Investigating the Role of Antibiotics on Induction, Inhibition and Eradication of Biofilms of Poultry Associated Escherichia coli Isolated from Retail Chicken Meat
Abstract
:1. Introduction
2. Results
2.1. Prevalence of Antibiotic Resistance among E. coli
2.2. Detection of VF and ESBL Genes
2.3. Biofilm Formation Potential
2.4. Measurement of MIC-p and MIC-b
2.5. Determination of MBIC/MRC and MBEC
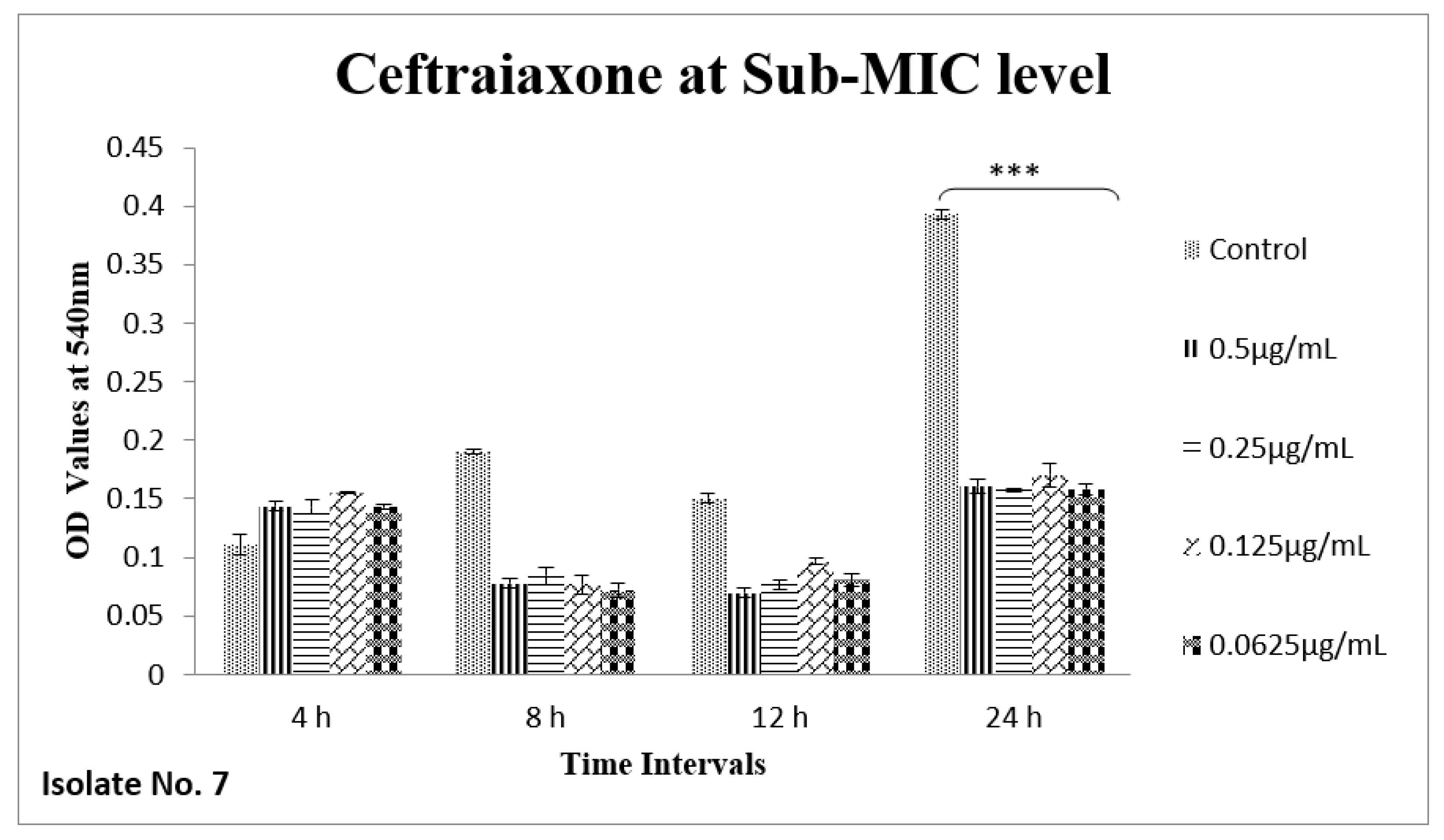
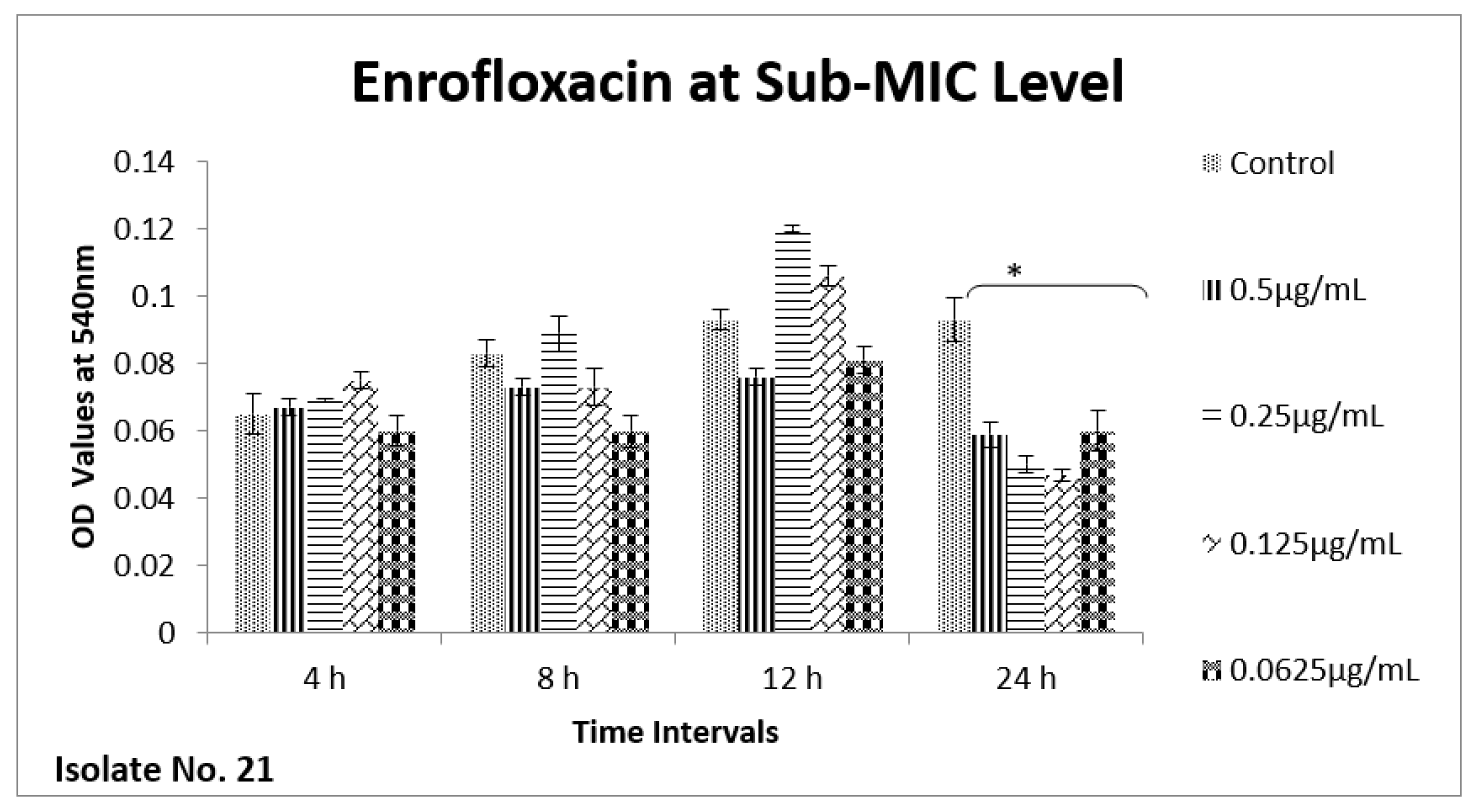
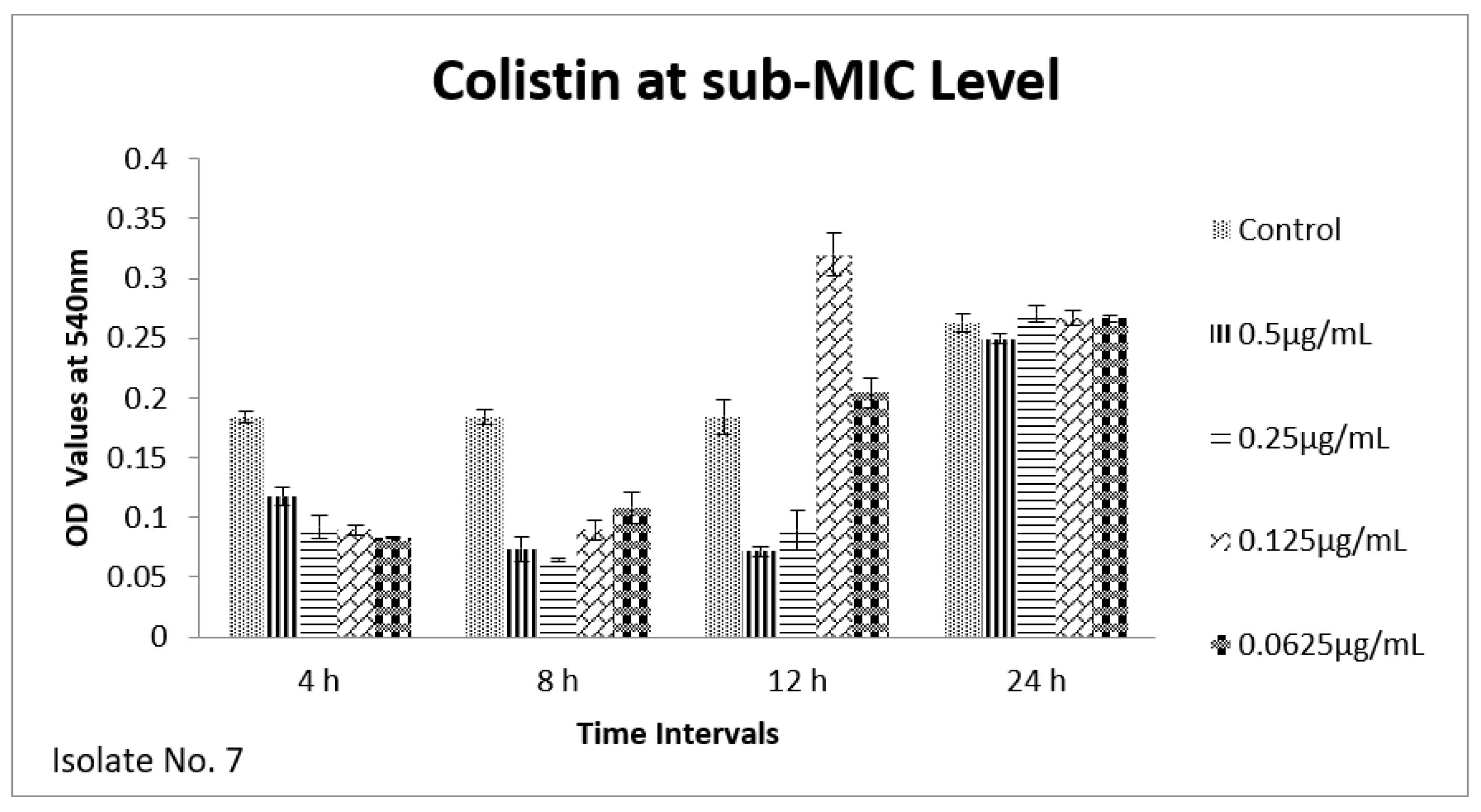
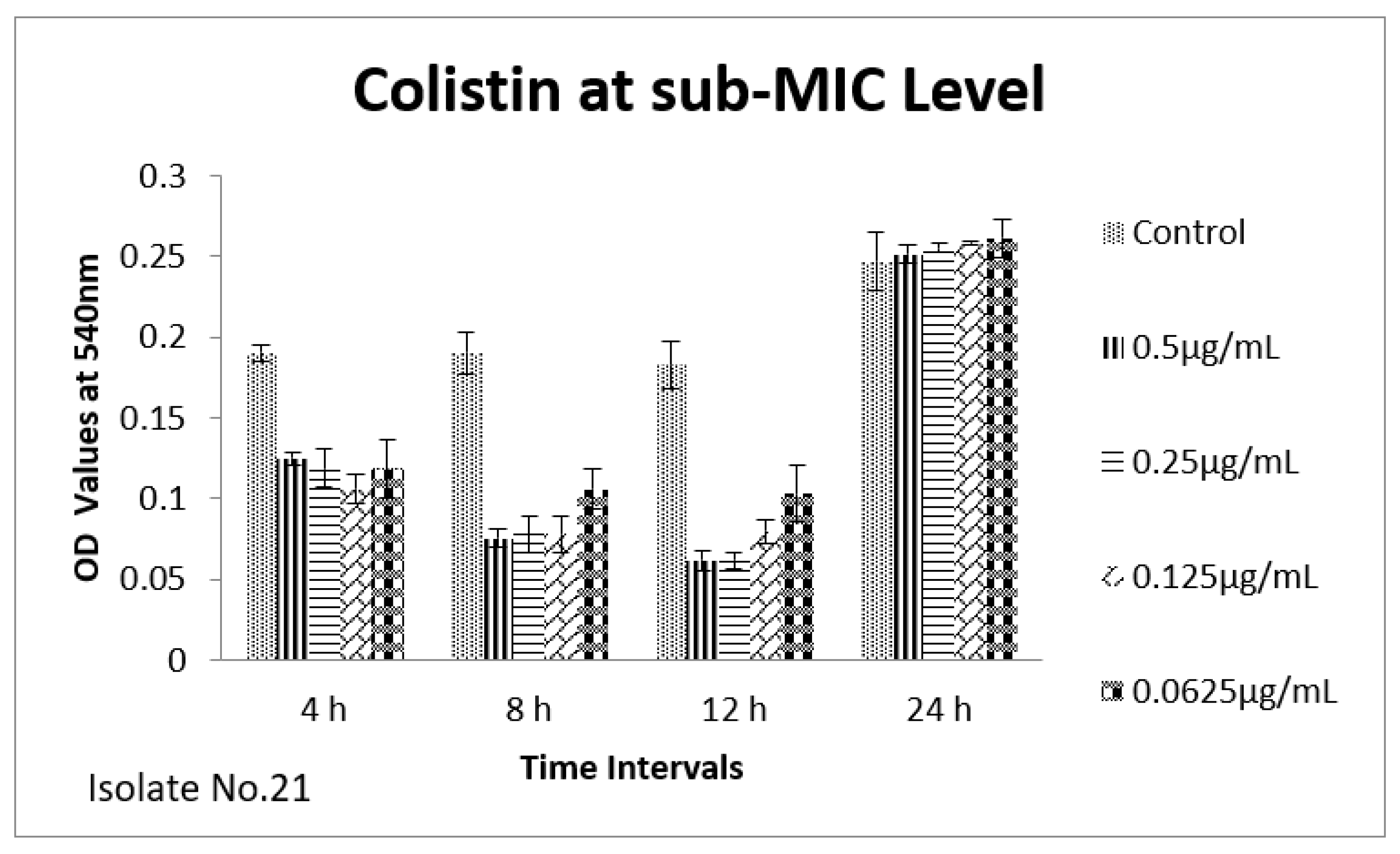
2.6. Effect of Sub-MICs on Biofilm
3. Discussion
4. Materials and Methods
4.1. Isolation and Identification of Bacterial Samples
4.2. Antibiotic Susceptibility Testing
4.3. PCR Detection of Virulence and ESBL Genes
4.4. Biofilm Formation Assay
4.5. Measurement of MIC-p and MBC
4.6. Determination of MIC-b, MBIC/MRC and MBEC
4.7. Sub-MIC Concentration of Ceftriaxone
4.8. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Alkeskas, A.; Ogrodzki, P.; Saad, M.; Masood, N.; Rhoma, N.R.; Moore, K.; Farbos, A.; Paszkiewicz, K.; Forsythe, S. The molecular characterisation of Escherichia coli K1 isolated from neonatal nasogastric feeding tubes. BMC Infect. Dis. 2015, 15, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Madappa, T.; Go, C.H.U. Escherichia coli(E coli) Infections: Background. Pathophysiology 2019. [Google Scholar]
- Barbieri, N.L.; Oliveira, A.L.d.; Tejkowski, T.M.; Pavanelo, D.B.; Matter, L.B.; Pinheiro, S.R.S.; Vaz, T.M.I.; Nolan, L.K.; Logue, C.M.; Brito, B.G.d. Molecular characterization and clonal relationships among Escherichia coli strains isolated from broiler chickens with colisepticemia. Foodborne Pathog. Dis. 2015, 12, 74–83. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, N.M.; Johnson, J.R.; Johnston, B.; Curtiss, R.; Mellata, M. Zoonotic Potential of Escherichia coli Isolates from Retail Chicken Meat Products and Eggs. Appl. Environ. Microbiol. 2015, 81, 1177–1187. