In-Silico Design, Synthesis and Evaluation of a Nanostructured Hydrogel as a Dimethoate Removal Agent
Abstract
:1. Introduction
2. .Results and Discussion
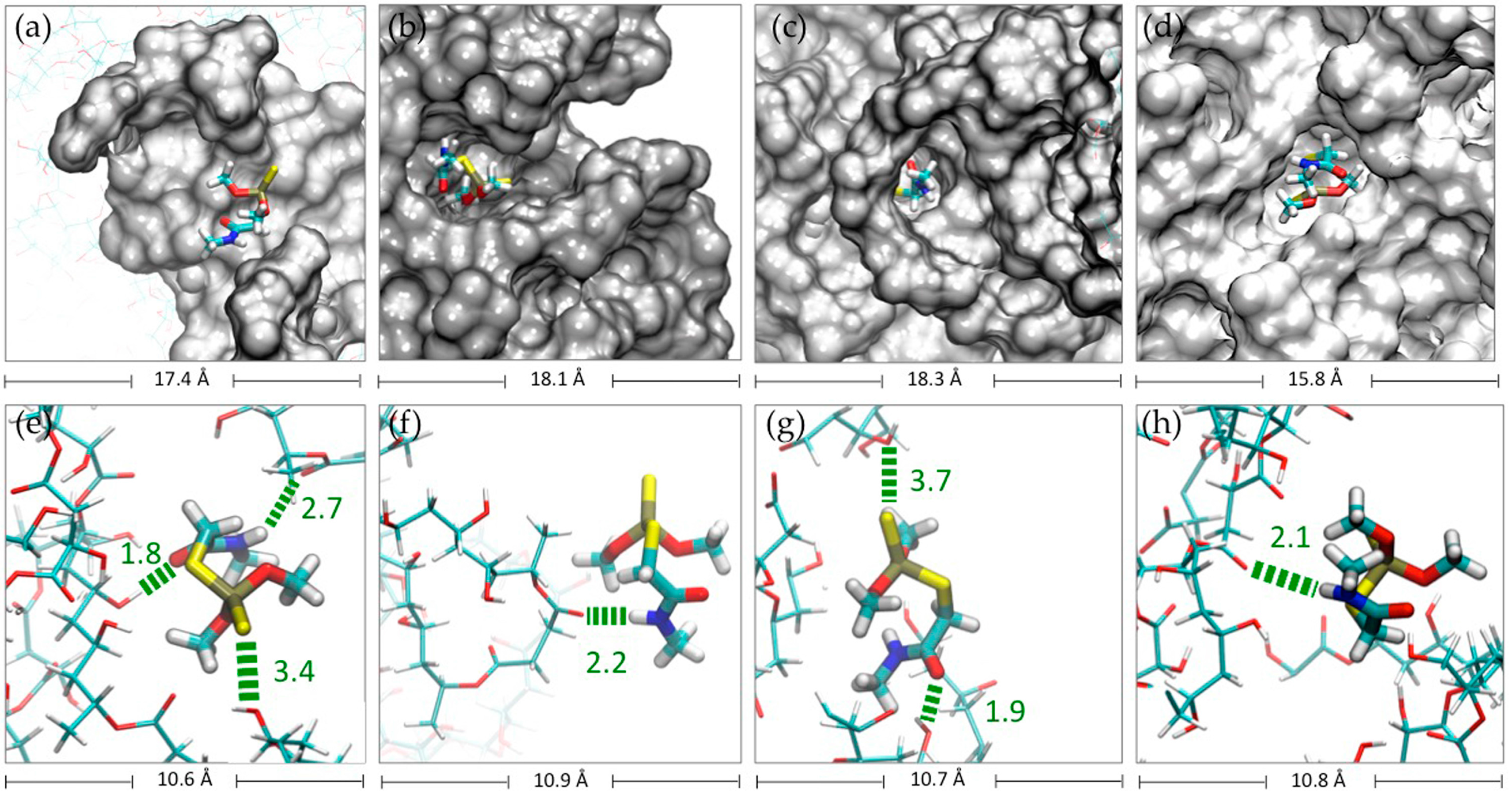
2.1. In-Silico Interaction Energy
2.2. Molecular Dynamics Simulations (MDS) Studies
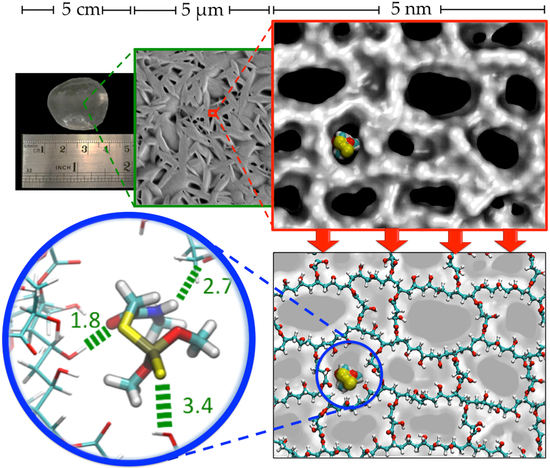
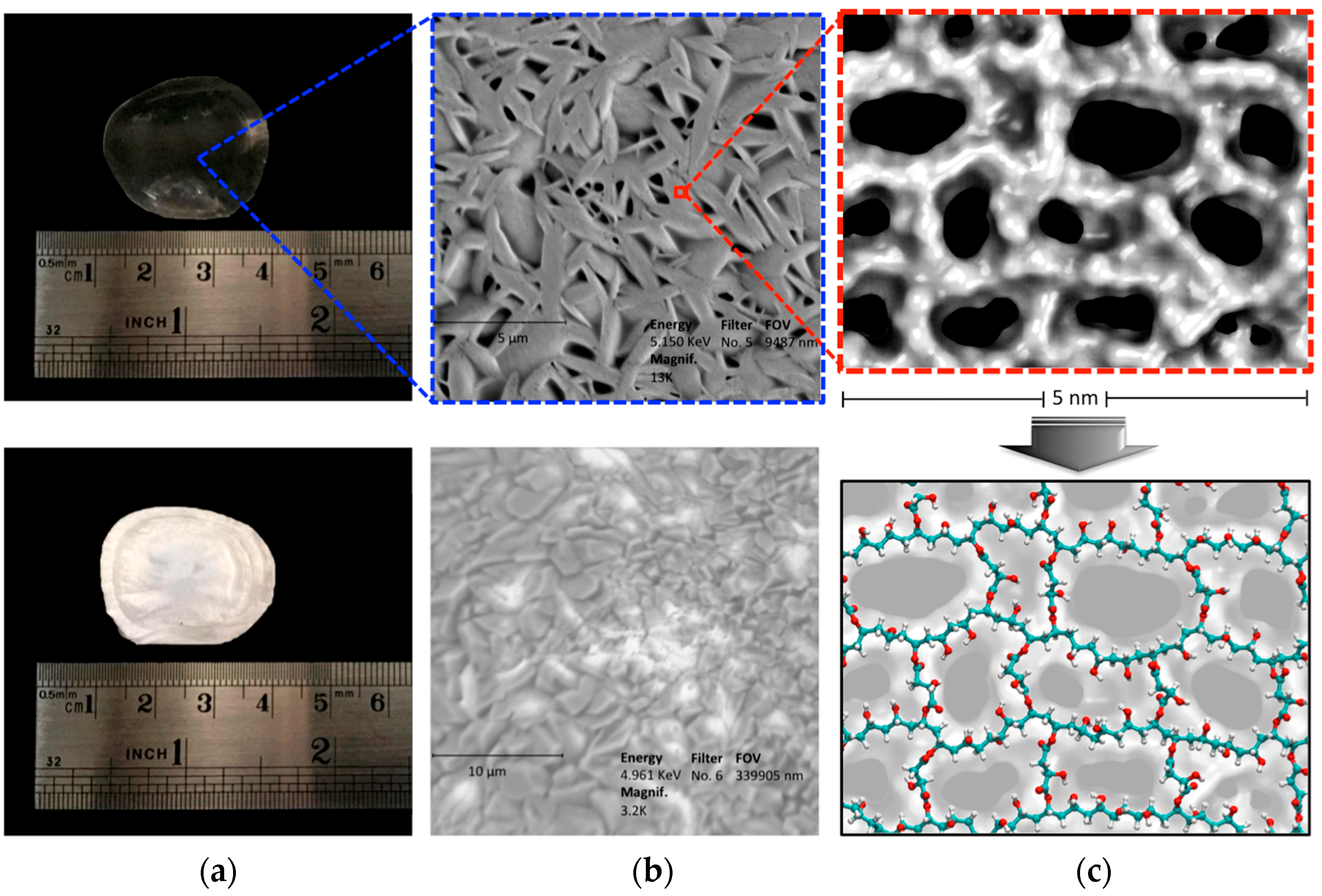
2.3. Characterization of CLPH-MA20 by Scanning Electron Microcopy (SEM)
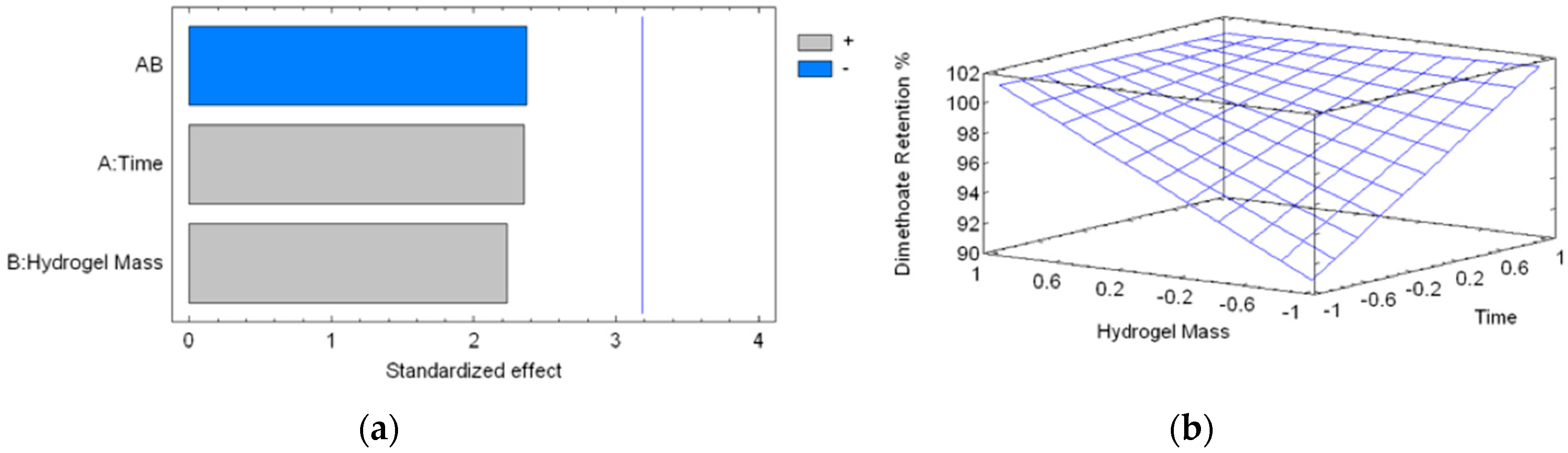
2.4. Absorption Kinetics of DMT
3. Materials and Methods
3.1. Theoretical Section
3.1.1. Building Molecular Structures
3.1.2. In-Silico Calculation of Interaction Energy
3.1.3. Molecular Dynamic Simulation (MDS)
3.2. Experimental Section
3.2.1. Materials
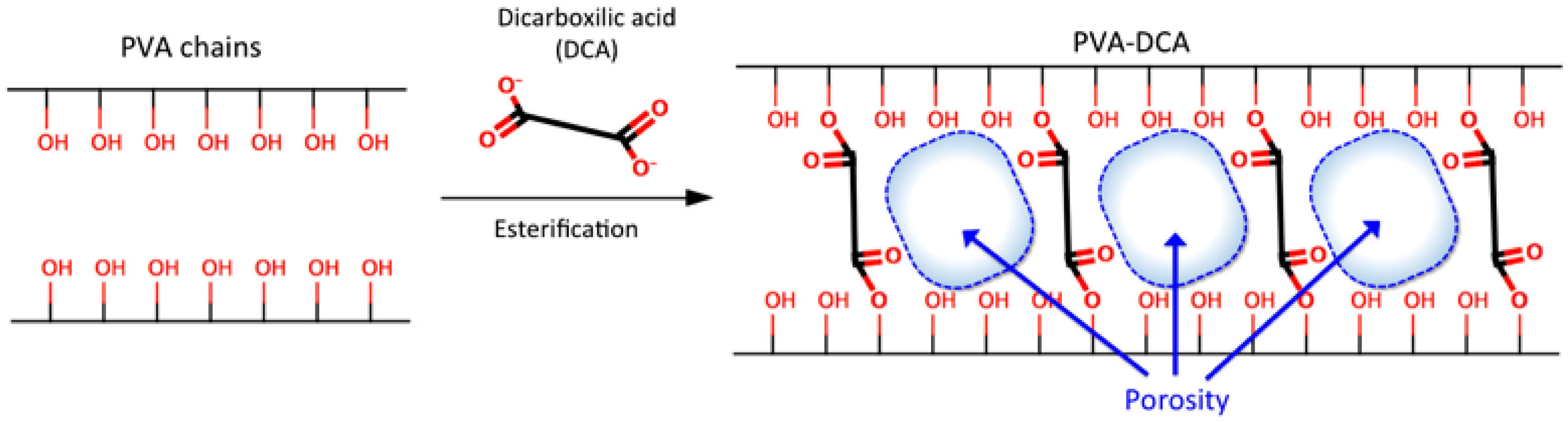
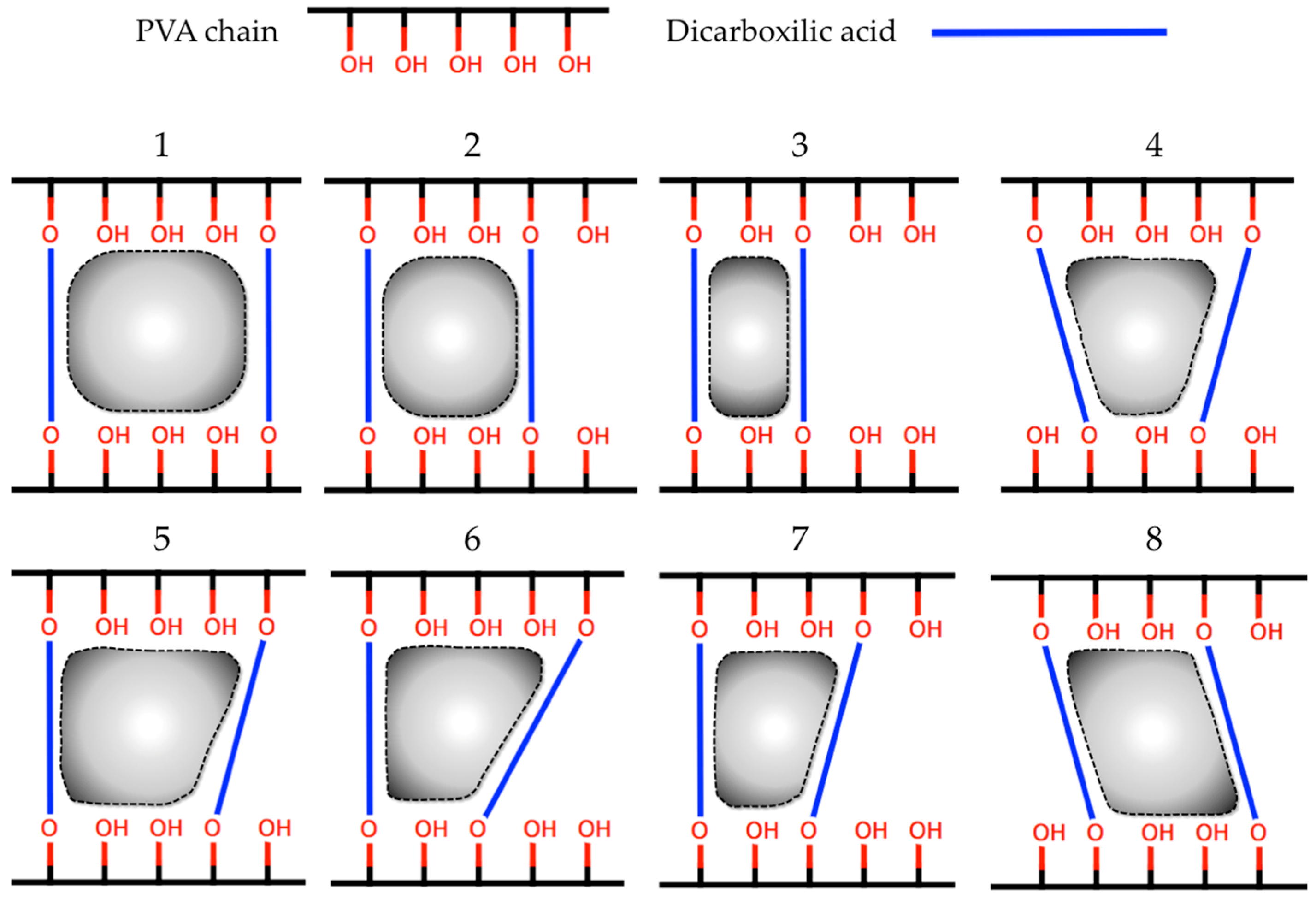
3.2.2. Synthesis and Characterization of CLPH-MA
3.2.3. SEM Analysis
3.2.4. Absorption Kinetic of DMT by CLPH-MA20 in Model Solutions
3.2.5. Statistical Analysis
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Delfino, R.T.; Ribeiro, T.S.; Figueroa-Villar, J.D. Organophosphorus Compounds as Chemical Warfare Wgents: A Review. J. Braz. Chem. Soc. 2009, 20, 407–428. [Google Scholar] [CrossRef]
- Durán-Lara, E.F.; Marple, J.L.; Giesen, J.A.; Fang, Y.; Jordan, J.H.; Godbey, W.T.; Marican, A.; Santos, L.S.; Grayson, S.M. Investigation of Lysine-Functionalized Dendrimers as Dichlorvos Detoxification Agents. Biomacromolecules 2015, 16, 3434–3444. [Google Scholar] [CrossRef] [PubMed]
- Lionetto, M.G.; Caricato, R.; Calisi, A.; Giordano, M.E.; Schettino, T. Acetylcholinesterase as a Biomarker in Environmental and Occupational Medicine: New Insights and Future Perspectives. BioMed Res. Int. 2013, 2013, 321213. [Google Scholar] [CrossRef] [PubMed]
