Genome-Wide Analysis of Innate Susceptibility Mechanisms of Escherichia coli to Colistin
Abstract
:1. Introduction
2. Results
2.1. Effect of Colistin Stress on the E. coli Mutant Library
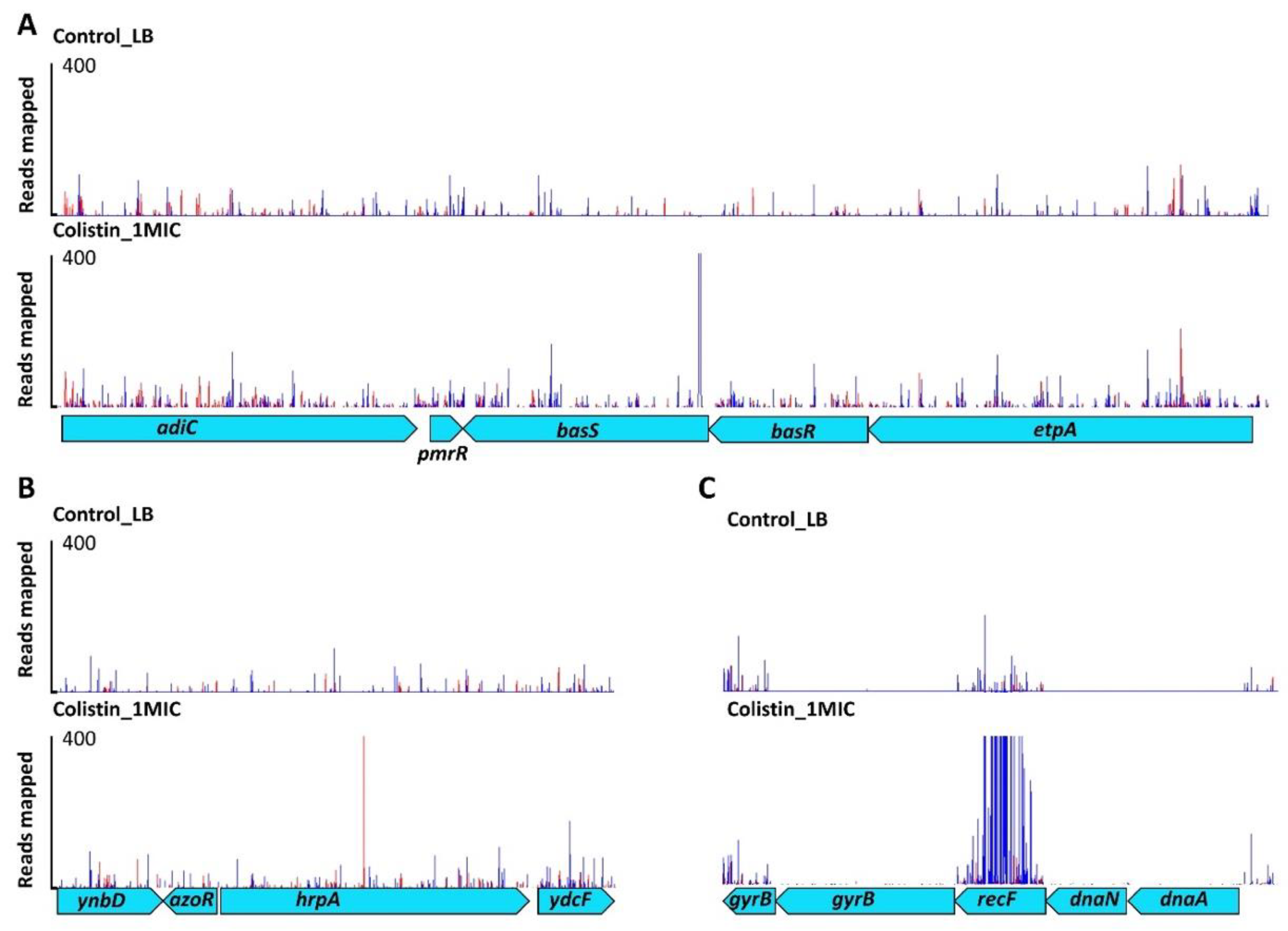
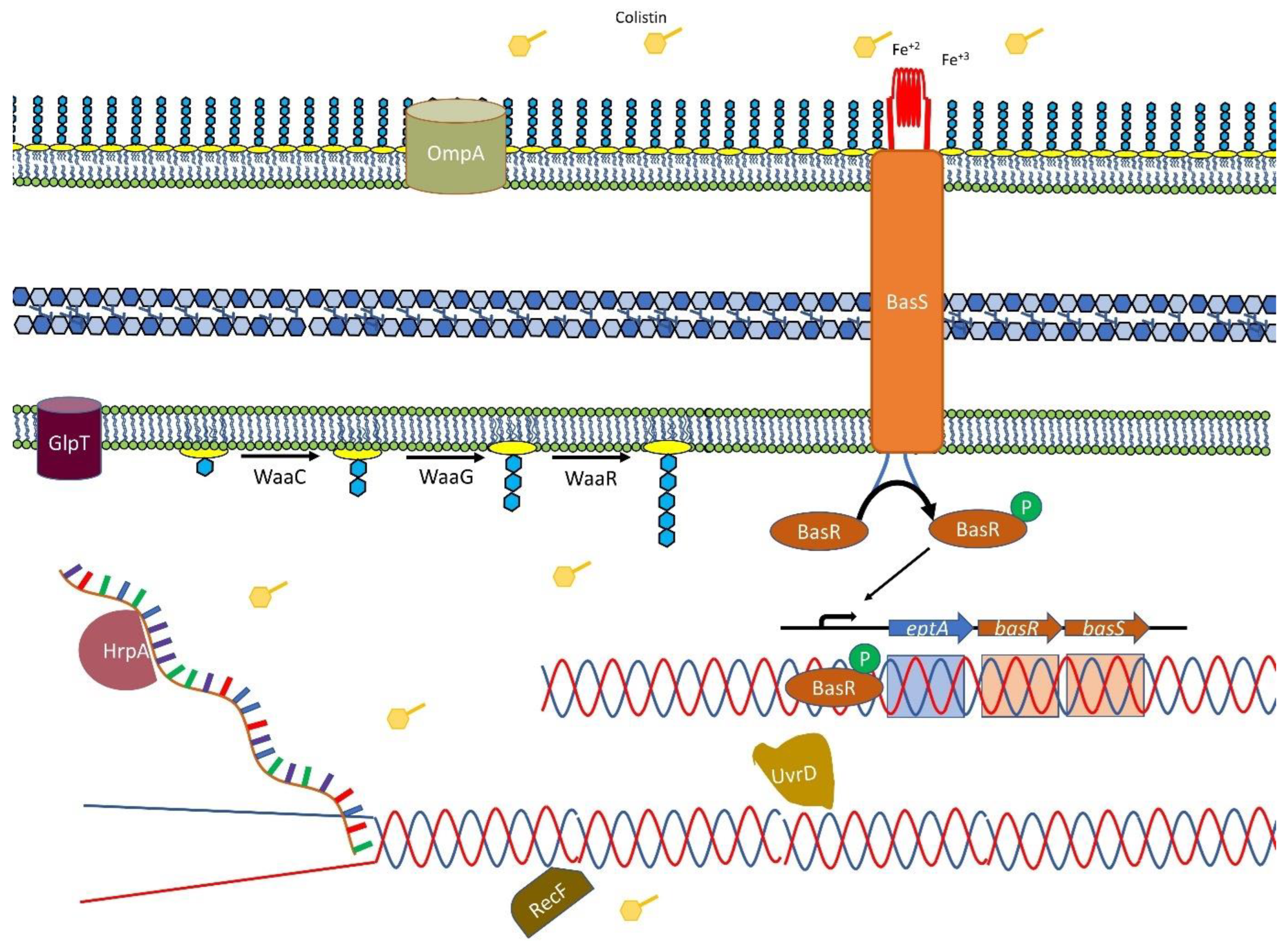
2.2. Effect of Colistin on Cell Membranes
2.3. DNA Housekeeping Genes Associated with Colistin Susceptibility
2.4. Small RNA Dependent Regulation
2.5. GlpT Transporter and Phosphonates
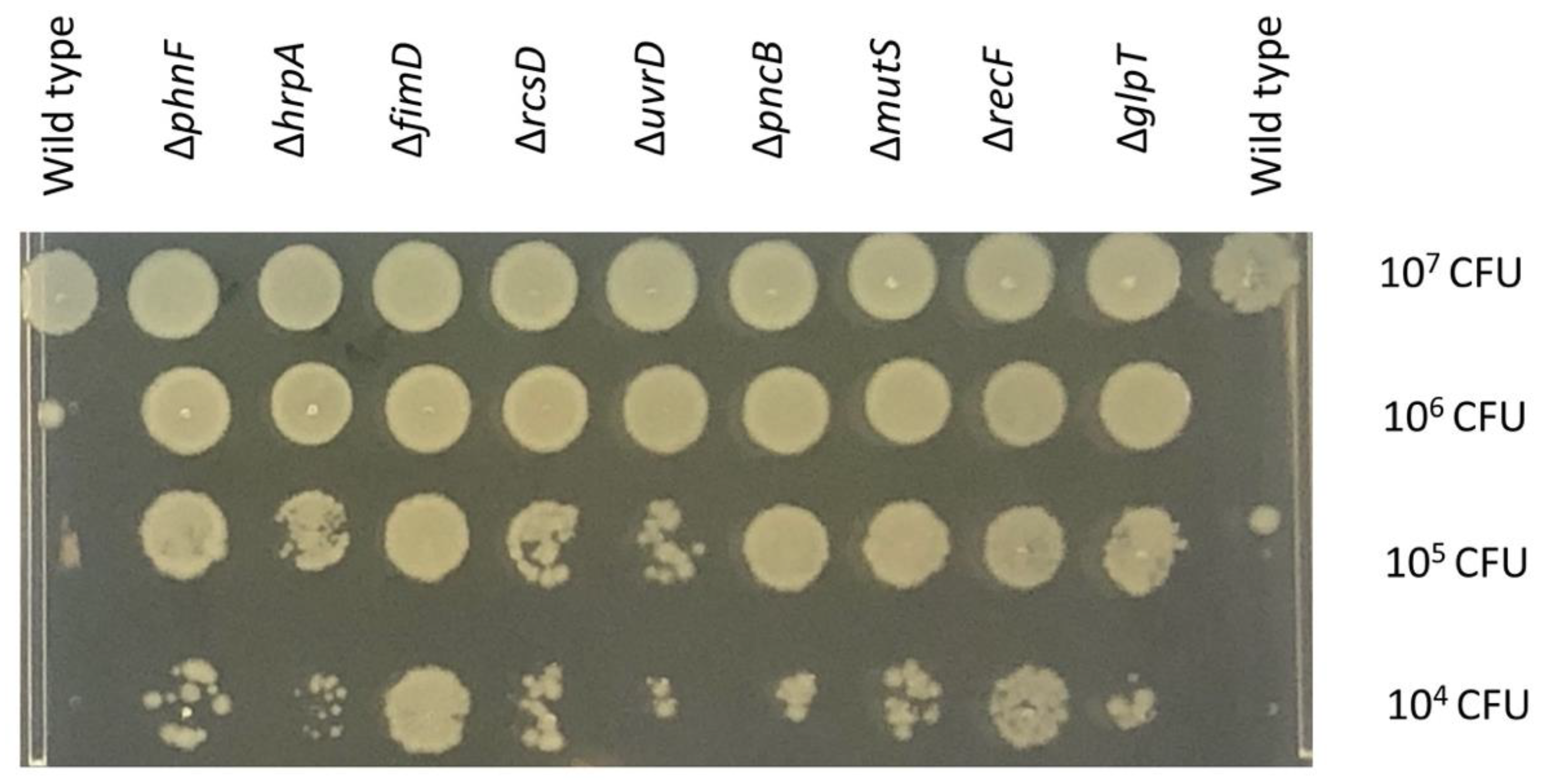
2.6. Validation of the Potential Genes
3. Discussion
4. Materials and Methods
4.1. TraDIS-Xpress Library
4.2. Colistin Exposure Conditions and TraDIS-Xpress Sequencing
4.3. Bioinformatics
4.4. Validation Experiments
4.5. Comparing Combinations of Two Antibiotics at Different Concentrations
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Koulenti, D.; Xu, E.; Mok, I.Y.S.; Song, A.; Karageorgopoulos, D.E.; Armaganidis, A.; Lipman, J.; Tsiodras, S. Novel Antibiotics for Multidrug-Resistant Gram-Positive Microorganisms. Microorganisms 2019, 7, 270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bassetti, M.; Peghin, M.; Vena, A.; Giacobbe, D.R. Treatment of Infections Due to MDR Gram-Negative Bacteria. Front. Med. 2019, 6, 74. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walia, K.; Sharma, M.; Vijay, S.; Shome, B.R. Understanding policy dilemmas around antibiotic use in food animals & offering potential solutions. Indian J. Med. Res. 2019, 149, 107–118. [Google Scholar] [CrossRef] [PubMed]
- Edrees, N.E.; Galal, A.A.A.; Abdel Monaem, A.R.; Beheiry, R.R.; Metwally, M.M.M. Curcumin alleviates colistin-induced nephrotoxicity and neurotoxicity in rats via attenuation of oxidative stress, inflammation and apoptosis. Chem.-Biol. Interact. 2018, 294, 56–64. [Google Scholar] [CrossRef]
- El-Sayed Ahmed, M.A.E.; Zhong, L.L.; Shen, C.; Yang, Y.; Doi, Y.; Tian, G.B. Colistin and its role in the Era of antibiotic resistance: An extended review (2000–2019). Emerg. Microbes Infect. 2020, 9, 868–885. [Google Scholar] [CrossRef] [Green Version]