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sarowska, J.; Futoma-Koloch, B.; Jama-Kmiecik, A.; Frej-Madrzak, M.; Ksiazczyk, M.; Bugla-Ploskonska, G.; Choroszy-Krol, I. Virulence factors, prevalence and potential transmission of extraintestinal pathogenic Escherichia coli isolated from different sources: Recent reports. Gut Pathog. 2019, 11, 1–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Belova, V.A.; Smutka, L.; Rosochatecká, E. World chicken meat market–its development and current status. Acta Univ. Agric. Et Silvic. Mendel. Brun. 2012, 60, 15–30. [Google Scholar] [CrossRef] [Green Version]
- Pozdniakova, E.; Pozdniakov, V.; Brench, A. Development trends and risk factors of meat global exports. Ukr. Food J. 2019, 8, 645–658. [Google Scholar] [CrossRef]
- Hafez, M.H.; Attia, Y.A. Challenges to the poultry industry: Current perspectives and strategic future after the COVID-19 outbreak. Front. Vet. Sci. 2020, 7, 516. [Google Scholar] [CrossRef]
- Swelum, A.A.; Elbestawy, A.R.; El-Saadony, M.T.; Hussein, E.O.; Alhotan, R.; Suliman, G.M.; Taha, A.E.; Ba-Awadh, H.; El-Tarabily, K.A.; El-Hack, M.E.A. Ways to minimize bacterial infections, with special reference to Escherichia coli, to cope with the first-week mortality in chicks: An updated overview. Poult. Sci. 2021, 100, 101039. [Google Scholar] [CrossRef]
- Agyare, C.; Boamah, V.E.; Zumbi, C.N.; Osei, F.B. Antibiotic use in poultry production and its effects on bacterial resistance. In Antimicrobial Resistance—A Global Threat; Kumar, Y., Ed.; IntechOpen: Rijeka, Croatia, 2018; pp. 33–51. [Google Scholar]
- Valiakos, G.; Kapna, I. Colistin resistant mcr genes prevalence in livestock animals (swine, bovine, poultry) from a multinational perspective. A Systematic Review. Vet. Sci. 2021, 8, 265. [Google Scholar] [CrossRef]
- Olsen, R.; Frantzen, C.; Christensen, H.; Bisgaard, M. An investigation on first-week mortality in layers. Avian Dis. 2012, 56, 51–57. [Google Scholar] [CrossRef] [PubMed]
- Benz, F.; Huisman, J.S.; Bakkeren, E.; Herter, J.A.; Stadler, T.; Ackermann, M.; Diard, M.; Egli, A.; Hall, A.R.; Hardt, W.-D. Plasmid-and strain-specific factors drive variation in ESBL-plasmid spread in vitro and in vivo. ISME J. 2021, 15, 862–878. [Google Scholar] [CrossRef] [PubMed]
