Evaluating the In Vivo Virulence of Environmental Pseudomonas aeruginosa Using Microinjection Model of Zebrafish (Danio rerio)
Abstract
:1. Introduction
2. Results
2.1. Method Development
2.2. Optimization of the Combined Zebrafish Microinjection Virulence Model
2.2.1. Determination of the Optimal Incubation Time
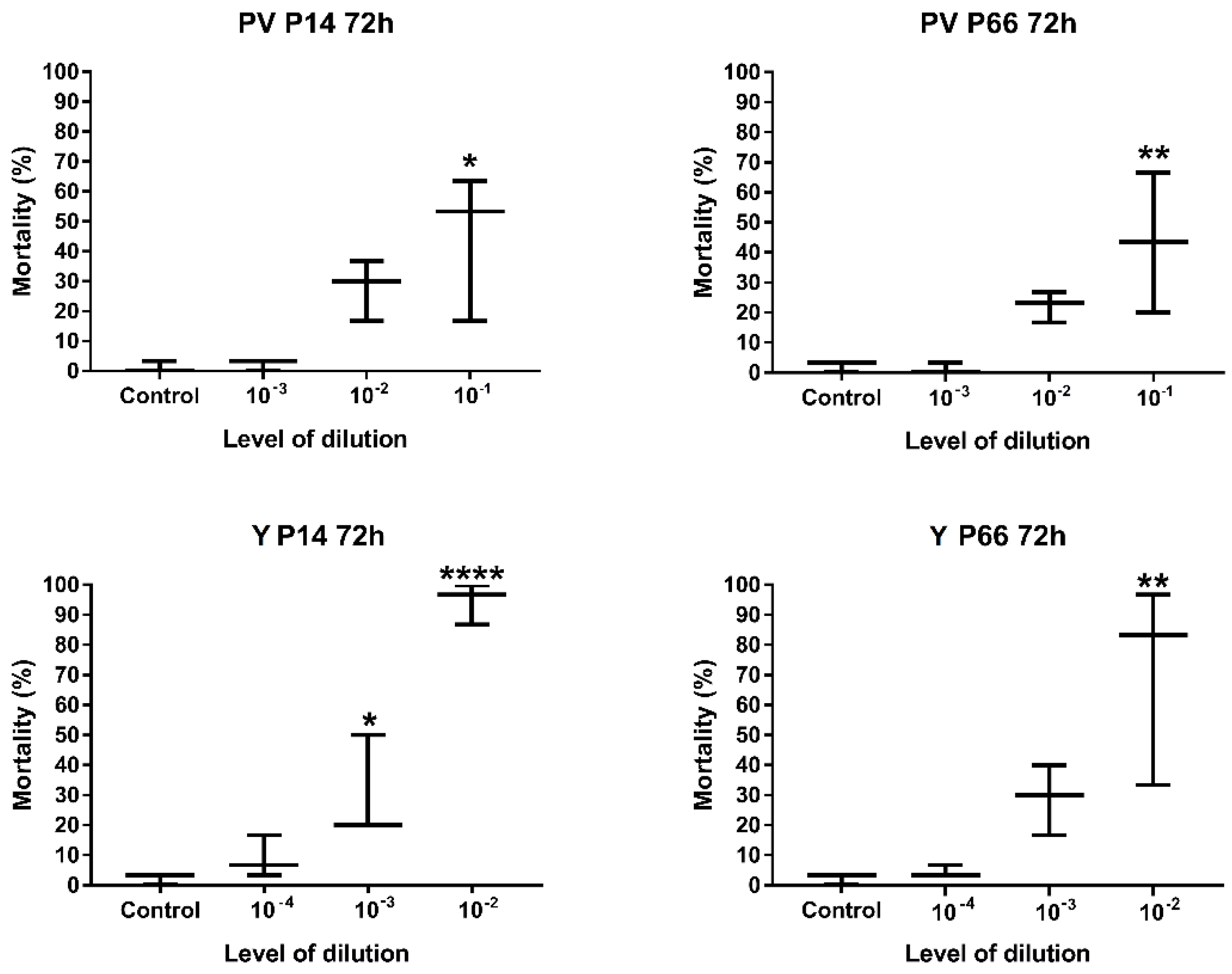
2.2.2. Determination of the Optimal Level of Dilution of P. aeruginosa
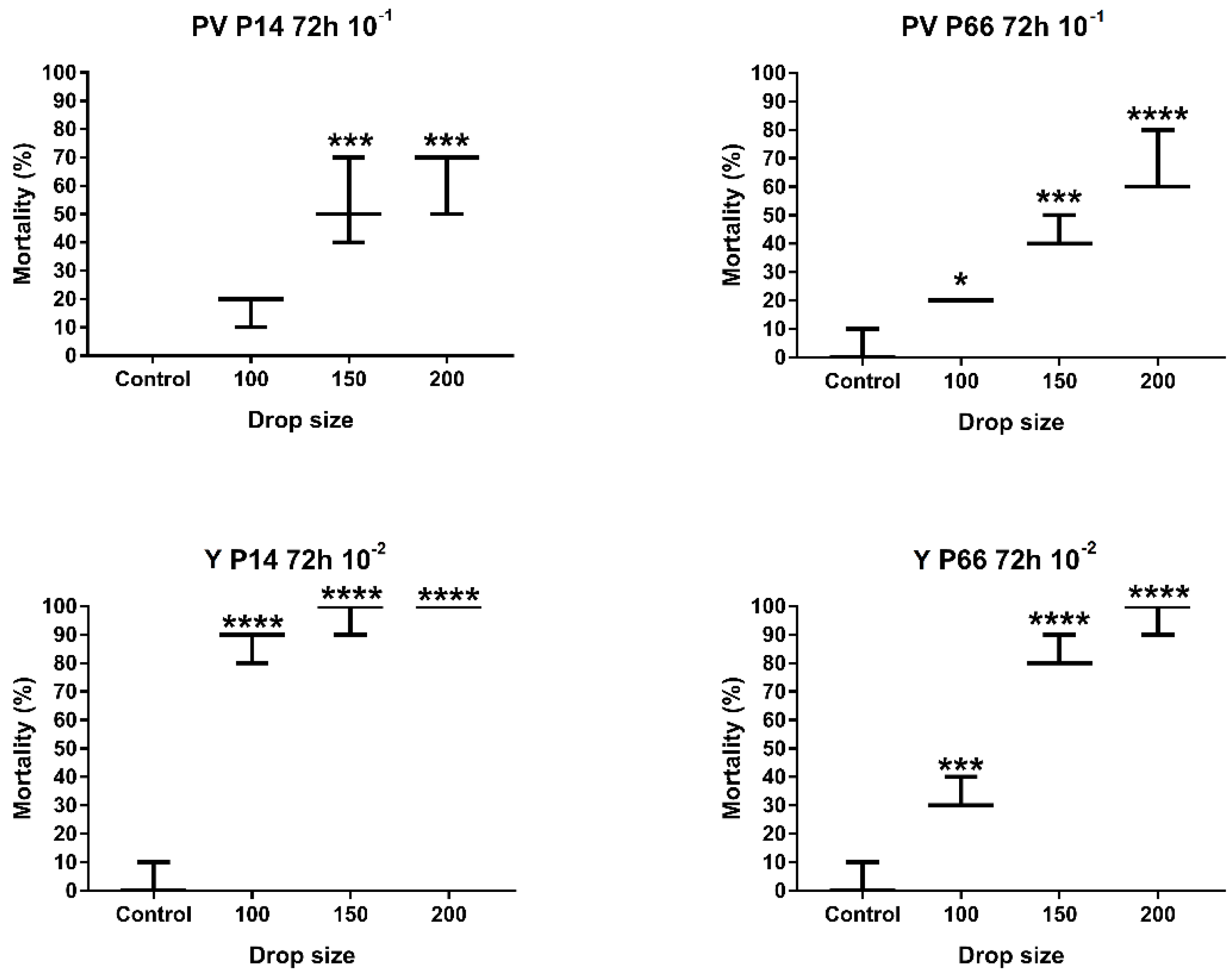
2.2.3. Determination of the Optimal Drop Size
2.2.4. Development of the Recommended Infection Model
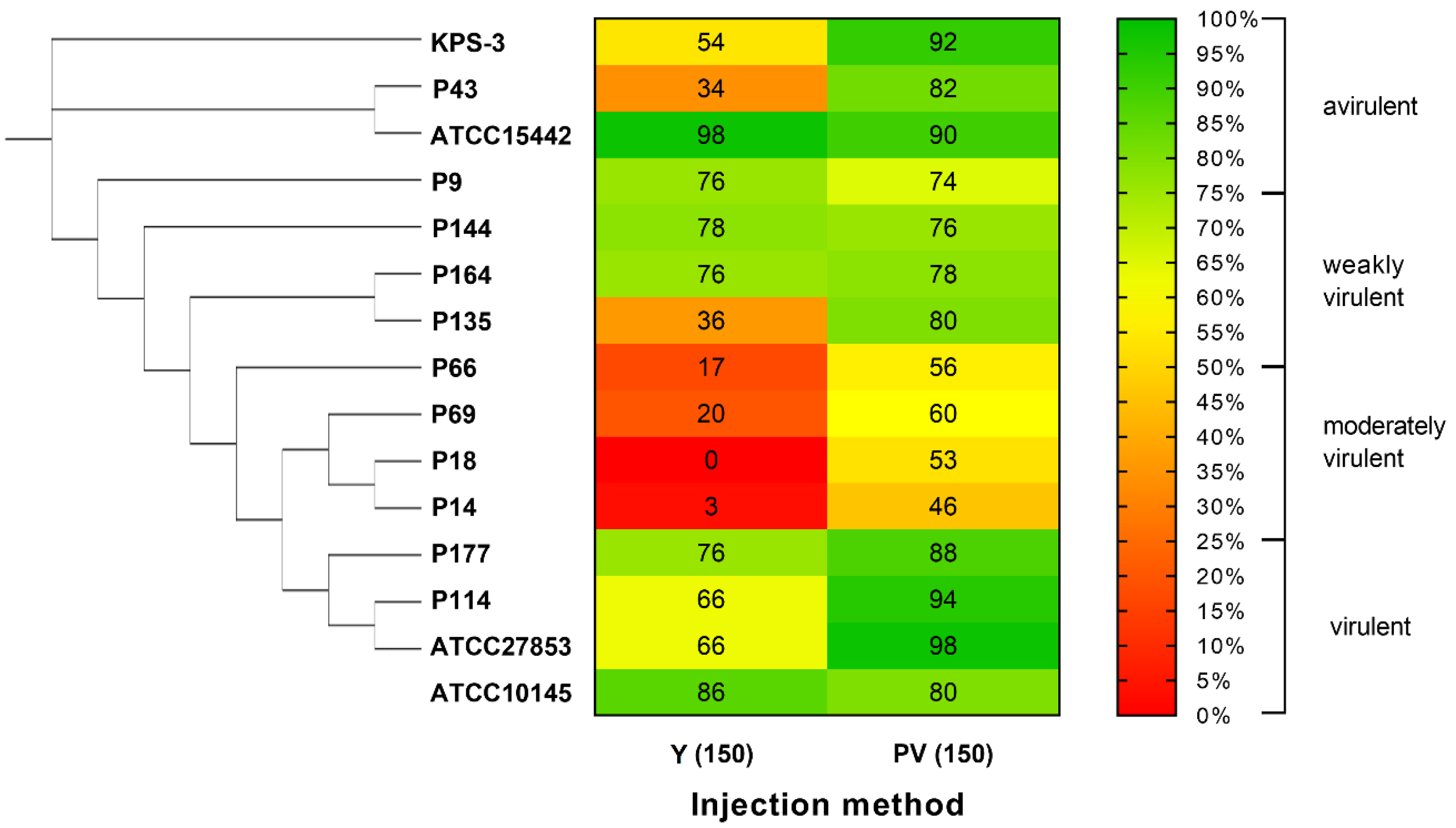
2.3. Validation of the Developed Microinjection Method with a Set of P. aeruginosa Isolates
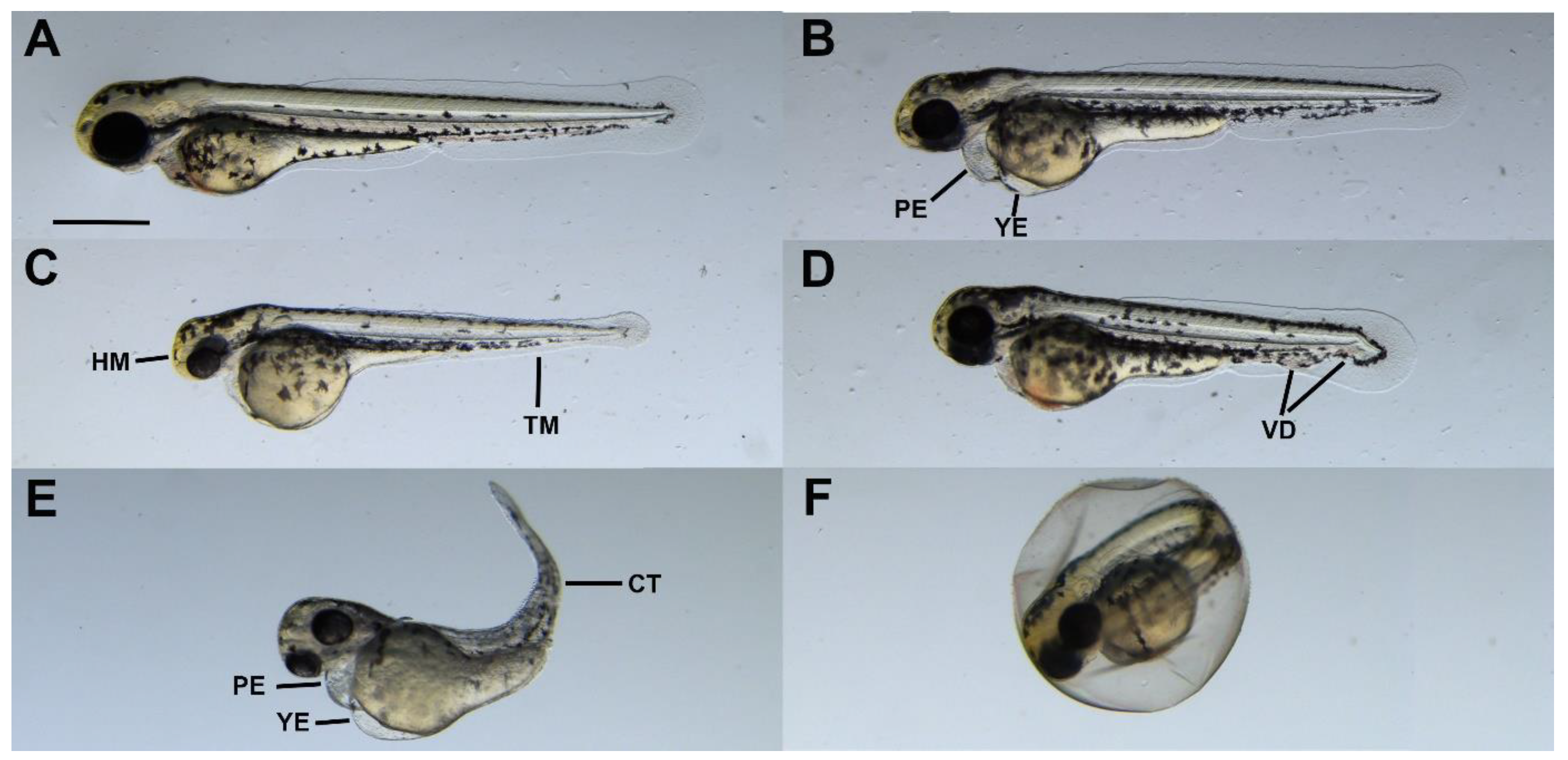
2.4. Sublethal Symptoms Detected at the End of Incubation
2.5. Methodological Summary of the Newly Developed, Combined Virulence Model
3. Discussion
4. Materials and Methods
4.1. Ethics Statement
4.2. Bacterial Strains
