Development of FRET Biosensor to Characterize CSK Subcellular Regulation
Abstract
:1. Introduction
2. Materials and Methods
2.1. DNA Constructs
2.2. Reagents and Cell Culture
2.3. Cell Transfection with DNA
2.4. Microscope and Image Acquisition
2.5. FRET Quantification
3. Results
3.1. Biosensor Design and Characterization of Sensitivity and Specificity of CSK Biosensor in Mammalian Cells
3.2. Detection of CSK Activity with FRET Biosensor at Membrane Microregions
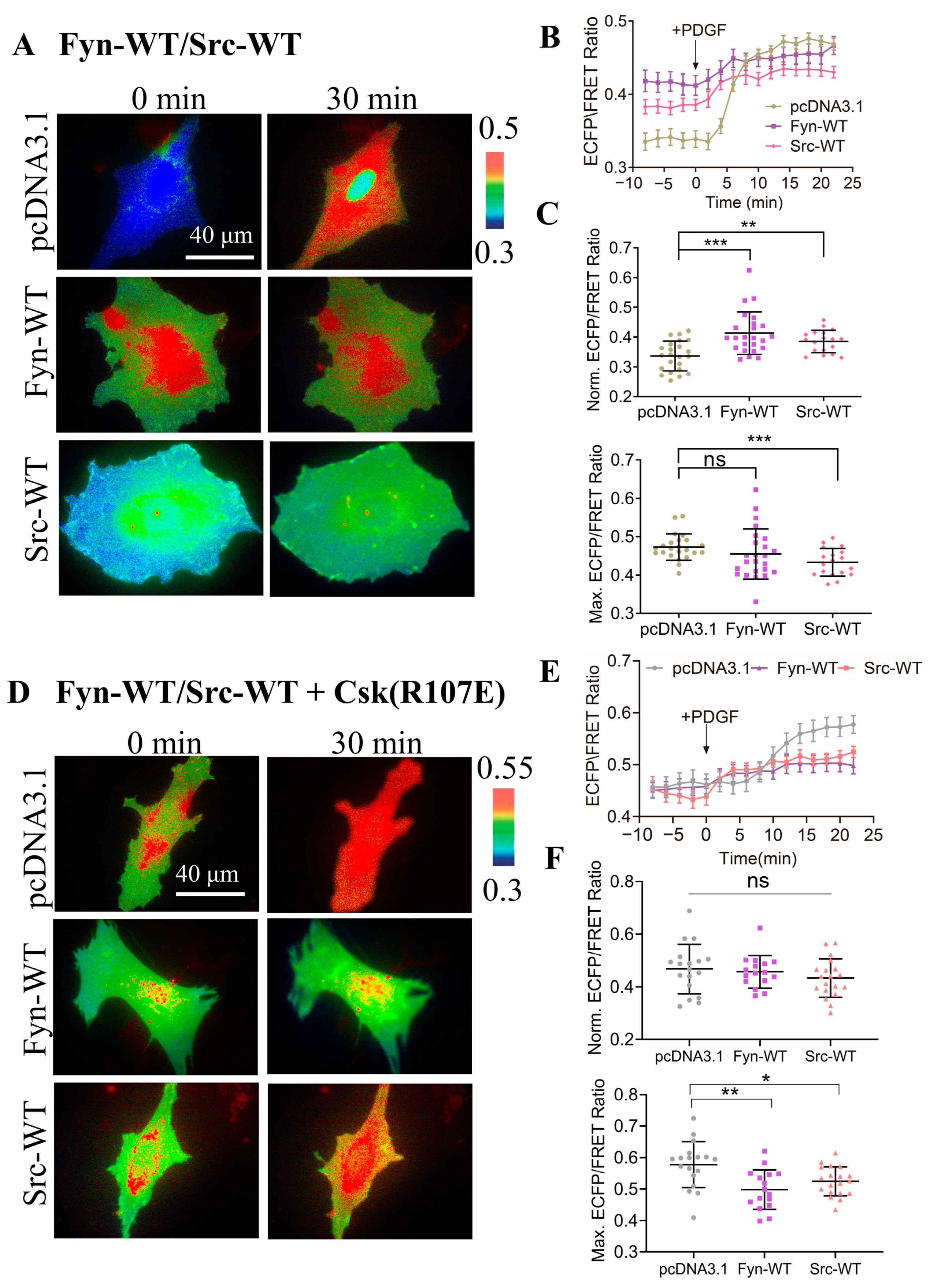
3.3. Demonstration of SH2 Domain Crucial for CSK Activation
3.4. Characterization of Specificity of CSK Biosensor to CSK Kinase
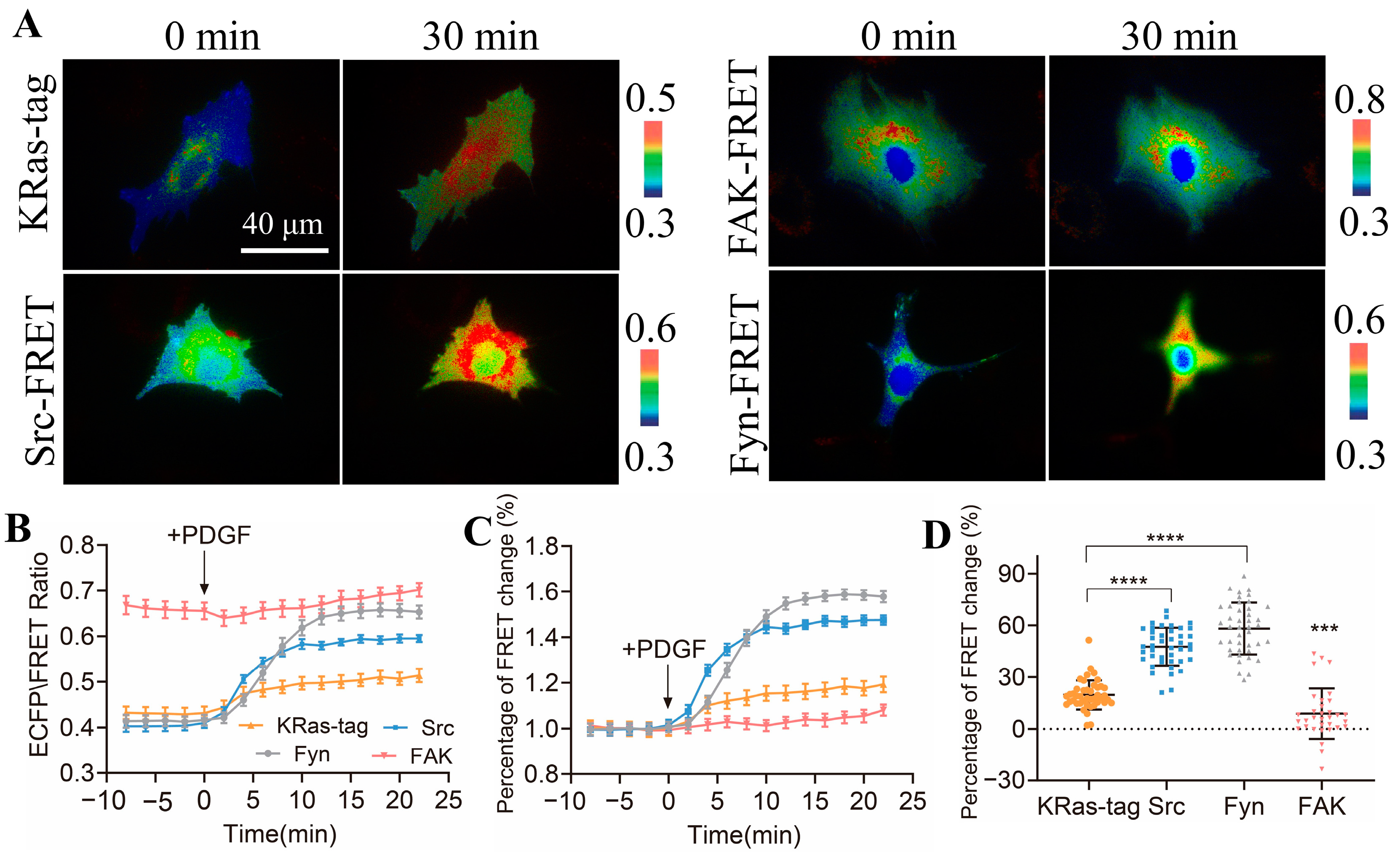
3.5. Comparison of Activity Levels of Different Kinases in ASM Cells
3.6. Protein Tyrosine Phosphatase PTPα in Regulating CSK Kinase Activity
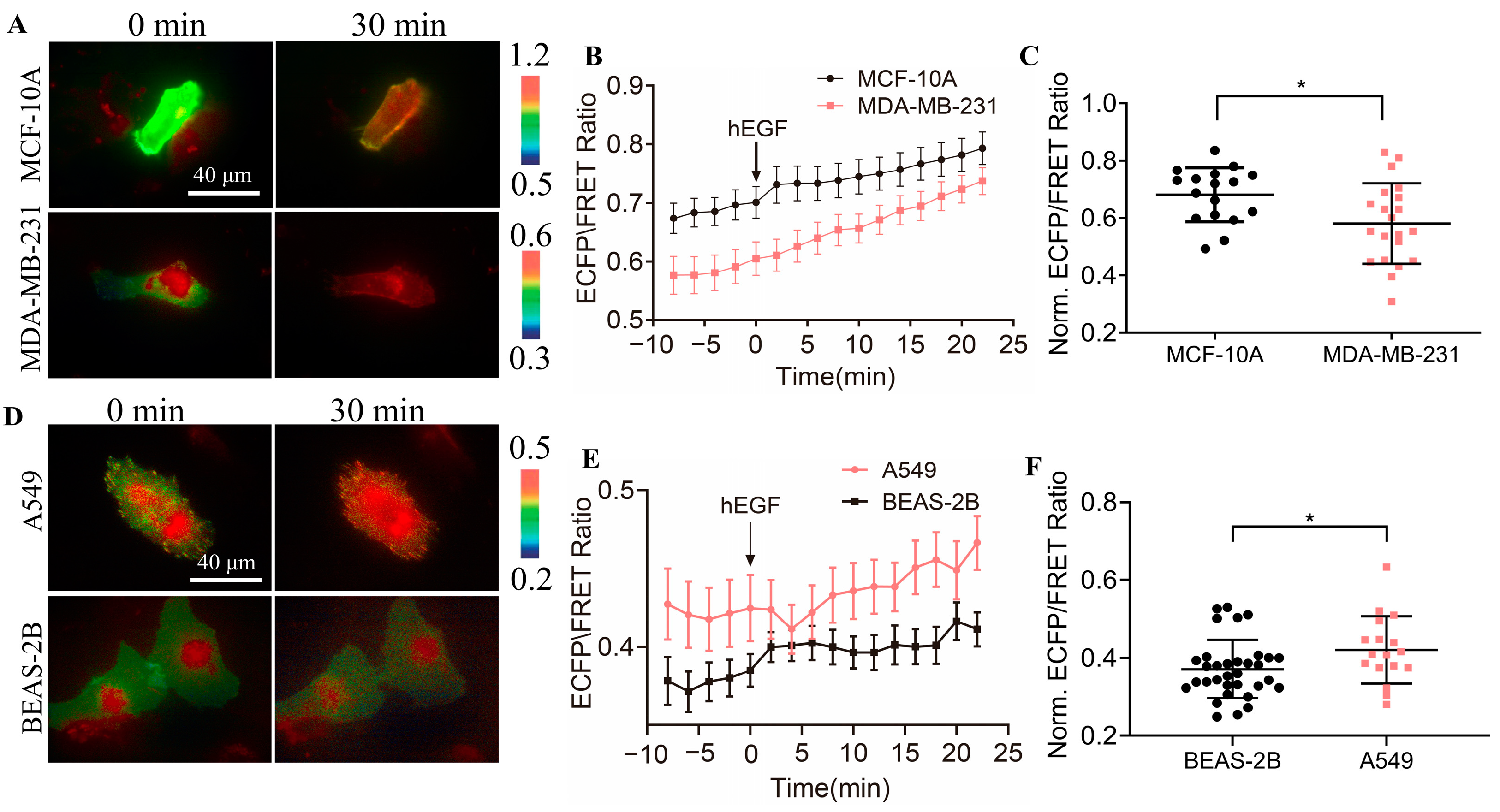
3.7. Detection of CSK Activation in Cancer Cells
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Coffin, J.M. Genes responsible for transformation by avian RNA tumor viruses. Cancer Res. 1976, 36, 4282–4288. [Google Scholar] [PubMed]
- Kawai, S.; Duesberg, P.H.; Hanafusa, H. Transformation-defective mutants of Rous sarcoma virus with src gene deletions of varying length. J. Virol. 1977, 24, 910–914. [Google Scholar] [CrossRef] [PubMed]
- Purchio, A.F.; Erikson, E.; Brugge, J.S.; Erikson, R.L. Identification of a polypeptide encoded by the avian sarcoma virus src gene. Proc. Natl. Acad. Sci. USA 1978, 75, 1567–1571. [Google Scholar] [CrossRef] [PubMed]
- Roskoski, R., Jr. Src protein-tyrosine kinase structure, mechanism, and small molecule inhibitors. Pharmacol. Res. 2015, 94, 9–25. [Google Scholar] [CrossRef] [PubMed]
- Roskoski, R., Jr. Src protein-tyrosine kinase structure and regulation. Biochem. Biophys. Res. Commun. 2004, 324, 1155–1164. [Google Scholar] [CrossRef] [PubMed]
- Okada, M.; Nakagawa, H. A protein tyrosine kinase involved in regulation of pp60c-src function. J. Biol. Chem. 1989, 264, 20886–20893. [Google Scholar] [CrossRef] [PubMed]
