scholar] [CrossRef]
- Marx, H.; Minogue, C.E.; Jayaraman, D.; Richards, A.L.; Kwiecien, N.W.; Siahpirani, A.F.; Rajasekar, S.; Maeda, J.; Garcia, K.; Del Valle-Echevarria, A.R.; et al. A proteomic atlas of the legume Medicago truncatula and its nitrogen-fixing endosymbiont Sinorhizobium meliloti. Nat. Biotechnol. 2016, 34, 1198. [Google Scholar] [CrossRef] [PubMed]
- Nie, L.; Zhang, H.; Wu, Z.; Wang, Y.; Liu, H.; Wu, X.; Wang, W. Modified TCA/acetone precipitation of plant proteins for proteomic analysis. PLoS ONE 2018, 13, e0202238. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Kachuk, C.; Stephen, K.; Doucette, A. Comparison of sodium dodecyl sulfate depletion techniques for proteome analysis by mass spectrometry. J Chromatogr. A 2015, 1418, 158–166. [Google Scholar] [CrossRef]
- Wisniewski, J.R.; Zougman, A.; Nagaraj, N.; Mann, M. Universal sample preparation method for proteome analysis. Nat. Methods 2009, 6, 359–360. [Google Scholar] [CrossRef]
- Gillette, M.A.; Satpathy, S.; Cao, S.; Dhanasekaran, S.M.; Vasaikar, S.V.; Krug, K.; Petralia, F.; Li, Y.Z.; Liang, W.W.; Reva, B.; et al. Proteogenomic Characterization Reveals Therapeutic Vulnerabilities in Lung Adenocarcinoma. Cell 2020, 182, 200. [Google Scholar] [CrossRef]


| Run Name | 126 | 127N | 128N | 129N | 130N | 131N | 132N | 133N | 134N |
|---|---|---|---|---|---|---|---|---|---|
| run1 | Ctrl-1 | Ctrl-2 | Ctrl-3 | N2-1 | N2-2 | N2-3 | N2-SF-1 | N2-SF-2 | N2-SF-3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Y.; Wang, J.; Chen, D.; Wang, B.; Wu, J.; Liu, R.; Li, Q. The Pd (II) Reduction Mechanisms in Bacillus megaterium Y-4 Revealed by Proteomic Analysis. Nanomaterials 2024, 14, 512. https://doi.org/10.3390/nano14060512
Chen Y, Wang J, Chen D, Wang B, Wu J, Liu R, Li Q. The Pd (II) Reduction Mechanisms in Bacillus megaterium Y-4 Revealed by Proteomic Analysis. Nanomaterials. 2024; 14(6):512. https://doi.org/10.3390/nano14060512
Chicago/Turabian StyleChen, Yuan, Jiaxing Wang, Daidi Chen, Boxi Wang, Jinchuan Wu, Rongrong Liu, and Qingxin Li. 2024. "The Pd (II) Reduction Mechanisms in Bacillus megaterium Y-4 Revealed by Proteomic Analysis" Nanomaterials 14, no. 6: 512. https://doi.org/10.3390/nano14060512






