High Carriage of Extended-Spectrum, Beta Lactamase-Producing, and Colistin-Resistant Enterobacteriaceae in Tibetan Outpatients with Diarrhea
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection, Bacterial Isolates, and Isolate Characterization
2.2. ARG Screening
2.3. DNA Extraction and Genome Sequencing
2.4. Molecular Typing, Virulence Genes, ARGs, and Plasmid Identification
2.5. Antimicrobial Susceptibility Testing
2.6. Conjugation and Transformation Analysis
3. Results
3.1. ESBL-Producing and Colistin-Resistant Enterobacteriaceae Isolates Screened from Tibetan Outpatients with Diarrhea
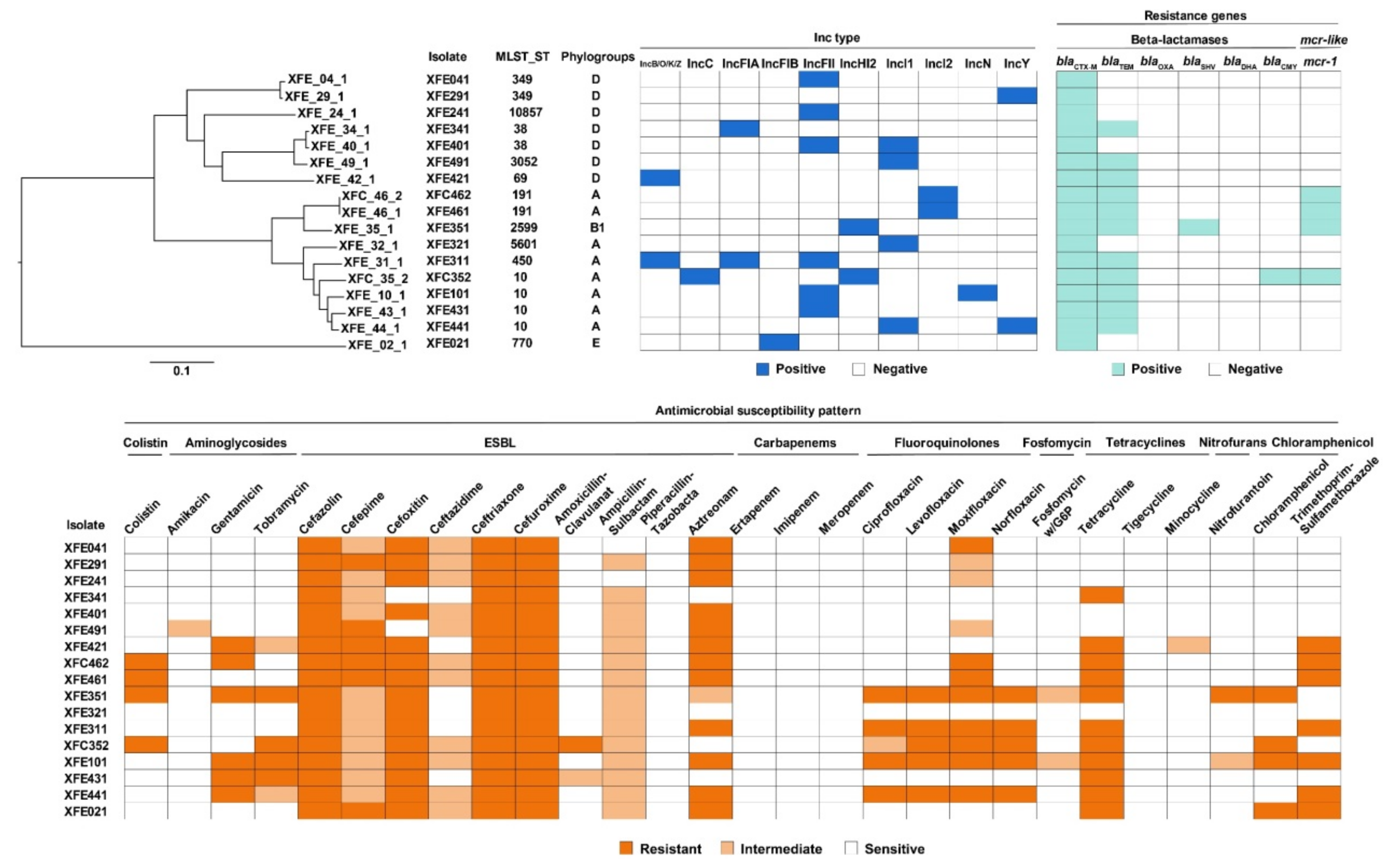
3.2. Most ESBL-Producing and mcr-1-Carrying E. coli Isolates Were MDR and Carried ARGs
3.3. All ESBL-Producing and mcr-1-Carrying E. coli Isolates Carried Plasmids, and all mcr-1 Plasmids Were Transferable
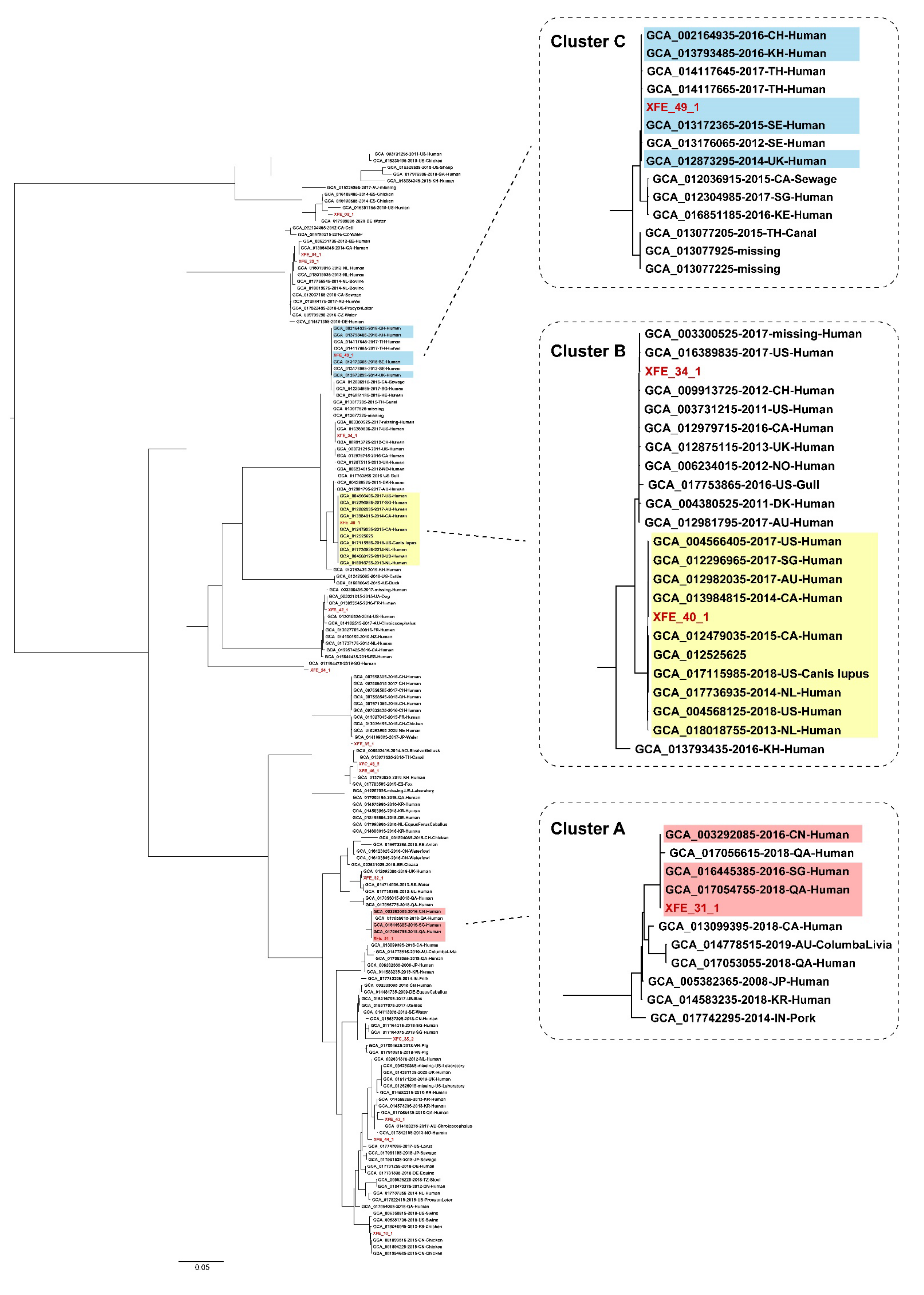
3.4. ESBL-Producing and mcr-1-Carrying E. coli Isolates from Outpatients in Lhasa Had Clonality with Strains from Other Regions and Countries
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Poeta, P.; Costa, D.; Sáenz, Y.; Klibi, N.; Ruiz-Larrea, F.; Rodrigues, J.; Torres, C. Characterization of antibiotic resistance genes and virulence factors in faecal enterococci of wild animals in Portugal. J. Vet. Med. Ser. B Infect. Dis. Vet. Public Health 2005, 52, 396–402. [Google Scholar] [CrossRef] [PubMed]
- Skandalis, N.; Maeusli, M.; Papafotis, D.; Miller, S.; Lee, B.; Theologidis, I.; Luna, B. Environmental Spread of Antibiotic Resistance. Antibiotics 2021, 10, 640. [Google Scholar] [CrossRef]
- Fish, D.N.; Ohlingerm, J. Antimicrobial resistance: Factors and outcomes. Crit. Care Clin. 2006, 22, 291–311. [Google Scholar] [CrossRef]
- Ahmad, I.; Malak, H.A.; Abulreesh, H.H. Environmental antimicrobial resistance and its drivers: A potential threat to public health. J. Glob. Antimicrob. Resist. 2021, 27, 101–111. [Google Scholar]
- Mingji, C.; Onakpoya, I.J.; Perera, R.; Ward, A.M.; Heneghan, C.J. Relationship between altitude and the prevalence of hypertension in Tibet: A systematic review. Heart 2015, 101, 1054–1060. [Google Scholar] [CrossRef] [Green Version]