- Durán-Lara, E.F.; Ávila-Salas, F.; Galaz, S.; John, A.; Maricán, A.; Gutiérrez, M.; Nachtigall, F.M.; Gonzalez-Nilo, F.D.; Santos, L.S. Nano-detoxification of Organophosphate Agents by PAMAM Derivatives. J. Braz. Chem. Soc. 2015, 26, 580–591. [Google Scholar] [CrossRef]
- Tian, F.; Liu, W.; Guo, G.; Qiang, Z.; Zhang, C. Kinetics and Mechanism of Dimethoate Chlorination during Drinking Water Treatment. Chemosphere 2014, 103, 181–187. [Google Scholar] [CrossRef] [PubMed]
- Momić, T.; Pašti, T.L.; Bogdanović, U.; Vodnik, V.; Mraković, A.; Rakočević, Z.; Pavlović, V.B.; Vasić, V. Adsorption of Organophosphate Pesticide Dimethoate on Gold Nanospheres and Nanorods. J. Nanomater. 2016, 2016, 8910271. [Google Scholar] [CrossRef]
- Du, J.-J.; Gao, R.-X.; Yu, H.; Li, X.-J.; Mu, H. Selective Extraction of Dimethoate from Cucumber Samples by Use of Molecularly Imprinted Microspheres. J. Pharm. Anal. 2015, 5, 200–206. [Google Scholar] [CrossRef]
- Gupta, V.K.; Ali, I.; Saleh, T.A.; Nayak, A.; Agarwal, S. Chemical Treatment Technologies for Waste-Water Recycling—An Overview. RSC Adv. 2012, 2, 6380–6388. [Google Scholar] [CrossRef]
- Sharma, S.; Bhattacharya, A. Drinking Water Contamination and Treatment Techniques. Appl. Water Sci. 2017, 7, 1043–1067. [Google Scholar] [CrossRef]
- Abd El-Mohdy, H.; Hegazy, E.; El-Nesr, E.; El-Wahab, M. Removal of Some Pesticides from Aqueous Solutions using PVP/(AAc-co-Sty) Hydrogels Prepared by Gamma Radiation. J. Macromol. Sci. Part A 2012, 49, 814–827. [Google Scholar] [CrossRef]
- Vishnubhakthula, S.; Elupula, R.; Durán-Lara, E.F. Recent Advances in Hydrogel-Based Drug Delivery for Melanoma Cancer Therapy: A Mini Review. J. Drug Deliv. 2017, 2017, 7275985. [Google Scholar] [CrossRef] [PubMed]