- Silva, O.V.; Soares, L.O.; Júnior, A.S.; Mantovani, H.C.; Chang, Y.-F.; Moreira, M.A.S. Biofilm formation on biotic and abiotic surfaces in the presence of antimicrobials by Escherichia coli isolates from cases of bovine mastitis. Appl. Environ. Microbiol. 2014, 80, 6136–6145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stępień-Pyśniak, D.; Hauschild, T.; Kosikowska, U.; Dec, M.; Urban-Chmiel, R. Biofilm formation capacity and presence of virulence factors among commensal Enterococcus spp. from wild birds. Sci. Rep. 2019, 9, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Dong, G.; Li, J.; Chen, L.; Bi, W.; Zhang, X.; Liu, H.; Zhi, X.; Zhou, T.; Cao, J. Effects of sub-minimum inhibitory concentrations of ciprofloxacin on biofilm formation and virulence factors of Escherichia coli. Braz. J. Infect. Dis. 2019, 23, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Rafaque, Z.; Abid, N.; Liaqat, N.; Afridi, P.; Siddique, S.; Masood, S.; Kanwal, S.; Dasti, J.I. In-vitro investigation of antibiotics efficacy against uropathogenic Escherichia coli biofilms and antibiotic induced biofilm formation at sub-minimum inhibitory concentration of ciprofloxacin. Infect. Drug Resist. 2020, 13, 2801. [Google Scholar] [CrossRef] [PubMed]
- Ali, I.; Rafaque, Z.; Ahmed, S.; Malik, S.; Dasti, J.I. Prevalence of multi-drug resistant uropathogenic Escherichia coli in Potohar region of Pakistan. Asian Pac. J. Trop. Biomed. 2016, 6, 60–66. [Google Scholar] [CrossRef] [Green Version]
- Wanger, A.; Chávez, V. Antibiotic susceptibility Testing. In Practical Handbook of Microbiology; CRC Press: Boca Raton, FL, USA, 2021; pp. 119–128. [Google Scholar]
- Ali, I.; Rafaque, Z.; Ahmed, I.; Tariq, F.; Graham, S.E.; Salzman, E.; Foxman, B.; Dasti, J.I. Phylogeny, sequence-typing and virulence profile of uropathogenic Escherichia coli(UPEC) strains from Pakistan. BMC Infect. Dis. 2019, 19, 620. [Google Scholar] [CrossRef] [Green Version]