- Okada, M.; Nakagawa, H. Identification of a novel protein tyrosine kinase that phosphorylates pp60c-src and regulates its activity in neonatal rat brain. Biochem. Biophys. Res. Commun. 1988, 154, 796–802. [Google Scholar] [CrossRef] [PubMed]
- Okada, M. Regulation of the Src Family Kinases by Csk. Int. J. Biol. Sci. 2012, 8, 1385–1397. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Doshi, A.; Lei, M.; Eck, M.J.; Harrison, S.C. Crystal structures of c-Src reveal features of its autoinhibitory mechanism. Mol. Cell 1999, 3, 629–638. [Google Scholar] [CrossRef]
- Sun, G.; Ayrapetov, M.K. Dissection of the catalytic and regulatory structure-function relationships of Csk protein tyrosine kinase. Front. Cell Dev. Biol. 2023, 11, 1148352. [Google Scholar] [CrossRef]
- Nada, S.; Yagi, T.; Takeda, H.; Tokunaga, T.; Nakagawa, H.; Ikawa, Y.; Okada, M.; Aizawa, S. Constitutive activation of Src family kinases in mouse embryos that lack Csk. Cell 1993, 73, 1125–1135. [Google Scholar] [CrossRef] [PubMed]
- Kawabuchi, M.; Satomi, Y.; Takao, T.; Shimonishi, Y.; Nada, S.; Nagai, K.; Tarakhovsky, A.; Okada, M. Transmembrane phosphoprotein Cbp regulates the activities of Src-family tyrosine kinases. Nature 2000, 404, 999–1003. [Google Scholar] [CrossRef] [PubMed]
- Svec, A. Phosphoprotein associated with glycosphingolipid-enriched microdomains/Csk-binding protein: A protein that matters. Pathol. Res. Pract. 2008, 204, 785–792. [Google Scholar] [CrossRef] [PubMed]
- Cartwright, C.A.; Eckhart, W.; Simon, S.; Kaplan, P.L. Cell transformation by pp60c-src mutated in the carboxy-terminal regulatory domain. Cell 1987, 49, 83–91. [Google Scholar] [CrossRef] [PubMed]
- Cowan-Jacob, S.W.; Fendrich, G.; Manley, P.W.; Jahnke, W.; Fabbro, D.; Liebetanz, J.; Meyer, T. The crystal structure of a c-Src complex in an active conformation suggests possible steps in c-Src activation. Structure 2005, 13, 861–871. [Google Scholar] [CrossRef] [PubMed]
- Fortner, A.; Chera, A.; Tanca, A.; Bucur, O. Apoptosis regulation by the tyrosine-protein kinase CSK. Front. Cell Dev. Biol. 2022, 10, 1078180. [Google Scholar] [CrossRef] [PubMed]