- Valdes, O.; Avila-Salas, F.; Marican, A.; Fuentealba, N.; Villaseñor, J.; Arenas-Salinas, M.; Argandoña, Y.; Durán-Lara, E.F. Methamidophos removal from aqueous solutions using a super adsorbent based on crosslinked poly(vinyl alcohol) hydrogel. J. Appl. Polym. Sci. 2018, 135, 45964. [Google Scholar] [CrossRef]
- Ahmed, E.M.; Aggor, F.S.; Awad, A.M.; El-Aref, A.T. An Innovative Method for Preparation of Nanometal Hydroxide Superabsorbent Hydrogel. Carbohydr. Polym. 2013, 91, 693–698. [Google Scholar] [CrossRef] [PubMed]
- Pooley, S.A.; Rivas, B.L.; Lillo, F.E.; Pizarro, G.C. Hydrogels From Acrylic Acid with N,N-dimethylacrylamide: Synthesis, Characterization, and Water Absorption Properties. J. Chil. Chem. Soc. 2010, 55, 19–24. [Google Scholar] [CrossRef]
- Bordi, F.; Paradossi, G.; Rinaldi, C.; Ruzicka, B. Chemical and Physical Hydrogels: Two Casesystems Studied by Quasi Elastic Light Scattering. Physics A 2002, 304, 119–128. [Google Scholar] [CrossRef]
- Hu, X.; Vatankhah-Varnoosfaderani, M.; Zhou, J.; Li, Q.; Sheiko, S.S. Weak Hydrogen Bonding Enables Hard, Strong, Tough, and Elastic Hydrogels. Adv. Mater. 2015, 27, 6899–6905. [Google Scholar] [CrossRef] [PubMed]
- Appel, E.A.; Tibbitt, M.W.; Greer, J.M.; Fenton, O.S.; Kreuels, K.; Anderson, D.G.; Langer, R. Exploiting Electrostatic Interactions in Polymer–Nanoparticle Hydrogels. ACS Macro Lett. 2015, 4, 848–852. [Google Scholar] [CrossRef]
- Dhotel, A.; Chen, Z.; Delbreilh, L.; Youssef, B.; Saiter, J.-M.; Tan, L. Molecular Motions in Functional Self-Assembled Nanostructures. Int. J. Mol. Sci. 2013, 14, 2303–2333. [Google Scholar] [CrossRef] [PubMed]
- Akhtar, M.F.; Hanif, M.; Ranjha, N.M. Methods of Synthesis of Hydrogels … A Review. Saudi Pharm. J. 2016, 24, 554–559. [Google Scholar] [CrossRef] [PubMed]
- Maitra, J.; Shukla, V.K. Cross-linking in Hydrogels—A Review. Am. J. Polym. Sci. 2014, 4, 25–31. [Google Scholar] [CrossRef]