| Biofilm-Forming Ability (48 h) [35] | Origin | G. mellonella Virulence (Survial %, 48 h) [35] | Multilocus Sequence Type (ST) [72,73] | Virulence Factors [35] | Biofilm-Forming Ability [33] | Antibiotic Resistance Phenotype [74] | Serotype [35] | Swimming [35] | Swarming [35] | Twitching [35] | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| exoS | exoU | lasB | algD | aprA | plcH | Hemolysis | ||||||||||
| ATCC 10145 | Type strain, unknown source | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. |
| ATCC 15442 | Water bottle in animal room | n.d. | 252 | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. |
| ATCC 27853 | Clinical | 15% (virulent) | 155 | + | – | + | + | + | + | + | ++ | Sensitive (10/0) | n.d. | n.d. | n.d. | n.d. |
| KPS-3 | Clinical | 35% (moderately virulent) | 253 | − | + | + | + | + | + | + | +++ | Sensitive (10/0) | n.d. | n.d. | n.d. | n.d. |
| P9 | Hydrocarbon-contaminated groundwater | 75% (avirulent) | 377 | − | + | + | + | + | + | − | ++ | Resistant (10/1) | ONT | NM | NM | M |
| P14 | Hydrocarbon-contaminated soil | 95% (avirulent) | 2586 * | − | + | + | + | + | + | +/− | +++ | Resistant (10/5) | O3 | HM | NM | M |
| P18 | Hydrocarbon-contaminated soil | 90% (avirulent) | 3243 * | − | + | + | + | + | + | +/− | + | Sensitive (10/0) | O3 | M | M | M |
| P43 | Hydrocarbon-contaminated groundwater | 90% (avirulent) | 253 | + | – | + | + | + | + | + | + | Multidrug-resistant (10/6) | ONT | M | M | M |
| P66 | Hydrocarbon-contaminated groundwater | 5% (virulent) | 455 | + | – | + | + | + | + | +++ | +++ | Sensitive (10/0) | O11 | HM | M | HM |
| P69 | Hydrocarbon-contaminated soil | 50% (weakly virulent) | 439 | − | + | + | + | + | + | ++ | − | Resistant (10/5) | O6 | M | M | M |
| P114 | Compost | 15% (virulent) | 3255 * | + | – | + | + | + | + | +++ | + | Resistant (10/4) | O1 | HM | HM | HM |
| P135 | Hydrocarbon-contaminated soil | 70% (weakly virulent) | 3257 * | − | – | + | + | + | + | +++ | + | Intermediate (10/1) | O3 | HM | M | HM |
| P144 | Hydrocarbon-contaminated soil | 0% (virulent) | 1411 | − | + | + | − | − | − | + | − | Sensitive (10/0) | O1 | HM | M | HM |