- Roskoski, R., Jr. Src kinase regulation by phosphorylation and dephosphorylation. Biochem. Biophys. Res. Commun. 2005, 331, 1–14. [Google Scholar] [CrossRef]
- Pallen, C.J. Protein tyrosine phosphatase α (PTPα): A Src family kinase activator and mediator of multiple biological effects. Curr. Top. Med. Chem. 2003, 3, 821–835. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.M.; Wang, Y.; Pallen, C.J. Cell transformation and activation of pp60c-src by overexpression of a protein tyrosine phosphatase. Nature 1992, 359, 336–339. [Google Scholar] [CrossRef]
- Zheng, X.M.; Resnick, R.J.; Shalloway, D. A phosphotyrosine displacement mechanism for activation of Src by PTPα. EMBO J. 2000, 19, 964–978. [Google Scholar] [CrossRef]
- Bajar, B.; Wang, E.; Zhang, S.; Lin, M.; Chu, J. A Guide to Fluorescent Protein FRET Pairs. Sensors 2016, 16, 1488. [Google Scholar] [CrossRef]
- Iruela, G.; Fernández, A.; Sagar, A.; Carvajal, F.J.; Bernadó, P.; Pons, M. A FRET-Based Biosensor for the Src N-Terminal Regulatory Element. Biosensors 2022, 12, 96. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.J.; Jiang, Y.P.; Wu, C.X.; Zhou, D.; Gong, J.F.; Zhao, T.J.; Jin, Z.G. Development of FRET and Stress Granule Dual-Based System to Screen for Viral 3C Protease Inhibitors. Molecules 2023, 28, 3020. [Google Scholar] [CrossRef]
- Ouyang, M.; Sun, J.; Chien, S.; Wang, Y. Determination of hierarchical relationship of Src and Rac at subcellular locations with FRET biosensors. Proc. Natl. Acad. Sci. USA 2008, 105, 14353–14358. [Google Scholar] [CrossRef]
- Ouyang, M.; Wan, R.; Qin, Q.; Peng, Q.; Wang, P.; Wu, J.; Allen, M.; Shi, Y.; Laub, S.; Deng, L.; et al. Sensitive FRET Biosensor Reveals Fyn Kinase Regulation by Submembrane Localization. ACS Sens. 2018, 4, 76–86. [Google Scholar] [CrossRef]
- Komatsu, N.; Aoki, K.; Yamada, M.; Yukinaga, H.; Fujita, Y.; Kamioka, Y.; Matsuda, M. Development of an optimized backbone of FRET biosensors for kinases and GTPases. Mol. Biol. Cell 2011, 22, 4647–4656. [Google Scholar] [CrossRef] [PubMed]