- Hoare, T.R.; Kohane, D.S. Hydrogels in Drug Delivery: Progress and Challenges. Polymer 2008, 49, 1993–2007. [Google Scholar] [CrossRef]
- Oliveira, R.N.; McGuinness, G.B.; Ramos, M.E.; Kajiyama, C.E.; Thiré, R.M. Properties of PVA Hydrogel Wound-Care Dressings Containing UK Propolis. Macromol. Symp. 2016, 368, 122–127. [Google Scholar] [CrossRef]
- Escobar-Sierra, D.M.; Perea-Mesa, Y.P. Manufacturing and Evaluation of Chitosan, PVA and Aloe Vera hydrogels for Skin Applications. DYNA 2017, 84, 134–142. [Google Scholar] [CrossRef]
- Schanuel, F.S.; Santos, K.S.R.; Monte-Alto-Costa, A.; de Oliveira, M.G. Combined Nitric Oxide-releasing Poly(vinyl alcohol) Film/F127 Hydrogel for Accelerating Wound Healing. Colloids Surf. B Biointerfaces 2015, 130, 182–191. [Google Scholar] [CrossRef] [PubMed]
- Buwalda, S.J.; Vermonden, T.; Hennink, W.E. Hydrogels for Therapeutic Delivery: Current Developments and Future Directions. Biomacromolecules 2017, 18, 316–330. [Google Scholar] [CrossRef] [PubMed]
- Avila-Salas, F.; Sandoval, C.; Caballero, J.; Guiñez-Molinos, S.; Santos, L.S.; Cachau, R.E.; González-Nilo, F.D. Study of Interaction Energies Between the PAMAM Dendrimer and Nonsteroidal Anti-inflammatory Drug Using a Distributed Computational Strategy and Experimental Analysis by ESI-MS/MS. J. Phys. Chem. B 2012, 116, 2031–2039. [Google Scholar] [CrossRef] [PubMed]
- Durán-Lara, E.F.; López-Cortés, X.A.; Castro, R.I.; Avila-Salas, F.; González-Nilo, F.D.; Laurie, V.F.; Santos, L.S. Experimental and Theoretical Binding Affinity Between Polyvinylpolypyrrolidone and Selected Phenolic Compounds From Food Matrices. Food Chem. 2015, 168, 464–470. [Google Scholar] [CrossRef] [PubMed]