| P164 | Sewage | 0% (virulent) | 3260 * | + | − | + | + | + | − | ++ | − | Sensitive (10/0) | O11 | M | NM | M |
| P177 | Hydrocarbon-contaminated groundwater | 30% (moderately virulent) | 3262 * | + | − | + | + | + | + | ++ | + | Sensitive (10/0) | O6 | HM | M | HM |
4.3. Preparation of Bacterial Suspensions
4.4. Zebrafish Maintenance and Egg Collection
4.5. Microinjection
4.6. Endpoints of the Virulence Test/Examination of Injected Embryos
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hardalo, C.; Edberg, S.C. Pseudomonas Aeruginosa: Assessment of Risk from Drinking Water. Crit. Rev. Microbiol. 2008, 23, 47–75. [Google Scholar] [CrossRef]
- Aliaga, L.; Mediavilla, J.D.; Cobo, F. A Clinical Index Predicting Mortality with Pseudomonas Aeruginosa Bacteraemia. J. Med. Microbiol. 2002, 51, 615–619. [Google Scholar] [CrossRef]
- Thaden, J.T.; Park, L.P.; Maskarinec, S.A.; Ruffin, F.; Fowler, V.G.; Van Duin, D. Results from a 13-Year Prospective Cohort Study Show Increased Mortality Associated with Bloodstream Infections Caused by Pseudomonas Aeruginosa Compared to Other Bacteria. Antimicrob. Agents Chemother. 