An Optimized Workflow for the Discovery of New Antimicrobial Compounds Targeting Bacterial RNA Polymerase Complex Formation
Abstract
:1. Introduction

2. Results and Discussion
2.1. Drug-Discovery Workflow
- (i)
- (ii)
- The ICBMS-Lyon 1 University in-house library, containing 3120 original synthetic and natural compounds, selected for their scaffold diversity and physicochemical properties.
- (iii)
- The PKRC library, containing 1200 synthetic and natural compounds, focused on protein kinase inhibitors [29].
2.1.1. yBRET Screening
2.1.2. In Vitro Competitive ELISA Assay
2.1.3. Bacterial Growth Inhibition Assay
2.2. Hit Characterization
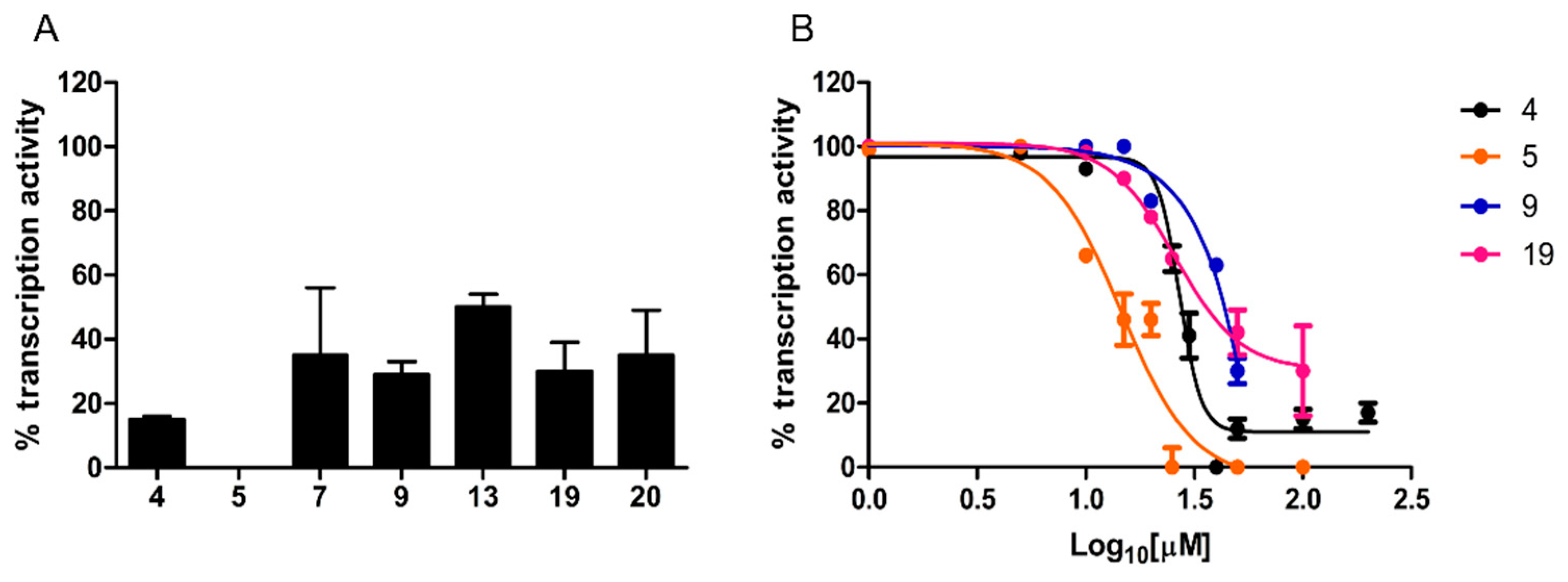
2.2.1. Antimicrobial Activity
2.2.2. RNA Polymerase Inhibitory Activity
2.2.3. β’ CH Region–σ70 PPI Inhibition Characterization
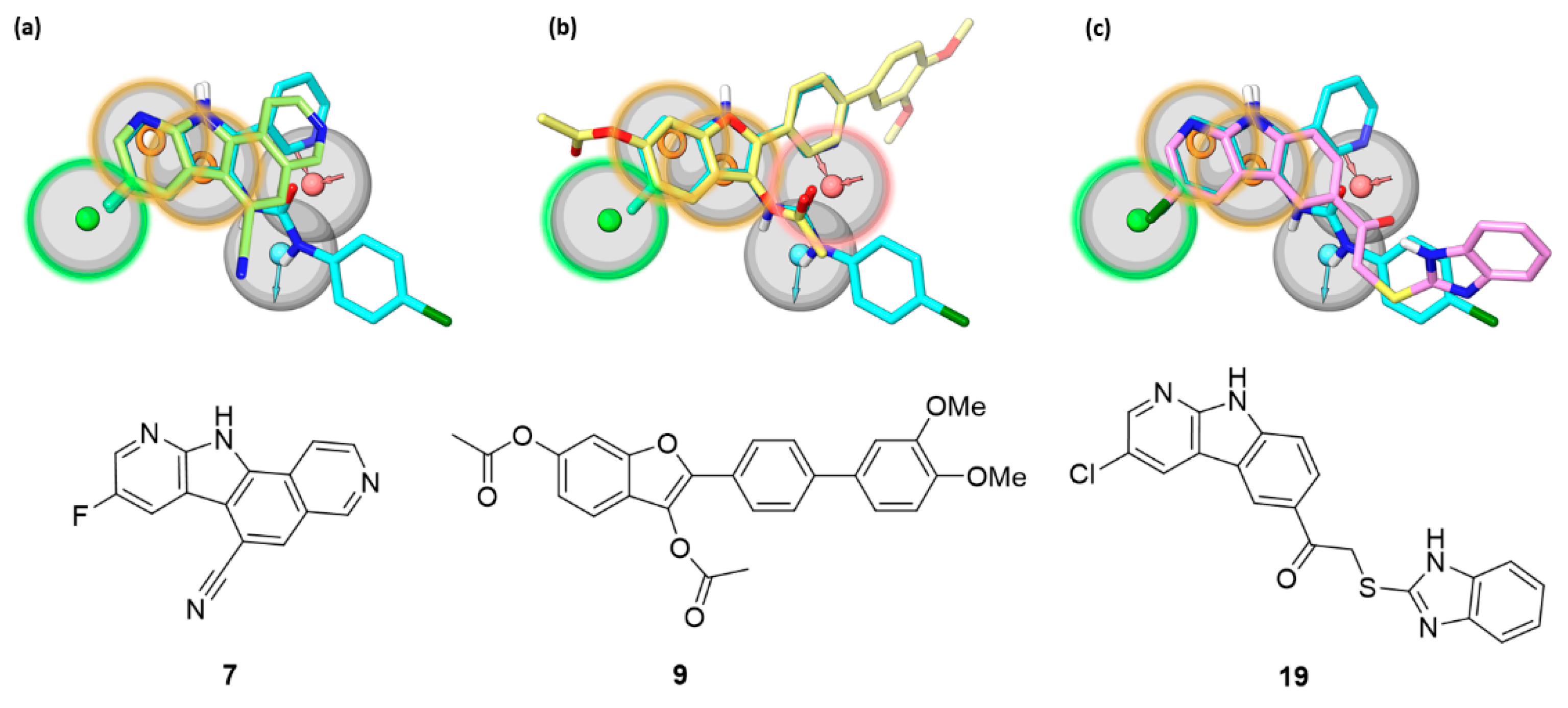
2.3. Similarity with Indolyl-Urea Inhibitors and Pharmacophore Model Matching
3. Conclusions
4. Materials and Methods
4.1. Chemicals
4.2. yBRET Assay
4.3. Competitive ELISA Assay
4.4. Antibacterial Activity Assays
4.5. In Vitro Transcription Assay
4.6. Alignment of New Compounds on Indolyl-Urea 4
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References and Notes
- Munita, J.M.; Arias, C.A. Mechanisms of Antibiotic Resistance. Annu. Rep. Med. Chem. 1982, 17, 119–127. [Google Scholar] [CrossRef]
- Coates, A.R.; Halls, G.; Hu, Y. Novel Classes of Antibiotics or More of the Same? Br. J. Pharm. 2011, 163, 184–194. [Google Scholar] [CrossRef] [Green Version]
- Cossar, P.J.; Lewis, P.J.; McCluskey, A. Protein-Protein Interactions as Antibiotic Targets: A Medicinal Chemistry Perspective. Med. Res. Rev. 2020, 40, 469–494. [Google Scholar] [CrossRef]
- Kohanski, M.A.; Dwyer, D.J.; Collins, J.J. How Antibiotics Kill Bacteria: From Targets to Networks. Nat. Rev. Microbiol. 2010, 8, 423–435. [Google Scholar] [CrossRef] [Green Version]
- Kahan, R.; Worm, D.J.; De Castro, G.V.; Ng, S.; Barnard, A. Modulators of Protein-Protein Interactions as Antimicrobial Agents. RSC Chem. Biol. 2021, 2, 387–409. [Google Scholar] [CrossRef]