- Gil, M.; Avila-Salas, F.; Santos, L.S.; Iturmendi, N.; Moine, V.; Cheynier, V.; Saucier, C. Rosé Wine Fining Using Polyvinylpolypyrrolidone: Colorimetry, Targeted Polyphenomics, and Molecular Dynamics Simulations. J. Agric. Food Chem. 2017, 65, 10591–10597. [Google Scholar] [CrossRef] [PubMed]
- Dennington, R., II; Keith, T.; Milliam, J.; Eppinnett, K.; Hovell, W.L.; Gilliland, R. GaussView, version 3.09; Semichem, Inc.: Shawnee Mission, KS, USA, 2003. [Google Scholar]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Montgomery, J.A., Jr.; Vreven, T.; Kudin, K.; Burant, J. Gaussian 03; revision C.02; Gaussian, Inc.: Wallingford, CT, USA, 2004. [Google Scholar]
- Liu, K.; Steed, J.W. Triggered Formation of Thixotropic Hydrogels by Balancing Competitive Supramolecular Synthons. Soft Matter 2013, 9, 11699–11705. [Google Scholar] [CrossRef] [Green Version]
- Glasser, W.G.; Jain, R.K. Method of Making Ester-Crosslinked Chitosan Support Materials and Products Thereof. U.S. Patent 5,874,551, 23 February 1999. [Google Scholar]
- Saraydin, D.; Karadağ, E.; Sahiner, N.; GüVen, O. Incorporation of Malonic Acid Into Acrylamide Hydrogel by Radiation Technique and Its Effect on Swelling Behavior. J. Mater. Sci. 2002, 37, 3217–3223. [Google Scholar] [CrossRef]
- Tsao, C.T.; Chang, C.H.; Li, Y.D.; Wu, M.F.; Lin, C.P.; Han, J.L.; Chen, S.H.; Hsieh, K.H. Development of Chitosan/dicarboxylic Acid Hydrogels as Wound Dressing Materials. J. Bioact. Compat. Polym. 2011, 26, 519–536. [Google Scholar] [CrossRef]
- Valderruten, N.E.; Valverde, J.D.; Zuluaga, F.; Ruiz-Durántez, E. Synthesis and Characterization of Chitosan Hydrogels Cross-linked with Dicarboxylic Acids. React. Funct. Polym. 2014, 84, 21–28. [Google Scholar] [CrossRef]