- Gharaibeh, M.H.; Shatnawi, S.Q. An overview of colistin resistance, mobilized colistin resistance genes dissemination, global responses, and the alternatives to colistin: A review. Vet. World 2019, 12, 1735–1746. [Google Scholar] [CrossRef] [Green Version]
- Romano, K.P.; Warrier, T.; Poulsen, B.E.; Nguyen, P.H.; Loftis, A.R.; Saebi, A.; Pentelute, B.L.; Hung, D.T. Mutations in pmrB Confer Cross-Resistance between the LptD Inhibitor POL7080 and Colistin in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2019, 63, e00511-19. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.Y.; Wang, Y.; Walsh, T.R.; Yi, L.X.; Zhang, R.; Spencer, J.; Doi, Y.; Tian, G.; Dong, B.; Huang, X.; et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: A microbiological and molecular biological study. Lancet Infect. Dis. 2016, 16, 161–168. [Google Scholar] [CrossRef]
- Rebelo, A.R.; Bortolaia, V.; Kjeldgaard, J.S.; Pedersen, S.K.; Leekitcharoenphon, P.; Hansen, I.M.; Guerra, B.; Malorny, B.; Borowiak, M.; Hammerl, J.A.; et al. Multiplex PCR for detection of plasmid-mediated colistin resistance determinants, mcr-1, mcr-2, mcr-3, mcr-4 and mcr-5 for surveillance purposes. Eurosurveillance 2018, 23, 17-00672. [Google Scholar] [CrossRef] [Green Version]
- Yap, P.S.-X.; Cheng, W.-H.; Chang, S.-K.; Lim, S.-H.E.; Lai, K.-S. MgrB Mutations and Altered Cell Permeability in Colistin Resistance in Klebsiella pneumoniae. Cells 2022, 11, 2995. [Google Scholar] [CrossRef]
- Nordmann, P.; Jayol, A.; Poirel, L. Rapid Detection of Polymyxin Resistance in Enterobacteriaceae. Emerg. Infect. Dis. 2016, 22, 1038–1043. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Olaitan, A.O.; Morand, S.; Rolain, J.M. Mechanisms of polymyxin resistance: Acquired and intrinsic resistance in bacteria. Front. Microbiol. 2014, 5, 643. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cain, A.K.; Boinett, C.J.; Barquist, L.; Dordel, J.; Fookes, M.; Mayho, M.; Ellington, M.J.; Goulding, D.; Pickard, D.; Wick, R.R.; et al. Morphological, genomic and transcriptomic responses of Klebsiella pneumoniae to the last-line antibiotic colistin. Sci. Rep. 2018, 8, 9868. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jana, B.; Cain, A.K.; Doerrler, W.T.; Boinett, C.J.; Fookes, M.C.; Parkhill, J.; Guardabassi, L. The secondary resistome of multidrug-resistant Klebsiella pneumoniae. Sci. Rep. 2017, 7, 42483. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yasir, M.; Turner, A.K.; Bastkowski, S.; Baker, D.; Page, A.J.; Telatin, A.; Phan, M.D.; Monahan, L.; Savva, G.M.; Darling, A.; et al. TraDIS-Xpress: A high-resolution whole-genome assay identifies novel mechanisms of triclosan action and resistance. Genome Res. 2020, 30, 239–249. [Google Scholar] [CrossRef] [PubMed]
- Kohanski, M.A.; Dwyer, D.J.; Hayete, B.; Lawrence, C.A.; Collins, J.J. A common mechanism of cellular death induced by bactericidal antibiotics. Cell 2007, 130, 797–810. [Google Scholar] [CrossRef] [Green Version]
- Aghapour, Z.; Gholizadeh, P.; Ganbarov, K.; Bialvaei, A.Z.; Mahmood, S.S.; Tanomand, A.; Yousefi, M.; Asgharzadeh, M.; Yousefi, B.; Kafil, H.S. Molecular mechanisms related to colistin resistance in Enterobacteriaceae. Infect. Drug Resist. 2019, 12, 965–975. [Google Scholar] [CrossRef] [Green Version]
- Bertani, B.R.; Taylor, R.J.; Nagy, E.; Kahne, D.; Ruiz, N. A cluster of residues in the lipopolysaccharide exporter that selects substrate variants for transport to the outer membrane. Mol. Microbiol. 2018, 109, 541–554. [Google Scholar] [CrossRef] [Green Version]
- Jayol, A.; Poirel, L.; Brink, A.; Villegas, M.V.; Yilmaz, M.; Nordmann, P. Resistance to colistin associated with a single amino acid change in protein PmrB among Klebsiella pneumoniae isolates of worldwide origin. Antimicrob. Agents Chemother. 2014, 58, 4762–4766. [Google Scholar] [CrossRef] [Green Version]
- Hecht, A.; Glasgow, J.; Jaschke, P.R.; Bawazer, L.A.; Munson, M.S.; Cochran, J.R.; Endy, D.; Salit, M. Measurements of translation initiation from all 64 codons in E. coli. Nucleic Acids Res. 2017, 45, 3615–3626. [Google Scholar] [CrossRef]