- Ma, C.; Yang, X.; Lewis, P.J. Bacterial Transcription as a Target for Antibacterial Drug Development. Microbiol. Mol. Biol. Rev. 2016, 80, 139–160. [Google Scholar] [CrossRef] [Green Version]
- Goldstein, B.P. Resistance to Rifampicin: A Review. J. Antibiot. 2014, 67, 625–630. [Google Scholar] [CrossRef] [Green Version]
- Haupenthal, J.; Kautz, Y.; Elgaher, W.A.M.; Pätzold, L.; Röhrig, T.; Laschke, M.W.; Tschernig, T.; Hirsch, A.K.H.; Molodtsov, V.; Murakami, K.S.; et al. Evaluation of Bacterial RNA Polymerase Inhibitors in a Staphylococcus Aureus-Based Wound Infection Model in SKH1 Mice. ACS Infect. Dis. 2020, 6, 2573–2581. [Google Scholar] [CrossRef]
- Mukinda, F.K.; Theron, D.; Van Der Spuy, G.D.; Jacobson, K.R.; Roscher, M. Europe PMC Funders Group Rise in Rifampicin-Monoresistant Tuberculosis in Western Cape, South Africa. Int. J. Tuberc. Lung Dis. 2013, 16, 196–202. [Google Scholar] [CrossRef] [Green Version]
- Thach, O.; Mielczarek, M.; Ma, C.; Kutty, S.K.; Yang, X.; Black, D.S.C.; Griffith, R.; Lewis, P.J.; Kumar, N. From Indole to Pyrrole, Furan, Thiophene and Pyridine: Search for Novel Small Molecule Inhibitors of Bacterial Transcription Initiation Complex Formation. Bioorg. Med. Chem. 2016, 24, 1171–1182. [Google Scholar] [CrossRef]
- Murakami, K.S. X-Ray Crystal Structure of Escherichia coli RNA Polymerase σ70 Holoenzyme. J. Biol. Chem. 2013, 288, 9126–9134. [Google Scholar] [CrossRef] [Green Version]
- Murakami, K.S.; Masuda, S.; Darst, S.A. Structural Basis of Transcription Initiation: RNA Polymerase Holoenzyme at 4 Å Resolution. Science 2002, 296, 1280–1284. [Google Scholar] [CrossRef]
- Yokoyama, S.; Yokoyama, S.; Vassylyeva, M.N.; Yokoyama, S. Crystal Structure of a Bacterial RNA Polymerase Holoenzyme at 2.6. Å Resolution. Nature 2002, 417, 712–719. [Google Scholar] [CrossRef]
- Bae, B.; Feklistov, A.; Lass-Napiorkowska, A.; Landick, R.; Darst, S.A. Structure of a Bacterial RNA Polymerase Holoenzyme Open Promoter Complex. Elife 2015, 4, e08504. [Google Scholar] [CrossRef]