- Saito, H.; Taguchi, T.; Aoki, H.; Murabayashi, S.; Mitamura, Y.; Tanaka, J.; Tateishi, T. pH-responsive Swelling Behavior of Collagen Gels Prepared by Novel Crosslinkers Based on Naturally Derived di- or Tricarboxylic Acids. Acta Biomater. 2007, 3, 89–94. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.S.; Im, J.S.; Baek, S.T.; Lee, J.O.; Azuma, Y.; Yoshinaga, K. Synthesis and Characterization of Crosslinked Hyperbranched Polyglycidol Hydrogel Films. J. Macromol. Sci. A 2006, 43, 829–839. [Google Scholar] [CrossRef]
- Lawal, O.S.; Storz, J.; Storz, H.; Lohmann, D.; Lechner, D.; Kulicke, W.M. Hydrogels Based on Barboxymethyl Cassava Starch Cross-linked with di- or Polyfunctional Carboxylic Acids: Synthesis, Water Absorbent Behavior and Rheological Characterizations. Eur. Polym. J. 2009, 45, 3399–3408. [Google Scholar] [CrossRef]
- Burchacka, E.; Potaczek, P.; Paduszyński, P.; Karłowicz-Bodalska, K.; Han, T.; Han, S. New Effective Azelaic Acid Liposomal Gel Formulation of Enhanced Pharmaceutical Bioavailability. Biomed. Pharmacother. 2016, 83, 771–775. [Google Scholar] [CrossRef] [PubMed]
- Fan, C.F.; Olafson, B.D.; Blanco, M. Application of Molecular Simulation To Derive Phase Diagrams of Binary Mixtures. Macromolecules 1992, 25, 3667–3676. [Google Scholar] [CrossRef]
- Stewart, J.J.P. Optimization of Parameters for Semiempirical Methods VI: More Modifications to the NDDO Approximations and Re-optimization of Parameters. J. Mol. Model. 2013, 19, 1–32. [Google Scholar] [CrossRef] [PubMed]
- Stewart, J.J.P. MOPAC2016 Computational Chemistry, version 16.111L (LINUX); Stewart Computational Chemistry: Colorado Springs, CO, USA, 2016; Available online: http://openmopac.net/downloads.html (accessed on 21 November 2017).
- Case, D.A.; Cerutti, D.S.; Cheatham, T.E., III; Darden, T.A.; Duke, R.E.; Giese, T.J.; Gohlke, H.; Goetz, A.W.; Greene, D.; Homeyer, N.; et al. AMBER 2017; University of California: San Francisco, CA, USA, 2017; Available online: http://ambermd.org/#AmberTools (accessed on 16 November 2017).