- Abd El Tawab, A.A.; Ammar, A.M.; Nasef, S.A.; Reda, R.M. Antibacterial resistance and resistance gene detriments of E. coli isolated from chicken. Benha Vet. Med. J. 2015, 28, 231–240. [Google Scholar] [CrossRef] [Green Version]
- Mohamed, M.A.; Shehata, M.A.; Rafeek, E. Virulence genes content and antimicrobial resistance in Escherichia coli from broiler chickens. Vet. Med. Int. 2014, 2014, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Mund, M.D.; Khan, U.H.; Tahir, U.; Mustafa, B.-E.; Fayyaz, A. Antimicrobial drug residues in poultry products and implications on public health: A review. Int. J. Food Prop. 2017, 20, 1433–1446. [Google Scholar] [CrossRef]
- Mohsin, M.; van Boeckel, T.P.; Saleemi, M.K.; Umair, M.; Naseem, M.N.; He, C.; Khan, A.; Laxminarayan, R. Excessive use of medically important antimicrobials in food animals in Pakistan: A five-year surveillance survey. Glob. Health Action 2019, 12, 1697541. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khan, G.J.; Khan, R.A.; Majeed, I.; Siddiqui, F.A.; Khan, S. Ciprofloxacin: The frequent use in poultry and its consequences on human health. Prof. Med. J. 2015, 22, 001–005. [Google Scholar] [CrossRef]
- Choe, H.-S.; Lee, S.-J.; Yang, S.S.; Hamasuna, R.; Yamamoto, S.; Cho, Y.-H.; Matsumoto, T. The Committee for Development of the UAA-AAUS Guidelines for UTI and STI Summary of the UAA-AAUS guidelines for urinary tract infections. Int. J. Urol. 2018, 25, 175–185. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choe, H.-S.; Lee, S.-J.; Cho, Y.-H.; Çek, M.; Tandoğdu, Z.; Wagenlehner, F.; Bjerklund-Johansen, T.E.; Naber, K.; Nikfallah, A.; Kassem, A.M. Aspects of urinary tract infections and antimicrobial resistance in hospitalized urology patients in Asia: 10-Year results of the Global Prevalence Study of Infections in Urology (GPIU). J. Infect.Chemother. 2018, 24, 278–283. [Google Scholar] [CrossRef] [Green Version]