- Glyde, R.; Ye, F.; Jovanovic, M.; Kotta-Loizou, I.; Buck, M.; Zhang, X. Structures of Bacterial RNA Polymerase Complexes Reveal the Mechanism of DNA Loading and Transcription Initiation. Mol. Cell 2018, 70, 1111–1120.e3. [Google Scholar] [CrossRef] [Green Version]
- Ma, C.; Yang, X.; Kandemir, H.; Mielczarek, M.; Johnston, E.B.; Griffith, R.; Kumar, N.; Lewis, P.J. Inhibitors of Bacterial Transcription Initiation Complex Formation. ACS Chem. Biol. 2013, 8, 1972–1980. [Google Scholar] [CrossRef]
- Ma, C.; Yang, X.; Lewis, P.J. Bacterial Transcription Inhibitor of RNA Polymerase Holoenzyme Formation by Structure-Based Drug Design: From in Silico Screening to Validation. ACS Infect. Dis. 2016, 2, 39–46. [Google Scholar] [CrossRef]
- Sartini, S.; Levati, E.; Maccesi, M.; Guerra, M.; Spadoni, G.; Bach, S.; Benincasa, M.; Scocchi, M.; Ottonello, S.; Rivara, S.; et al. New Antimicrobials Targeting Bacterial RNA Polymerase Holoenzyme Assembly Identified with an in Vivo BRET-Based Discovery Platform. ACS Chem. Biol. 2019, 14, 1727–1736. [Google Scholar] [CrossRef]
- Wenholz, D.S.; Miller, M.; Dawson, C.; Bhadbhade, M.; Black, D.S.C.; Griffith, R.; Dinh, H.; Cain, A.; Lewis, P.; Kumar, N. Inhibitors of Bacterial RNA Polymerase Transcription Complex. Bioorg. Chem. 2022, 118, 105481. [Google Scholar] [CrossRef]
- Campbell, E.A.; Pavlova, O.; Zenkin, N.; Leon, F.; Irschik, H.; Jansen, R.; Severinov, K.; Darst, S.A. Structural, Functional, and Genetic Analysis of Sorangicin Inhibition of Bacterial RNA Polymerase. EMBO J. 2005, 24, 674–682. [Google Scholar] [CrossRef]
- Cao, X.; Boyaci, H.; Chen, J.; Bao, Y.; Landick, R.; Campbell, E.A. Basis of Narrow-Spectrum Activity of Fidaxomicin on Clostridioides difficile. Nature 2022, 604, 541–545. [Google Scholar] [CrossRef]
- Mielczarek, M.; Thomas, R.V.; Ma, C.; Kandemir, H.; Yang, X.; Bhadbhade, M.; Black, D.S.; Griffith, R.; Lewis, P.J.; Kumar, N. Synthesis and Biological Activity of Novel Mono-Indole and Mono-Benzofuran Inhibitors of Bacterial Transcription Initiation Complex Formation. Bioorg. Med. Chem. 2015, 23, 1763–1775. [Google Scholar] [CrossRef]