- Grass, L.M.; Wollenhaupt, J.; Barthel, T.; Parfentev, I.; Urlaub, H.; Loll, B.; Klauck, E.; Antelmann, H.; Wahl, M.C. Large-scale ratcheting in a bacterial DEAH/RHA-type RNA helicase that modulates antibiotics susceptibility. Proc. Natl. Acad. Sci. USA 2021, 118, e2100370118. [Google Scholar] [CrossRef] [PubMed]
- Turner, A.K.; Yasir, M.; Bastkowski, S.; Telatin, A.; Page, A.; Webber, M.; Charles, I. Chemical biology-whole genome engineering datasets predict new antibacterial combinations. Microb. Genom. 2021, 7, 000718. [Google Scholar] [CrossRef] [PubMed]
- Janssen, A.B.; Bartholomew, T.L.; Marciszewska, N.P.; Bonten, M.J.M.; Willems, R.J.L.; Bengoechea, J.A.; van Schaik, W. Nonclonal Emergence of Colistin Resistance Associated with Mutations in the BasRS Two-Component System in Escherichia coli Bloodstream Isolates. mSphere 2020, 5, e00143-20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Douglas, F.; Browning, J.L.H.; Stephen, J.W. Busby, Laboratory strains of Escherichia coli K-12: Not such perfect role models after all. BioRxiv 2022, 32, 1109–1111. [Google Scholar] [CrossRef]
- Barquist, L.; Mayho, M.; Cummins, C.; Cain, A.K.; Boinett, C.J.; Page, A.J.; Langridge, G.C.; Quail, M.A.; Keane, J.A.; Parkhill, J. The TraDIS toolkit: Sequencing and analysis for dense transposon mutant libraries. Bioinformatics 2016, 32, 1109–1111. [Google Scholar] [CrossRef] [Green Version]
- Page, A.J.; Bastkowski, S.; Yasir, M.; Turner, A.K.; Le Viet, T.; Savva, G.M.; Webber, M.A.; Charles, I.G. AlbaTraDIS: Comparative analysis of large datasets from parallel transposon mutagenesis experiments. PloS Comput. Biol. 2020, 16, e1007980. [Google Scholar] [CrossRef]
- Robinson, M.D.; Oshlack, A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010, 11, R25. [Google Scholar] [CrossRef] [Green Version]
- Benjamini, Y.; Drai, D.; Elmer, G.; Kafkafi, N.; Golani, I. Controlling the false discovery rate in behavior genetics research. Behav. Brain Res. 2001, 125, 279–284. [Google Scholar] [CrossRef] [Green Version]