- Kranz, J.; Schmidt, S.; Lebert, C.; Schneidewind, L.; Mandraka, F.; Kunze, M.; Helbig, S.; Vahlensieck, W.; Naber, K.; Schmiemann, G. The 2017 update of the German clinical guideline on epidemiology, diagnostics, therapy, prevention, and management of uncomplicated urinary tract infections in adult patients. Part II: Therapy and prevention. Urol. Int. 2018, 100, 271–278. [Google Scholar] [CrossRef]
- Mead, A.; Richez, P.; Azzariti, S.; Pelligand, L. Pharmacokinetics of Colistin in the Gastrointestinal Tract of Poultry Following Dosing via Drinking Water and Its Bactericidal Impact on Enteric Escherichia coli. Front. Vet. Sci. 2021, 8, 634. [Google Scholar] [CrossRef]
- Imtiaz, W.; Syed, Z.; Rafaque, Z.; Andrews, S.C.; Dasti, J.I. Analysis of antibiotic resistance and virulence traits (genetic and phenotypic) in Klebsiella pneumoniae clinical isolates from Pakistan: Identification of significant levels of carbapenem and colistin resistance. Infect. Drug Resist. 2021, 14, 227. [Google Scholar] [CrossRef]
- Vaara, M. Polymyxins and their potential next generation as therapeutic antibiotics. Front. Microbiol. 2019, 10, 1689. [Google Scholar] [CrossRef]
- Pour, N.K.; Dusane, D.H.; Dhakephalkar, P.K.; Zamin, F.R.; Zinjarde, S.S.; Chopade, B.A. Biofilm formation by Acinetobacter baumannii strains isolated from urinary tract infection and urinary catheters. FEMS Immunol. Med. Microbiol. 2011, 62, 328–338. [Google Scholar] [CrossRef] [Green Version]
- Sato, Y.; Unno, Y.; Ubagai, T.; Ono, Y. Sub-minimum inhibitory concentrations of colistin and polymyxin B promote Acinetobacter baumannii biofilm formation. PLoS ONE 2018, 13, e0194556. [Google Scholar] [CrossRef]
- Nucleo, E.; Steffanoni, L.; Fugazza, G.; Migliavacca, R.; Giacobone, E.; Navarra, A.; Pagani, L.; Landini, P. Growth in glucose-based medium and exposure to subinhibitory concentrations of imipenem induce biofilm formation in a multidrug-resistant clinical isolate of Acinetobacter baumannii. BMC Microbiol. 2009, 9, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, X.; Lu, F.; Yuan, F.; Jiang, D.; Zhao, P.; Zhu, J.; Cheng, H.; Cao, J.; Lu, G. Biofilm formation caused by clinical Acinetobacter baumannii isolates is associated with overexpression of the AdeFGH efflux pump. Antimicrob. Agents Chemother. 