- André, E.; Bastide, L.; Villain-Guillot, P.; Latouche, J.; Rouby, J.; Leonetti, J.P. A Multiwell Assay to Isolate Compounds Inhibiting the Assembly of the Prokaryotic RNA Polymerase. Assay Drug Dev. Technol. 2004, 2, 629–635. [Google Scholar] [CrossRef] [PubMed]
- Glaser, B.T.; Bergendahl, V.; Thompson, N.E.; Olson, B.; Burgess, R.R. LRET-Based HTS of a Small-Compound Library for Inhibitors of Bacterial RNA Polymerase. Assay Drug Dev. Technol. 2007, 5, 759–768. [Google Scholar] [CrossRef]
- Bergendahl, V.; Heyduk, T.; Burgess, R.R. Luminescence Resonance Energy Transfer-Based High-Throughput Screening Assay for Inhibitors of Essential Protein-Protein Interactions in Bacterial RNA Polymerase. Appl. Environ. Microbiol. 2003, 69, 1492–1498. [Google Scholar] [CrossRef] [Green Version]
- Corbel, C.; Sartini, S.; Levati, E.; Colas, P.; Maillet, L.; Couturier, C.; Montanini, B.; Bach, S. Screening for Protein-Protein Interaction Inhibitors Using a Bioluminescence Resonance Energy Transfer (BRET)–Based Assay in Yeast. SLAS Discov. 2017, 22, 751–759. [Google Scholar] [CrossRef] [Green Version]
- Labrière, C.; Lozach, O.; Blairvacq, M.; Meijer, L.; Guillou, C. Further Investigation of Paprotrain: Towards the Conception of Selective and Multi-Targeted CNS Kinase Inhibitors. Eur. J. Med. Chem. 2016, 124, 920–934. [Google Scholar] [CrossRef]
- Le Cann, F.; Delehouz, C.; Leverrier-Penna, S.; Filliol, A.; Comte, A.; Delalande, O.; Desban, N.; Baratte, B.; Gallais, I.; Piquet-Pellorce, C.; et al. Sibiriline a New Small Chemical Inhibitor of Receptor-interacting Protein Kinase. FEBS J. 2017, 284, 3050–3068. [Google Scholar] [CrossRef] [Green Version]
- The PKRC library was established during the European Commission funded project Protein kinases—Novel drug targets of postgenomic era, ProteinKinaseResearch (LSHB-CT-2004-50367). Contact Arnaud Comte.