- Seong, J.; Ouyang, M.; Kim, T.; Sun, J.; Wen, P.-C.; Lu, S.; Zhuo, Y.; Llewellyn, N.M.; Schlaepfer, D.D.; Guan, J.-L.; et al. Detection of focal adhesion kinase activation at membrane microdomains by fluorescence resonance energy transfer. Nat. Commun. 2011, 2, 406. [Google Scholar] [CrossRef]
- Yao, H.; Wang, L.; Guo, J.; Liu, W.; Li, J.; Wang, Y.; Deng, L.; Ouyang, M. Genetically Encoded FRET Biosensor Detects the Enzymatic Activity of Prostate-Specific Antigen. Mol. Cell. Biomech. 2020, 17, 101. [Google Scholar] [CrossRef]
- Qin, Q.; Laub, S.; Shi, Y.; Ouyang, M.; Peng, Q.; Zhang, J.; Wang, Y.; Lu, S. Fluocell for Ratiometric and High-Throughput Live-Cell Image Visualization and Quantitation. Front. Phys. 2019, 7, 154. [Google Scholar] [CrossRef] [PubMed]
- Weijland, A.; Williams, J.C.; Neubauer, G.; Courtneidge, S.A.; Wierenga, R.K.; Superti-Furga, G. Src regulated by C-terminal phosphorylation is monomeric. Proc. Natl. Acad. Sci. USA 1997, 94, 3590–3595. [Google Scholar] [CrossRef]
- Lee, S.; Lin, X.; Nam, N.H.; Parang, K.; Sun, G. Determination of the substrate-docking site of protein tyrosine kinase C-terminal Src kinase. Proc. Natl. Acad. Sci. USA 2003, 100, 14707–14712. [Google Scholar] [CrossRef] [PubMed]
- Waksman, G. Crystal structure of the phosphotyrosine recognition domain SH2 of the Src oncogene product complexed with tyrosine-phosphorylated peptides. Cell. Mol. Biol. 1994, 40, 611–618. [Google Scholar] [PubMed]
- Hamamura, K.; Tsuji, M.; Hotta, H.; Ohkawa, Y.; Takahashi, M.; Shibuya, H.; Nakashima, H.; Yamauchi, Y.; Hashimoto, N.; Hattori, H.; et al. Functional activation of Src family kinase yes protein is essential for the enhanced malignant properties of human melanoma cells expressing ganglioside GD3. J. Biol. Chem. 2011, 286, 18526–18537. [Google Scholar] [CrossRef] [PubMed]
- Manz, B.N.; Tan, Y.X.; Courtney, A.H.; Rutaganira, F.; Palmer, E.; Shokat, K.M.; Weiss, A. Small molecule inhibition of Csk alters affinity recognition by T cells. eLife 2015, 4, e08088. [Google Scholar] [CrossRef] [PubMed]