- Li, K.; Peng, W.; Zhou, Y.; Ren, Y.; Zhao, J.; Fu, X.; Nie, Y. Host Genetic and Environmental Factors Shape the Composition and Function of Gut Microbiota in Populations Living at High Altitude. Biomed Res. Int. 2020, 2020, 1482109. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Yuan, K.; Chen, X.; Yang, Y.; Zhang, T.; Wang, Y.; Luan, T.; Zou, S.; Li, X. Metagenomic Analysis Revealing Antibiotic Resistance Genes (ARGs) and Their Genetic Compartments in the Tibetan Environment. Environ. Sci. Technol. 2016, 50, 6670–6679. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Su, X.; Su, J.; Zhu, Y.; Ding, K. Profiling the antibiotic resistome in soils between pristine and human-affected sites on the Tibetan Plateau. J. Environ. Sci. 2022, 111, 442–451. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Zou, W.; Zeng, J.; Xie, S.; An, T.; Luo, X.; Chen, D.; Feng, L.; Cheng, G.; Cai, R.; et al. Prevalence of antimicrobial resistance and integron gene cassettes in Escherichia coli isolated from yaks (Poephagus grunniens) in Aba Tibetan Autonomous Prefecture, China. Microb. Pathog. 2017, 111, 274–279. [Google Scholar] [CrossRef]
- Paterson, D.L.; Bonomo, R.A. Extended-spectrum beta-lactamases: A clinical update. Clin. Microbiol. Rev. 2005, 18, 657–686. [Google Scholar] [CrossRef] [Green Version]
- Ahmad, T.A.; El-Sayed, L.H.; Haroun, M.; Hussein, A.A.; El Sayed, H. Development of immunization trials against Klebsiella pneumoniae. Vaccine 2012, 30, 2411–2420. [Google Scholar] [CrossRef]
- Li, D.X.; Sick-Samuels, A.C.; Suwantarat, N.; Same, R.G.; Simner, P.J.; Tamma, P.D. Risk Factors for Extended-Spectrum Beta-Lactamase-Producing Enterobacteriaceae Carriage Upon Pediatric Intensive Care Unit Admission. Infect. Control. Hosp. Epidemiol. 2018, 39, 116–118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seo, Y.B.; Lee, J.; Kim, Y.K.; Lee, S.S.; Lee, J.; Kim, H.Y.; Uh, Y.; Kim, H.S.; Song, W. Randomized controlled trial of piperacillintazobactam, cefepime and ertapenem for the treatment of urinary tract infection caused by extended-spectrum betalactamase-producing Escherichia coli. BMC Infect. Dis. 2017, 17, 404. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caniaux, I.; Van Belkum, A.; Zambardi, G.; Poirel, L.; Gros, M.F. MCR: Modern colistin resistance. Eur. J. Clin. Microbiol. Infect. Dis. 2017, 36, 415–420. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.Y.; Wang, Y.; Walsh, T.R.; Yi, L.X.; Zhang, R.; Spencer, J.; Doi, Y.; Tian, G.; Dong, B.; Huang, X.; et al. Emergence of plasmid-mediated colistin resistance mechanism mcr-1 in animals and human beings in China: A microbiological and molecular biological study. Lancet Infect. Dis. 2016, 16, 161–168. [Google Scholar] [CrossRef]
- Li, Z.; Guan, H.; Wang, W.; Gao, H.; Feng, W.; Li, J.; Diao, B.; Zhao, H.; Kan, B.; Zhang, J. Development of a Rapid and Fully Automated Multiplex Real-Time PCR Assay for Identification and Differentiation of Vibrio cholerae and Vibrio parahaemolyticus on the BD MAX Platform. Front. Cell. Infect. Microbiol. 2021, 11, 72. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Zeng, M.; Xu, J.; Zhou, H.; Gu, B.; Li, Z.; Jin, H.; Wang, X.; Zhang, W.; Hu, Y.; et al. Epidemiologic and genomic insights on mcr-1-harbouring Salmonella from diarrhoeal outpatients in Shanghai, China, 2006–2016. EBiomedicine 2019, 42, 133–144. [Google Scholar] [CrossRef] [Green Version]
- Guo, S. The Study of Antimicrobial Resistance and Resistance Genes among Escherichia coli and Salmonella Isolated from Swine in Different Areas of Shanghai. Master’s Thesis, Nanjing Agricultural University, Nanjing, China, 2011. (In Chinese). [Google Scholar]
- Chen, L.; Zhang, J.; Wang, J.; Butaye, P.; Kelly, P.; Li, M.; Yang, F.; Gong, J.; Yassin, A.K.; Guo, W.; et al. Newly identified colistin resistance genes, mcr-4 and mcr-5, from upper and lower alimentary tract of pigs and poultry in China. PLoS ONE 2018, 13, e0193957. [Google Scholar] [CrossRef] [Green Version]
- Rebelo, A.R.; Bortolaia, V.; Kjeldgaard, J.S.; Pedersen, S.K.; Leekitcharoenphon, P.; Hansen, I.M.; Guerra, B.; Malorny, B.; Borowiak, M.; Hammerl, J.A.; et al. Multiplex PCR for detection of plasmid-mediated colistin resistance determinants, mcr-1, mcr-2, mcr-3, mcr-4 and mcr-5 for surveillance purposes. Eurosurveillance 2018, 23, 29–39. [Google Scholar] [CrossRef]