- Martínez, L.; Andrade, R.; Birgin, E.G.; Martínez, J.M. Software News and Update Packmol: A Package for Building Initial Configurations for Molecular Dynamics Simulations. J. Comput. Chem. 2009, 30, 2157–2164. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wang, W.; Kollman, P.A.; Case, D.A. Automatic atom type and bond type perception in molecular mechanical calculations. J. Mol. Graph. Model. 2006, 25, 247–260. [Google Scholar] [CrossRef] [PubMed]
- O’Boyle, N.M.; Banck, M.; James, C.A.; Morley, C.; Vandermeersch, T.; Hutchison, G.R. Open Babel: An Open Chemical Toolbox. J. Cheminform. 2011, 3, 33. [Google Scholar] [CrossRef] [PubMed]
- Schrödinger Release: Maestro-Desmond, Molecular Dynamics System, version 4.4; DE Shaw Research: New York, NY, USA, 2015.
- Vergara-Jaque, A.; Comer, J.; Monsalve, L.; González-Nilo, F.D.; Sandoval, C. Computationally Efficient Methodology for Atomic-level Characterization of Dendrimer–Drug Complexes: A Comparison of Amine-and Acetyl-terminated PAMAM. J. Phys. Chem. B 2013, 117, 6801–6813. [Google Scholar] [CrossRef] [PubMed]
- Han, M.; Chen, P.; Yang, X. Molecular Dynamics Simulation of PAMAM Dendrimer in Aqueous Solution. Polymer 2015, 46, 3481–3488. [Google Scholar] [CrossRef]
- Filipe, L.C.; Machuqueiro, M.; Baptista, A.M. Unfolding the Conformational Behavior of Peptide Dendrimers: Insights from Molecular Dynamics Simulations. J. Am. Chem. Soc. 2011, 133, 5042–5052. [Google Scholar] [CrossRef] [PubMed]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual Molecular Dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Williams, T.; Kelley, C. Gnuplot 5.0: An Interactive Plotting Program, Official Gnuplot Documentation. 2015. Available online: http://sourceforge.net/projects/gnuplot (accessed on 22 November 2017).