2015, 59, 4817–4825. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klinger-Strobel, M.; Stein, C.; Forstner, C.; Makarewicz, O.; Pletz, M.W. Effects of colistin on biofilm matrices of Escherichia coli and Staphylococcus aureus. Int. J. Antimicrob. Agents 2017, 49, 472–479. [Google Scholar] [CrossRef] [PubMed]
- Magiorakos, A.P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef]




| Resistance Traits | Total Isolates n (%) | MDR (n = 70) | XDR (n = 12) | p Value |
|---|---|---|---|---|
| Ampicillin | 80 (96.38) | 67 | 12 | 0.4650 |
| Amoxicillin | 75 (90.36) | 62 | 12 | 0.1179 |
| Levofloxacin | 51 (61.44) | 40 | 10 | 0.0857 |
| Ciprofloxacin | 51 (61.44) | 42 | 8 | 0.6618 |
| Tobramycin | 5 (6.02) | 2 | 3 | 0.0031 |
| Gentamycin | 7 (8.43) | 3 | 4 | 0.0009 |
| Neomycin | 44 (53.01) | 32 | 12 | 0.0005 |
| Streptomycin | 67 (80.72) | 55 | 11 | 0.2902 |
| Tigecycline | 0 | 0 | 0 | - |
| Tetracycline | 75 (90.36) | 65 | 9 | 0.0541 |
| Oxytetracycline | 80 (90.38) | 68 | 11 | 0.3505 |
| Doxycycline | 50 (60.24) | 40 | 10 | 0.0857 |
| Nitrofurantoin | 7 (8.43) | 4 | 3 | 0.0272 |
| Chloramphenicol | 53 (63.85) | 42 | 11 | 0.0340 |
| Cefotaxime | 1 (1.20) | 1 | 0 | 0.6770 |
| Cefixime | 6 (7.22) | 2 | 4 | 0.0002 |
| Cephalothin | 36 (43.37) | 26 | 10 | 0.0029 |
| Ceftazidime | 6 (7.22) | 4 | 2 | 0.1783 |
| Cefepime | 4 (4.81) | 1 | 3 | 0.0005 |
| Imipenem | 4 (4.81) | 2 | 2 | 0.0402 |
| Meropenem | 8 (9.63) | 5 | 3 | 0.0541 |
| TMP-SMX | 64 (77.10) | 52 | 11 | 0.1873 |
| Lincomycin | 83 (100) | 70 | 12 | 0.1945 |
| Augmentin | 4 (4.81) | 2 | 10 | 0.0001 |
| Polymyxin B | 1 (1.20) | 1 | 0 | 0.6770 |
| Colistin (Polymyxin E) | 22 (26.50) | 16 | 6 | 0.0499 |
| Virulence Genes | ||||
| fimH | 32 (38.55) | 25 | 6 | 0.3457 |
| papC | 21 (25.30) | 19 | 2 | 0.4424 |
| iutA | 34 (40.96) | 26 | 7 | 0.1667 |
| kpsMT-II | 23 (27.71 | 18 | 4 | 0.5821 |
| papEF | 9 (10.84) | 6 | 3 | 0.0925 |
| papGII | 22 (26.50) | 17 | 5 | 0.2093 |