- Potuckova, L.; Draberova, L.; Halova, I.; Paulenda, T.; Draber, P. Positive and Negative Regulatory Roles of C-Terminal Src Kinase (CSK) in FcεRI-Mediated Mast Cell Activation, Independent of the Transmembrane Adaptor PAG/CSK-Binding Protein. Front. Immunol. 2018, 9, 1771. [Google Scholar] [CrossRef] [PubMed]
- Davidson, D.; Bakinowski, M.; Thomas, M.L.; Horejsi, V.; Veillette, A. Phosphorylation-dependent regulation of T-cell activation by PAG/Cbp, a lipid raft-associated transmembrane adaptor. Mol. Cell. Biol. 2003, 23, 2017–2028. [Google Scholar] [CrossRef] [PubMed]
- Torgersen, K.M.; Vang, T.; Abrahamsen, H.; Yaqub, S.; Horejsí, V.; Schraven, B.; Rolstad, B.; Mustelin, T.; Taskén, K. Release from tonic inhibition of T cell activation through transient displacement of C-terminal Src kinase (Csk) from lipid rafts. J. Biol. Chem. 2001, 276, 29313–29318. [Google Scholar] [CrossRef] [PubMed]
- Zacharias, D.A.; Violin, J.D.; Newton, A.C.; Tsien, R.Y. Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science 2002, 296, 913–916. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Zhang, J. Spatiotemporal analysis of differential Akt regulation in plasma membrane microdomains. Mol. Biol. Cell 2008, 19, 4366–4373. [Google Scholar] [CrossRef]
- Kazi, J.U.; Vaapil, M.; Agarwal, S.; Bracco, E.; Påhlman, S.; Rönnstrand, L. The tyrosine kinase CSK associates with FLT3 and c-Kit receptors and regulates downstream signaling. Cell. Signal. 2013, 25, 1852–1860. [Google Scholar] [CrossRef]
- Yaqub, S.; Abrahamsen, H.; Zimmerman, B.; Kholod, N.; Torgersen, K.M.; Mustelin, T.; Herberg, F.W.; Taskén, K.; Vang, T. Activation of C-terminal Src kinase (Csk) by phosphorylation at serine-364 depends on the Csk-Src homology 3 domain. Biochem. J. 2003, 372, 271–278. [Google Scholar] [CrossRef] [PubMed]
- Seong, J.; Lu, S.; Ouyang, M.; Huang, H.; Zhang, J.; Frame, M.C.; Wang, Y. Visualization of Src Activity at Different Compartments of the Plasma Membrane by FRET Imaging. Chem. Biol. 2009, 16, 48–57. [Google Scholar] [CrossRef] [PubMed]