2017, 61, 2671–2687. [Google Scholar] [CrossRef] [PubMed]
- Sood, U.; Hira, P.; Kumar, R.; Bajaj, A.; Rao, D.L.N.; Lal, R.; Shakarad, M. Comparative Genomic Analyses Reveal Core-Genome-Wide Genes under Positive Selection and Major Regulatory Hubs in Outlier Strains of Pseudomonas Aeruginosa. Front. Microbiol. 2019, 10, 53. [Google Scholar] [CrossRef] [PubMed]
- Cabot, G.; Zamorano, L.; Moyà, B.; Juan, C.; Navas, A.; Blázquez, J.; Oliver, A. Evolution of Pseudomonas Aeruginosa Antimicrobial Resistance and Fitness under Low and High Mutation Rates. Antimicrob. Agents Chemother. 2016, 60, 1767–1778. [Google Scholar] [CrossRef] [PubMed]
- Stover, C.K.; Pham, X.Q.; Erwin, A.L.; Mizoguchi, S.D.; Warrener, P.; Hickey, M.J.; Brinkman, F.S.L.; Hufnagle, W.O.; Kowallk, D.J.; Lagrou, M.; et al. Complete Genome Sequence of Pseudomonas Aeruginosa PAO1, an Opportunistic Pathogen. Nature 2000, 406, 959–964. [Google Scholar] [CrossRef]