- Xu, Y.; Zhong, L.L.; Srinivas, S.; Sun, J.; Huang, M.; Paterson, D.L.; Lei, S.; Lin, J.; Li, X.; Tang, Z.; et al. Spread of MCR-3 Colistin Resistance in China: An Epidemiological, Genomic and Mechanistic Study. EBiomedicine 2018, 34, 139–157. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Wang, Y.; Zhou, Y.; Li, J.; Yin, W.; Wang, S.; Zhang, S.; Shen, J.; Shen, Z.; Wang, Y. Emergence of a novel mobile colistin resistance gene, mcr-8, in NDM-producing Klebsiella pneumoniae. Emerg. Microbes Infect. 2018, 7, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vila, J.; Ruiz, J.; Goni, P.; De Anta, M.T. Detection of mutations in parC in quinolone-resistant clinical isolates of Escherichia coli. Antimicrob. Agents Chemother. 1996, 40, 491–493. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Furlan, J.P.; Stehling, E.G. Detection of beta-lactamase encoding genes in feces, soil and water from a Brazilian pig farm. Environ. Monit. Assess. 2018, 190, 76. [Google Scholar] [CrossRef] [PubMed]
- Gopalakrishnan, S.; Kamalanathan, A.; Rajan, S.; Bhagat, V.M.; Ali, M.S. Emergence of armA and rmtB genes among VIM, NDM, and IMP metallo-β-lactamase-producing multidrug-resistant Gram-negative pathogens. Acta Microbiol. Immunol. Hung. 2018, 65, 107–118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, X.; Zeng, M.; Zhang, N.; Wang, M.; Gu, B.; Li, J.; Jin, H.; Xiao, W.; Li, Z.; Zhao, H.; et al. Prevalence of 16S rRNA Methylation Enzyme Gene armA in Salmonella From Outpatients and Food. Front. Microbiol. 2021, 12, 969. [Google Scholar] [CrossRef] [PubMed]
- Von Baum, H.; Marre, R. Antimicrobial resistance of Escherichia coli and therapeutic implications. Int. J. Med. Microbiol. 2005, 295, 503–511. [Google Scholar] [CrossRef]
- Sawant, A.A.; Hegde, N.V.; Straley, B.A.; Donaldson, S.C.; Love, B.C.; Knabel, S.J.; Jayarao, B.M. Antimicrobial-resistant enteric bacteria from dairy cattle. Appl. Environ. Microbiol. 2007, 73, 156–163. [Google Scholar] [CrossRef] [Green Version]
- Rasmussen, M.M.; Opintan, J.A.; Frimodt-Møller, N.; Styrishave, B. Beta-Lactamase Producing Escherichia coli Isolates in Imported and Locally Produced Chicken Meat from Ghana. PLoS ONE 2015, 10, e0139706. [Google Scholar] [CrossRef] [Green Version]
- Schaumburg, F.; Sertic, S.M.; Correa-Martinez, C.; Mellmann, A.; Köck, R.; Becker, K. Acquisition and colonization dynamics of antimicrobial-resistant bacteria during international travel: A prospective cohort study. Clin. Microbiol. Infect. 2019, 25, 1287.e1–1287.e7. [Google Scholar] [CrossRef]