- Carver, T.; Harris, S.R.; Berriman, M.; Parkhill, J.; McQuillan, J.A. Artemis: An integrated platform for visualization and analysis of high-throughput sequence-based experimental data. Bioinformatics 2012, 28, 464–469. [Google Scholar] [CrossRef] [Green Version]
- Baba, T.; Ara, T.; Hasegawa, M.; Takai, Y.; Okumura, Y.; Baba, M.; Datsenko, K.A.; Tomita, M.; Wanner, B.L.; Mori, H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: The Keio collection. Mol. Syst. Biol. 2006, 2, 2006.0008. [Google Scholar] [CrossRef]
- Agarwal, M.; Rathore, R.S.; Chauhan, A. A Rapid and High Throughput MIC Determination Method to Screen Uranium Resistant Microorganisms. Methods Protoc. 2020, 3, 21. [Google Scholar] [CrossRef] [PubMed]
| Pathway | Gene | Annotation | Multiple of MIC | |||
|---|---|---|---|---|---|---|
| 2× | 1× | 0.5× | 0.25× | |||
| Lipid biosynthesis and regulation | basR | response regulator in two-component regulatory system with BasS | 4.03 | 0.00 | 0.00 | 0.00 |
| basS | sensory histidine kinase in two-component regulatory system with BasR | 17.44 | 5.65 | 1.94 | 5.45 | |
| waaB | UDP-D-galactose:(glucosyl)lipopolysaccharide-1 and 6-D-galactosyltransferase | 0.00 | −1.82 | 0.00 | 0.00 | |
| waaC | ADP-heptose:LPS heptosyl transferase I | 0.00 | −2.25 | −1.70 | 0.00 | |
| waaG | glucosyltransferase I | 0.00 | −3.06 | −1.98 | −1.61 | |
| waaP | kinase that phosphorylates core heptose of lipopolysaccharide | 0.00 | −2.46 | −1.92 | 0.00 | |
| waaR | UDP-D-galactose:(glucosyl)lipopolysaccharide-alpha-1 and 3-D-galactosyltransferase | 0.00 | −3.83 | −2.01 | −1.11 | |
| waaS | lipopolysaccharide core biosynthesis protein | 0.00 | −1.96 | −1.63 | −1.60 | |
| waaU | lipopolysaccharide core biosynthesis | 0.00 | −1.10 | 0.00 | 0.00 | |
| lptC | lipopolysaccharide export and IM-tethered periplasmic protein of the LptBFGC export complex | 0.00 | 1.57 | 1.50 | 1.42 | |
| waaD | ADP-L-glycero-D-mannoheptose-6-epimerase and NAD(P)-binding | 0.00 | −2.66 | −2.16 | 0.00 | |
| wbbH | O-antigen polymerase | 0.00 | −1.14 | −1.16 | −1.17 | |
| wbbI | d-Galf:alpha-d-Glc beta-1 and 6-galactofuranosyltransferase | 0.00 | 0.00 | −1.08 | 0.00 | |