- Zhang, Y.; Degen, D.; Ho, M.X.; Sineva, E.; Ebright, K.Y.; Ebright, Y.W.; Mekler, V.; Vahedian-Movahed, H.; Feng, Y.; Yin, R.; et al. GE23077 Binds to the RNA Polymerase “i” and “I+1” Sites and Prevents the Binding of Initiating Nucleotides. Elife 2014, 2014, e02450. [Google Scholar] [CrossRef]
- Maffioli, S.I.; Zhang, Y.; Degen, D.; Carzaniga, T.; Del Gatto, G.; Serina, S.; Monciardini, P.; Mazzetti, C.; Guglierame, P.; Candiani, G.; et al. Antibacterial Nucleoside-Analog Inhibitor of Bacterial RNA Polymerase. Cell 2017, 169, 1240–1248.e23. [Google Scholar] [CrossRef]
- Simpson, M.; Poulsen, S.A. An Overview of Australia’s Compound Management Facility: The Queensland Compound Library. ACS Chem. Biol. 2014, 9, 28–33. [Google Scholar] [CrossRef] [PubMed]
- Mologni, L.; Tardy, S.; Zambon, A.; Orsato, A.; Schneider, C.; Bisson, W.H.; Ceccon, M.; Vitaldi, M.; Goyard, D.; Garcia, P.; et al. Discovery of Novel Putative A-Carboline Inhibitors of Anaplastic Lymphoma Kinase. ACS Omega 2022, 7, 17083–17097. [Google Scholar] [CrossRef] [PubMed]
- Mologni, L.; Orsato, A.; Zambon, A.; Tardy, S.; Bisson, W.H.; Schneider, C.; Ceccon, M.; Viltadi, M.; D’Attoma, J.; Pannilunghi, S.; et al. Identification of Non-ATP-Competitive α-Carboline Inhibitors of the Anaplastic Lymphoma Kinase. Eur. J. Med. Chem. 2022, 238, 114488. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Zhou, Q.; He, J.; Jiang, Z.; Peng, C.; Tong, R.; Shi, J. Recent Advances in the Development of Protein–Protein Interactions Modulators: Mechanisms and Clinical Trials. Signal Transduct. Target. Ther. 2020, 5, 213. [Google Scholar] [CrossRef]
- Thomas, B.J.; Rothstein, R. Elevated Recombination Rates in Transcriptionally Active DNA. Cell 1989, 56, 619–630. [Google Scholar] [CrossRef]
- Armas, F.; Pacor, S.; Ferrari, E.; Guida, F.; Pertinhez, T.A.; Romani, A.A.; Scocchi, M.; Benincasa, M. Design, Antimicrobial Activity and Mechanism of Action of Arg-Rich Ultra-Short Cationic Lipopeptides. PLoS ONE 2019, 14, e0212447. [Google Scholar] [CrossRef] [Green Version]
- Rivetti, C.; Guthold, M.; Bustamante, C. Wrapping of DNA around the E. coli RNA Polymerase Open Promoter Complex. EMBO J. 1999, 18, 4464–4475. [Google Scholar] [CrossRef] [Green Version]
- Schrödinger. Schrödinger Release 2021-1: Phase; Schrödinger, LLC: New York, NY, USA, 2021. [Google Scholar]
- Dixon, S.L.; Smondyrev, A.M.; Knoll, E.H.; Rao, S.N.; Shaw, D.E.; Friesner, R.A. PHASE: A New Engine for Pharmacophore Perception, 3D QSAR Model Development, and 3D Database Screening: 1. Methodology and Preliminary Results. J. Comput. Aided Mol. Des. 2006, 20, 647–671. [Google Scholar] [CrossRef]