| fyuA | 13 (15.66) | 10 | 3 | 0.3478 |
| papGIII | 0 | 0 | 0 | - |
| Resistance Genes | ||||
| blaTEM-1 | 22 (26.50) | 17 | 5 | 0.2093 |
| blaOXA | 8 (9.63) | 4 | 4 | 0.0029 |
| blaSHV | 4 (4.81) | 1 | 3 | 0.0005 |
| blaPSE | 0 | 0 | 0 | - |
| Resistance Traits | Total Isolates n = 83 (%) | ESBL Producers n (%) | Non-ESBL n (%) | p Value |
|---|---|---|---|---|
| Ampicillin | 80 (96.38) | 38 (47.50) | 42 (52.5) | 0.5142 |
| Amoxicillin | 75 (90.36) | 35 (46.66) | 40 (53.33) | 0.3942 |
| Levofloxacin | 51 (61.44) | 26 (50.98) | 25 (49.01) | 0.5211 |
| Ciprofloxacin | 51 (61.44) | 31 (60.78) | 20 (39.2) | 0.0038 |
| Tobramycin | 5 (6.02) | 3 (60) | 2 (40) | 0.5857 |
| Gentamycin | 7 (8.43) | 4 (57.14) | 3 (42.85) | 0.6204 |
| Neomycin | 44 (53.01) | 27 (61.36) | 17 (38.63) | 0.0108 |
| Streptomycin | 67 (80.72) | 32 (47.76) | 35 (52.23) | 0.8721 |
| Tigecycline | 0 | 0 | 0 | - |
| Tetracycline | 75 (90.36) | 38 (50.66) | 37 (49.33) | 0.1673 |
| Oxytetracycline | 80 (90.38) | 38 (47.5) | 42 (52.5) | 0.5142 |
| Doxycycline | 50 (60.24) | 25 (50) | 25 (50) | 0.6850 |
| Nitrofurantoin | 7 (8.43) | 4 (57.1) | 3 (42.85) | 0.6204 |
| Chloramphenicol | 53 (63.85) | 28 (52.83) | 25 (47.16) | 0.2611 |
| Cefotaxime | 1 (1.20) | 1 (100) | 0 | 0.2969 |
| Cefixime | 6 (7.22) | 6 (100) | 0 | 0.0084 |
| Cephalothin | 36 (43.37) | 22 (61.11) | 14 (38.88) | 0.0393 |
| Ceftazidime | 6 (7.22) | 4 (60) | 2 (40) | 0.5804 |
| Cefepime | 4 (4.81) | 4 (100) | 0 | 0.0335 |
| Imipenem | 4 (4.81) | 3 (75) | 1 (25) | 0.2714 |
| Meropenem | 8 (9.63) | 6 (75) | 2 (25) | 0.1104 |
| TMP-SMX | 64 (77.10) | 32 (50) | 32 (50) | 0.5453 |
| Lincomycin | 83 (100) | 40 (48.19) | 43 (51.80) | >0.9999 |
| Augmentin | 4 (4.81) | 3 (75) | 1 (25) | 0.2714 |
| Polymyxin B | 1 (1) | 0 | 1 (100) | 0.2969 |
| Colistin (Polymyxin E) | 22 (26.50) | 13 (59.09) | 9 (40.90) | 0.2327 |
| fimH | 32 (38.55) | 17 (53.12) | 15 (46.87) | 0.4763 |
| papC | 21 (25.30) | 8 (38.09) | 13 (61.90) | 0.2840 |
| iutA | 34 (40.96) | 16 (47.05) | 18 (52.94) | 0.0039 |
| kpsMT-II | 23 (27.71 | 7 (30.43) | 16 (69.56) | 0.0450 |