| wbbK | lipopolysaccharide biosynthesis protein | 0.00 | −1.59 | −1.45 | −1.57 | |
| Transcription and translation regulation | aspU | tRNA-Asp | 0.00 | 0.00 | 0.00 | −2.70 |
| cmoB | tRNA (cmo5U34)-carboxymethyltransferase and carboxy-SAM-dependent | 0.00 | 1.30 | 0.00 | 0.00 | |
| valS | valyl-tRNA synthetase | 0.00 | 0.00 | 0.00 | −3.15 | |
| truA | tRNA pseudouridine(38–40) synthase | 0.00 | 1.11 | 1.32 | 1.23 | |
| hfq | global sRNA chaperone; HF-I and host factor for RNA phage Q beta replication | 0.00 | 1.41 | 1.55 | 1.55 | |
| ohsC | sRNA antisense regulator of shoB toxin | 0.00 | −2.49 | −2.81 | −2.55 | |
| oxyS | sRNA antisense regulator activates genes that detoxify oxidative damage and Hfq-dependent | 0.00 | 0.00 | −2.27 | 0.00 | |
| rpoN | RNA polymerase and sigma 54 (sigma N) factor | 0.00 | 1.99 | 0.00 | 1.81 | |
| rydC | sRNA antisense regulator of yejABEF and Hfq-dependent; over-expression causes a thermosensitive growth phenotype on minimal glycerol and maltose and or ribose media | 0.00 | −2.31 | 0.00 | −1.83 | |
| appY | global transcriptional activator; DLP12 prophage | 0.00 | −1.66 | −1.66 | −1.69 | |
| Membrane associated genes | bhsA | biofilm and cell surface and signaling protein | 0.00 | −1.86 | 0.00 | −2.21 |
| ariR | RcsB connector protein for regulation of biofilm and acid-resistance | 0.00 | −1.04 | −1.26 | −1.23 | |
| ompA | outer membrane protein A (3a;II*;G;d) | 0.00 | −1.32 | 0.00 | 0.00 | |
| surA | peptidyl-prolyl cis-trans isomerase (PPIase) | 0.00 | −3.29 | 0.00 | −1.38 | |
| flgA | assembly protein for flagellar basal-body periplasmic P ring | 0.00 | 1.22 | 0.00 | 0.00 | |
| Transporters | glpT | sn-glycerol-3-phosphate transporter | 3.26 | 1.08 | 1.15 | 1.14 |
| phnE | 4312259_4313047 | 3.84 | 1.44 | 1.35 | 1.29 | |
| phnF | putative DNA-binding transcriptional regulator of phosphonate uptake and biodegradation | 4.14 | 1.05 | 1.02 | 0.00 | |
| ygcS | putative transporter | 12.28 | 0.00 | 0.00 | 0.00 | |
| DNA associated proteins | hrpA | putative ATP-dependent helicase | 16.12 | 2.80 | 0.00 | 2.99 |