- Rehman, M.U.; Zhang, H.; Iqbal, M.K.; Mehmood, K.; Huang, S.; Nabi, F.; Luo, H.; Lan, Y.; Li, J. Antibiotic resistance, serogroups, virulence genes, and phylogenetic groups of Escherichia coli isolated from yaks with diarrhea in Qinghai Plateau, China. Gut Pathog. 2017, 9, 24. [Google Scholar] [CrossRef]
- Zhao, W.-H.; Hu, Z.-Q. Epidemiology and genetics of CTX-M extended-spectrum β-lactamases in Gram-negative bacteria. Crit. Rev. Microbiol. 2013, 39, 79–101. [Google Scholar] [CrossRef] [PubMed]
- Calbo, E.; Garau, J. The changing epidemiology of hospital outbreaks due to ESBL-producing Klebsiella pneumoniae: The CTX-M-15 type consolidation. Future Microbiol. 2015, 10, 1063–1075. [Google Scholar] [CrossRef] [PubMed]
- Dolejska, M.; Frolkova, P.; Florek, M.; Jamborova, I.; Purgertova, M.; Kutilova, I.; Cizek, A.; Guenther, S.; Literak, I. CTX-M-15-producing Escherichia coli clone B2-O25b-ST131 and Klebsiella spp. isolates in municipal wastewater treatment plant effluents. J. Antimicrob. Chemother. 2011, 66, 2784–2790. [Google Scholar] [CrossRef] [PubMed]
- Hiroi, M.; Yamazaki, F.; Harada, T.; Takahashi, N.; Iida, N.; Noda, Y.; Yagi, M.; Nishio, T.; Kanda, T.; Kawamori, F. Prevalence of Extended-Spectrum beta-Lactamase-Producing Escherichia coli and Klebsiella pneumoniae in Food-Producing Animals. J. Vet. Med. Sci. 2012, 74, 189–195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.; Zheng, B.; Zhao, L.; Wei, Z.; Ji, J.; Li, L.; Xiao, Y. Nationwide high prevalence of CTX-M and an increase of CTX-M-55 in Escherichia coli isolated from patients with community-onset infections in Chinese county hospitals. BMC Infect. Dis. 2014, 14, 659. [Google Scholar] [CrossRef]
- Prieto, A.; Bernabeu, M.; Sánchez-Herrero, J.F.; Pérez-Bosque, A.; Miró, L.; Bäuerl, C.; Collado, C.; Hüttener, M.; Juárez, A. Modulation of AggR levels reveals features of virulence regulation in enteroaggregative E. coli. Commun. Biol. 2021, 4, 1295. [Google Scholar] [CrossRef]