| papEF | 9 (10.84) | 5 (55.55) | 4 (44.44) | 0.6397 |
| papGII | 22 (26.50) | 11(50) | 11(50) | 0.8431 |
| fyuA | 13 (15.66) | 10 (76.92) | 3 (23.07) | 0.0240 |
| papGIII | 0 | 0 | 0 | - |
| blaTEM-1 | 22 (26.50) | 22 (100) | 0 | 0.0001 |
| blaOXA | 8 (9.63) | 8 (100) | 0 | 0.0020 |
| blaSHV | 4 (4.81) | 4 (100) | 0 | 0.0335 |
| blaPSE | 0 | 0 | 0 | - |
| Isolate No. | MIC-p µg/mL | MIC-b µg/mL | ||||||
|---|---|---|---|---|---|---|---|---|
| GEN | COL | CEF | ENR | GEN | COL | CEF | ENR | |
| 7 | 4 | 2 | 0.5 | 0.06 | 1 | 1 | 16 | 4 |
| 21 | 1 | 2 | 0.5 | 0.125 | 1 | 1 | 32 | 4 |
| Isolate No. | GEN | COL | CEF | ENR | ||||
|---|---|---|---|---|---|---|---|---|
| MBIC µg/mL | MBEC µg/mL | MBIC µg/mL | MBEC µg/mL | MBIC µg/mL | MBEC µg/mL | MBIC µg/mL | MBEC µg/mL | |
| 7 | 8 | 32 | 8 | 128 | 128 | >2048 | 256 | >2048 |
| 21 | 32 | 128 | 16 | 64 | 256 | >2048 | 64 | >2048 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Noreen, A.; Masood, H.; Zaib, J.; Rafaque, Z.; Fatima, A.; Shabbir, H.; Alam, J.; Habib, A.; Noor, S.; Dil, K.; et al. Investigating the Role of Antibiotics on Induction, Inhibition and Eradication of Biofilms of Poultry Associated Escherichia coli Isolated from Retail Chicken Meat. Antibiotics 2022, 11, 1663. https://doi.org/10.3390/antibiotics11111663
Noreen A, Masood H, Zaib J, Rafaque Z, Fatima A, Shabbir H, Alam J, Habib A, Noor S, Dil K, et al. Investigating the Role of Antibiotics on Induction, Inhibition and Eradication of Biofilms of Poultry Associated Escherichia coli Isolated from Retail Chicken Meat. Antibiotics. 2022; 11(11):1663. https://doi.org/10.3390/antibiotics11111663
Chicago/Turabian StyleNoreen, Aisha, Hamid Masood, Jaweria Zaib, Zara Rafaque, Areeta Fatima, Hira Shabbir, Javaria Alam, Aisha Habib, Saba Noor, Kinza Dil, and et al. 2022. "Investigating the Role of Antibiotics on Induction, Inhibition and Eradication of Biofilms of Poultry Associated Escherichia coli Isolated from Retail Chicken Meat" Antibiotics 11, no. 11: 1663. https://doi.org/10.3390/antibiotics11111663