- Sastry, G.M.; Dixon, S.L.; Sherman, W. Rapid Shape-Based Ligand Alignment and Virtual Screening Method Based on Atom/Feature-Pair Similarities and Volume Overlap Scoring. J. Chem. Inf. Model. 2011, 51, 2455–2466. [Google Scholar] [CrossRef]
- Lu, C.; Wu, C.; Ghoreishi, D.; Chen, W.; Wang, L.; Damm, W.; Ross, G.A.; Dahlgren, M.K.; Russell, E.; Von Bargen, C.D.; et al. OPLS4: Improving Force Field Accuracy on Challenging Regimes of Chemical Space. J. Chem. Theory Comput. 2021, 17, 4291–4300. [Google Scholar] [CrossRef] [PubMed]
- Schrödinger. Schrödinger Release 2021-1: Macromodel; Schrödinger, LLC: New York, NY, USA, 2021. [Google Scholar]
- Still, W.C.; Tempczyk, A.; Hawley, R.C.; Hendrickson, T. Semianalytical Treatment of Solvation for Molecular Mechanics and Dynamics. J. Am. Chem. Soc. 1990, 112, 6127–6129. [Google Scholar] [CrossRef]
- Polak, E.; Ribiere, G. Note Sur La Convergence de Méthodes de Directions Conjuguées. Rev. Française D’inform. Rech. Opérat. Sér. Rouge 1969, 3, 35–43. [Google Scholar] [CrossRef]


| Library | Compound | Growth Inhibition at 100 μM (8 h) | MIC (μM) | Binding Inhibition | |||
|---|---|---|---|---|---|---|---|
| B. subtilis | E. colib | S. cerevisiae | B. subtilis | E. colib | IC50 μM ± SD | ||
| Reference | 4 a | 73% | 91% | 0% | 50 | 25 | 8.09 ± 1.1 c |
| ReCC | 5 | 19% | 92% | 1% | >100 | 1.56 | 15.11 ± 1.96 |
| 7 | 13% | 66% | 7% | >100 | 0.78 | 8.56 ± 1.82 | |
| ICBMS | 9 | 24% | 24% | 0% | >100 | >100 | 13.93 ± 1.86 |
| 13 | 40% | 29% | 0% | 50 | 50 | 16.35 ± 1.8 | |
| PKRC | 19 | 37% | 0% | 0% | 100 | >100 | 40 ± 4.2 |
| 20 | 15% | 30% | 0% | 100 | 12.5 | 80 ± 6.56 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Caputo, A.; Sartini, S.; Levati, E.; Minato, I.; Elisi, G.M.; Di Stasi, A.; Guillou, C.; Goekjian, P.G.; Garcia, P.; Gueyrard, D.; et al. An Optimized Workflow for the Discovery of New Antimicrobial Compounds Targeting Bacterial RNA Polymerase Complex Formation. Antibiotics 2022, 11, 1449. https://doi.org/10.3390/antibiotics11101449
Caputo A, Sartini S, Levati E, Minato I, Elisi GM, Di Stasi A, Guillou C, Goekjian PG, Garcia P, Gueyrard D, et al. An Optimized Workflow for the Discovery of New Antimicrobial Compounds Targeting Bacterial RNA Polymerase Complex Formation. Antibiotics. 2022; 11(10):1449. https://doi.org/10.3390/antibiotics11101449
Chicago/Turabian StyleCaputo, Alessia, Sara Sartini, Elisabetta Levati, Ilaria Minato, Gian Marco Elisi, Adriana Di Stasi, Catherine Guillou, Peter G. Goekjian, Pierre Garcia, David Gueyrard, and et al. 2022. "An Optimized Workflow for the Discovery of New Antimicrobial Compounds Targeting Bacterial RNA Polymerase Complex Formation" Antibiotics 11, no. 10: 1449. https://doi.org/10.3390/antibiotics11101449