- Ponniah, S.; Wang, D.Z.; Lim, K.L.; Pallen, C.J. Targeted disruption of the tyrosine phosphatase PTPα leads to constitutive downregulation of the kinases Src and Fyn. Curr. Biol. CB 1999, 9, 535–538. [Google Scholar] [CrossRef] [PubMed]
- Su, J.; Muranjan, M.; Sap, J. Receptor protein tyrosine phosphatase α activates Src-family kinases and controls integrin-mediated responses in fibroblasts. Curr. Biol. CB 1999, 9, 505–511. [Google Scholar] [CrossRef] [PubMed]
- Krndija, D.; Schmid, H.; Eismann, J.L.; Lother, U.; Adler, G.; Oswald, F.; Seufferlein, T.; von Wichert, G. Substrate stiffness and the receptor-type tyrosine-protein phosphatase α regulate spreading of colon cancer cells through cytoskeletal contractility. Oncogene 2010, 29, 2724–2738. [Google Scholar] [CrossRef] [PubMed]
- Su, J.; Yang, L.T.; Sap, J. Association between receptor protein-tyrosine phosphatase RPTPα and the Grb2 adaptor. Dual Src homology (SH) 2/SH3 domain requirement and functional consequences. J. Biol. Chem. 1996, 271, 28086–28096. [Google Scholar] [CrossRef]
- den Hertog, J.; Tracy, S.; Hunter, T. Phosphorylation of receptor protein-tyrosine phosphatase α on Tyr789, a binding site for the SH3-SH2-SH3 adaptor protein GRB-2 in vivo. EMBO J. 1994, 13, 3020–3032. [Google Scholar] [CrossRef] [PubMed]
- Maksumova, L.; Wang, Y.; Wong, N.K.; Le, H.T.; Pallen, C.J.; Johnson, P. Differential function of PTPα and PTPα Y789F in T cells and regulation of PTPα phosphorylation at Tyr-789 by CD45. J. Biol. Chem. 2007, 282, 20925–20932. [Google Scholar] [CrossRef] [PubMed]
- Tracy, S.; van der Geer, P.; Hunter, T. The receptor-like protein-tyrosine phosphatase, RPTP α, is phosphorylated by protein kinase C on two serines close to the inner face of the plasma membrane. J. Biol. Chem. 1995, 270, 10587–10594. [Google Scholar] [CrossRef]
- Vacaru, A.M.; den Hertog, J. Serine dephosphorylation of receptor protein tyrosine phosphatase α in mitosis induces Src binding and activation. Mol. Cell. Biol. 2010, 30, 2850–2861. [Google Scholar] [CrossRef]