- Imuta, N.; Ooka, T.; Seto, K.; Kawahara, R.; Koriyama, T.; Kojyo, T.; Iguchi, A.; Tokuda, K.; Kawamura, H.; Yoshiie, K.; et al. Phylogenetic Analysis of Enteroaggregative Escherichia coli (EAEC) Isolates from Japan Reveals Emergence of CTX-M-14-Producing EAEC O25:H4 Clones Related to Sequence Type 131. J. Clin. Microbiol. 2016, 54, 2128–2134. [Google Scholar] [CrossRef] [Green Version]
- Zhong, L.L.; Phan, H.T.; Shen, C.; Vihta, K.D.; Sheppard, A.E.; Huang, X.; Zeng, K.J.; Li, H.Y.; Zhang, X.F.; Patil, S.; et al. High Rates of Human Fecal Carriage of mcr-1-Positive Multidrug-Resistant Enterobacteriaceae Emerge in China in Association with Successful Plasmid Families. Clin. Infect. Dis. 2018, 66, 676–685. [Google Scholar] [CrossRef]
- Shen, C.; Feng, S.; Chen, H.; Dai, M.; Paterson, D.L.; Zheng, X.; Wu, X.; Zhong, L.L.; Liu, Y.; Xia, Y.; et al. Transmission of mcr-1-Producing Multidrug-resistant Enterobacteriaceae in Public Transportation in Guangzhou, China. Clin. Infect. Dis. 2018, 67, S217–S224. [Google Scholar] [CrossRef]
- Ma, K.; Feng, Y.; Zong, Z. Fitness cost of a mcr-1-carrying IncHI2 plasmid. PLoS ONE 2018, 13, e0209706. [Google Scholar] [CrossRef]
- Yin, W.; Li, H.; Shen, Y.; Liu, Z.; Wang, S.; Shen, Z.; Zhang, R.; Walsh, T.R.; Shen, J.; Wang, Y. Novel Plasmid-Mediated Colistin Resistance Gene mcr-3 in Escherichia coli. mBio 2017, 8, e00543-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Target Gene | Primers | Sequences of Primers (5′ to 3′) |
|---|---|---|
| mcr-1 [17] | mcr-1-F | TCGGCTTTGTGCTGACGAT |
| mcr-1-R | AAATCAACACAGGCTTTAGCACATA | |
| mcr-1-P | (FAM)CTGTCGTGCTCTTTG(MGB) | |
| blaTEM [18] | blaTEM-F | GCATCTTACGGATGGCATGA |
| blaTEM-R | CCTCCGATCGTTGTCAGAAGT | |
| blaTEM-P | ATTATGCAGTGCTGCCATA ACCATGA | |
| mcr-2 [19] | mcr-2-F | AGCCGAGTCTAAGGACTTGATGAATTTG |
| mcr-2-R | GCGGTATCGACATCATAGTCATCTTG | |
| mcr-3 [19] | mcr-3-F | CCAATCAAAATGAGGCGTTAGCATAT |
| mcr-3-R | TGAGCAATTTCACTATCGAGGTCTTG | |
| mcr-4 [20] | mcr-4-F | TCACTTTCATCACTGCGTTG |
| mcr-4-R | TTGGTCCATGACTACCAATG | |
| mcr-5 [21] | mcr-5-F | ACTCGACTGCCACCAGATCATCG |
| mcr-5-R | CGCTGGAGTGTCAAGCCACTACTG | |
| mcr-6 [22] | mcr-6-F | GTCCGGTCAATCCCTATCTGT |
| mcr-6-R | ATCACGGGATTGACATAGCTAC | |
| mcr-7 [22] | mcr-7-F | TGCTCAAGCCCTTCTTTTCGT |
| mcr-7-R | TTCATCTGCGCCACCTCGT | |
| mcr-8 [22] | mcr-8-F | AACCGCCAGAGCACAGAATT |
| mcr-8-R | TTCCCCCAGCGATTCTCCAT | |
| blaCTX-M [23] | blaCTX-M-F | TTT GCG ATG TGC AGT ACC AGT AA |