- Discovery Studio Visualizer (DS Visualizer) Software, version 4.1; Accelrys Software Inc.: San Diego, CA, USA, 2014; Available online: http://www.accelrys.com (accessed on 20 November 2017).
- Byun, H.; Hong, B.; Nam, S.Y.; Jung, S.Y.; Rhim, J.W.; Lee, S.B.; Moon, G.Y. Swelling Behavior and Drug Release of Poly(vinyl alcohol) Hydrogel Cross-Linked with Poly(acrylic acid). Macromol. Res. 2008, 16, 189–193. [Google Scholar] [CrossRef]
- Hosny, W.M.; Khalaf-Alaa, P.A. Potentiometric Study and Biological Activity of Some Metal Ion Complexes of Polyvinyl Alcohol (PVA). Int. J. Electrochem. Sci. 2013, 8, 1520–1533. [Google Scholar]



| Chemical structure |  |
| Molecular formula | C5H12NO3PS2 |
| Appearance | White crystalline solid |
| Solubility in water (20–25 °C) | More than 5000 mg/L water |
| Mol. Wt. | 229.26 g·mol−1 |
| Melting point | 51–52 °C |
| Wavelength (λ, nm) | 280 nm |
| Id. | Hydrogel | Average Interaction Energy Kcal/mol | Id. | Hydrogel | Average Interaction Energy Kcal/mol |
|---|---|---|---|---|---|
| 1 | PVAnp-Oxalic acid | −1.861 | 8 | PVAnp-Itaconic acid | −1.988 |
| 2 | PVAnp-Malonic acid | −1.952 | 9 | PVAnp-Tartaric acid | −1.879 |
| 3 | PVAnp-Succinic acid | −1.955 | 10 | PVAnp-Glutaric acid | −1.961 |
| 4 | PVAnp-Malic acid | −1.998 | 11 | PVAnp-Adipic acid | −1.985 |
| 5 | PVAnp-Fumaric acid | −1.994 | 12 | PVAnp-Pimelic acid | −1.975 |
| 6 | PVAnp-Maleic acid | −1.964 | 13 | PVAnp-Suberic acid | −1.969 |
| 7 | PVAnp-Citraconic acid | −1.993 | 14 | PVAnp-Azelaic acid | −1.924 |
| Experiment | Time (min) | Hydrogel Mass (mg) | Dimethoate Retention (%) |
|---|---|---|---|
| 1 | 10 (−1) | 34.2 (−1.00024) | 89.28 |
| 2 | 90 (+1) | 34.3 (−0.99726) | 100 |
| 3 | 10 (−1) | 100.9 (0.98742) | 100 |
| 4 | 90 (+1) | 101.3 (0.99934) | 100 |
| 5 | 50 (0) | 65.1 (−0.07942) | 100 |
| 6 | 50 (0) | 60.1 (−0.22842) | 100 |
| 7 | 50 (0) | 65.6 (−0.06452) | 100 |
| Id. | Crosslinking Agent | Structure | Id. | Crosslinking Agent | Structure |
|---|---|---|---|---|---|
| 1 | Oxalic acid [31] |  | 8 | Itaconic acid [32] |  |
| 2 | Malonic acid [31,32,33] |  | 9 | l-(+)-Tartaric acid [31] |  |
| 3 | Succinic acid [31,32,34,35] |  | 10 | Glutaric acid [32,35,37,38] |  |
| 4 | DL-Malic acid [36] |  | 11 | Adipic acid [31,32,35,37] |  |
| 5 | Fumaric acid [31,32] |  | 12 | Pimelic acid [38] |  |
| 6 | Maleic acid [31,32] |  | 13 | Suberic acid [38] |  |
| 7 | Citraconic acid [32] |  | 14 | Azelaic acid [39] |  |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Avila-Salas, F.; Marican, A.; Villaseñor, J.; Arenas-Salinas, M.; Argandoña, Y.; Caballero, J.; Durán-Lara, E.F. In-Silico Design, Synthesis and Evaluation of a Nanostructured Hydrogel as a Dimethoate Removal Agent. Nanomaterials 2018, 8, 23. https://doi.org/10.3390/nano8010023
Avila-Salas F, Marican A, Villaseñor J, Arenas-Salinas M, Argandoña Y, Caballero J, Durán-Lara EF. In-Silico Design, Synthesis and Evaluation of a Nanostructured Hydrogel as a Dimethoate Removal Agent. Nanomaterials. 2018; 8(1):23. https://doi.org/10.3390/nano8010023
Chicago/Turabian StyleAvila-Salas, Fabian, Adolfo Marican, Jorge Villaseñor, Mauricio Arenas-Salinas, Yerko Argandoña, Julio Caballero, and Esteban F. Durán-Lara. 2018. "In-Silico Design, Synthesis and Evaluation of a Nanostructured Hydrogel as a Dimethoate Removal Agent" Nanomaterials 8, no. 1: 23. https://doi.org/10.3390/nano8010023