- Ishizawar, R.; Parsons, S.J. c-Src and cooperating partners in human cancer. Cancer Cell 2004, 6, 209–214. [Google Scholar] [CrossRef] [PubMed]
- Masaki, T.; Okada, M.; Tokuda, M.; Shiratori, Y.; Hatase, O.; Shirai, M.; Nishioka, M.; Omata, M. Reduced C-terminal Src kinase (Csk) activities in hepatocellular carcinoma. Hepatology 1999, 29, 379–384. [Google Scholar] [CrossRef] [PubMed]
- Oneyama, C.; Hikita, T.; Enya, K.; Dobenecker, M.W.; Saito, K.; Nada, S.; Tarakhovsky, A.; Okada, M. The lipid raft-anchored adaptor protein Cbp controls the oncogenic potential of c-Src. Mol. Cell 2008, 30, 426–436. [Google Scholar] [CrossRef] [PubMed]
- Silva, C.M. Role of STATs as downstream signal transducers in Src family kinase-mediated tumorigenesis. Oncogene 2004, 23, 8017–8023. [Google Scholar] [CrossRef] [PubMed]
- Harris, K.F.; Shoji, I.; Cooper, E.M.; Kumar, S.; Oda, H.; Howley, P.M. Ubiquitin-mediated degradation of active Src tyrosine kinase. Proc. Natl. Acad. Sci. USA 1999, 96, 13738–13743. [Google Scholar] [CrossRef]
- Miyawaki, A.; Tsien, R.Y. Monitoring protein conformations and interactions by fluorescence resonance energy transfer between mutants of green fluorescent protein. Methods Enzymol. 2000, 327, 472–500. [Google Scholar]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ouyang, M.; Xing, Y.; Zhang, S.; Li, L.; Pan, Y.; Deng, L. Development of FRET Biosensor to Characterize CSK Subcellular Regulation. Biosensors 2024, 14, 206. https://doi.org/10.3390/bios14040206
Ouyang M, Xing Y, Zhang S, Li L, Pan Y, Deng L. Development of FRET Biosensor to Characterize CSK Subcellular Regulation. Biosensors. 2024; 14(4):206. https://doi.org/10.3390/bios14040206
Chicago/Turabian StyleOuyang, Mingxing, Yujie Xing, Shumin Zhang, Liting Li, Yan Pan, and Linhong Deng. 2024. "Development of FRET Biosensor to Characterize CSK Subcellular Regulation" Biosensors 14, no. 4: 206. https://doi.org/10.3390/bios14040206