| recF | gap repair protein | 7.68 | 4.60 | 0.00 | 0.00 | |
| uvrD | DNA-dependent ATPase I and helicase II | 8.10 | 0.00 | 0.00 | 0.00 | |
| dinI | DNA damage-inducible protein I | 0.00 | 0.00 | 0.00 | −1.28 | |
| recG | ATP-dependent DNA helicase | 0.00 | 10.51 | 4.98 | 0.00 | |
| dcd | 2′-deoxycytidine 5′-triphosphate deaminase | 0.00 | 3.54 | 2.94 | 3.74 | |
| dnaG | DNA primase | 0.00 | −4.65 | −3.74 | 0.00 | |
| uspC | universal stress protein | 0.00 | −1.14 | −1.09 | 0.00 | |
| seqA | negative modulator of initiation of replication | 0.00 | 1.73 | 1.74 | 0.00 | |
| Miscellaneous | ybbC | putative immunity protein | 0.00 | −2.21 | −2.03 | −2.08 |
| ybcV | DLP12 prophage; uncharacterized protein | 0.00 | −1.74 | −1.37 | −1.48 | |
| ydcD | uncharacterized protein | 0.00 | −2.86 | 0.00 | 0.00 | |
| ygeN | 2988672_2989379 | 0.00 | −1.84 | −1.87 | −1.76 | |
| yjbL | uncharacterized protein | 0.00 | −2.44 | −2.44 | −2.48 | |
| yjbS | uncharacterized protein | 0.00 | −2.22 | −2.04 | −2.39 | |
| yfcZ | UPF0381 family protein | 0.00 | 2.99 | 0.00 | 0.00 | |
| ycjS | putative NADH-binding oxidoreductase | 0.00 | 1.19 | 1.09 | 1.19 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yasir, M.; Turner, A.K.; Bastkowski, S.; Lott, M.; Holden, E.R.; Telatin, A.; Page, A.J.; Webber, M.A.; Charles, I.G. Genome-Wide Analysis of Innate Susceptibility Mechanisms of Escherichia coli to Colistin. Antibiotics 2022, 11, 1668. https://doi.org/10.3390/antibiotics11111668
Yasir M, Turner AK, Bastkowski S, Lott M, Holden ER, Telatin A, Page AJ, Webber MA, Charles IG. Genome-Wide Analysis of Innate Susceptibility Mechanisms of Escherichia coli to Colistin. Antibiotics. 2022; 11(11):1668. https://doi.org/10.3390/antibiotics11111668
Chicago/Turabian StyleYasir, Muhammad, A. Keith Turner, Sarah Bastkowski, Martin Lott, Emma R. Holden, Andrea Telatin, Andrew J. Page, Mark A. Webber, and Ian G. Charles. 2022. "Genome-Wide Analysis of Innate Susceptibility Mechanisms of Escherichia coli to Colistin" Antibiotics 11, no. 11: 1668. https://doi.org/10.3390/antibiotics11111668