| blaCTX-M-R | CGA TAT CGT TGG TGG TGC CAT A | |
| blaOXA [24] | blaOXA-F | GGC ACC AGA TTC AAC TTT CAA G |
| blaOXA-R | GAC CCC AAG TTT CCT GTA AGT G | |
| blaSHV [25] | blaSHV-F | TTA TCT CCC TGT TAG CCA CC |
| blaSHV-R | GAT TTG CTG ATT TCG CTC GG | |
| blaCMY [26] | blaCMY-F | GAC AGC CTC TTT CTC CAC A |
| blaCMY-R | TGG AAC GAA GGC TAC GTA | |
| blaDHA [26] | blaDHA-F | CTG ATG AAA AAA TCG TTA TC |
| blaDHA-R | ATT CCA GTG CAC TCC AAA ATA |
| Source | Species | Genes Detected by PCR | |||
|---|---|---|---|---|---|
| Plate Type | No. of Strains | Species of Strains | No. of Strains | Resistant Genes or Genetic Elements Studied | No. of Strains |
| ESBLs | 19 | Escherichia coli | 15 | blaCTX-M | 6 |
| blaCTX-M + blaTEM | 7 | ||||
| blaCTX-M + blaTEM + blaSHV | 1 | ||||
| Klebsiella pneumoniae | 2 | blaCTX-M + blaTEM | 1 | ||
| blaCTX-M + blaTEM + blaSHV | 1 | ||||
| Klebsiella variicola | 1 | blaCTX-M | 1 | ||
| Raoultella ornithinolytica | 1 | blaCTX-M + blaCYM | 1 | ||
| Colistin | 29 | Enterobacter cloacae | 1 | ||
| Escherichia coli | 20 | mcr-1 | 2 | ||
| Klebsiella oxytoca | 1 | ||||
| Klebsiella pneumoniae | 5 | ||||
| Morganella morganii | 1 | ||||
| Salmonella spp. | 1 | ||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Z.; Li, J.; Liu, J.; Peng, Y.; Li, Z.; Wang, M.; Zhang, G.; Qu, G.; Zhang, J.; Fu, X.; et al. High Carriage of Extended-Spectrum, Beta Lactamase-Producing, and Colistin-Resistant Enterobacteriaceae in Tibetan Outpatients with Diarrhea. Antibiotics 2022, 11, 508. https://doi.org/10.3390/antibiotics11040508
Li Z, Li J, Liu J, Peng Y, Li Z, Wang M, Zhang G, Qu G, Zhang J, Fu X, et al. High Carriage of Extended-Spectrum, Beta Lactamase-Producing, and Colistin-Resistant Enterobacteriaceae in Tibetan Outpatients with Diarrhea. Antibiotics. 2022; 11(4):508. https://doi.org/10.3390/antibiotics11040508
Chicago/Turabian StyleLi, Zhe, Jiaqi Li, Jiaqi Liu, Yao Peng, Zhenpeng Li, Mengyu Wang, Ge Zhang, Geruo Qu, Jingyun Zhang, Xiuping Fu, and et al. 2022. "High Carriage of Extended-Spectrum, Beta Lactamase-Producing, and Colistin-Resistant Enterobacteriaceae in Tibetan Outpatients with Diarrhea" Antibiotics 11, no. 4: 508. https://doi.org/10.3390/antibiotics11040508