- Chaerun, S.K.; Tazaki, K.; Asada, R.; Kogure, K. Bioremediation of Coastal Areas 5 Years after the Nakhodka Oil Spill in the Sea of Japan: Isolation and Characterization of Hydrocarbon-Degrading Bacteria. Environ. Int. 2004, 30, 911–922. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Shao, X.; Xie, Y.; Wang, T.; Zhang, Y.; Wang, X.; Deng, X. An Integrated Genomic Regulatory Network of Virulence-Related Transcriptional Factors in Pseudomonas Aeruginosa. Nat. Commun. 2019, 10, 2931. [Google Scholar] [CrossRef]
- Liao, C.; Huang, X.; Wang, Q.; Yao, D.; Lu, W. Virulence Factors of Pseudomonas Aeruginosa and Antivirulence Strategies to Combat Its Drug Resistance. Front. Cell Infect. Microbiol. 2022, 12, 926758. [Google Scholar] [CrossRef]
- Strateva, T.; Mitov, I. Contribution of an Arsenal of Virulence Factors to Pathogenesis of Pseudomonas Aeruginosa Infections. Ann. Microbiol. 2011, 61, 717–732. [Google Scholar] [CrossRef]
- Algammal, A.M.; Mabrok, M.; Sivaramasamy, E.; Youssef, F.M.; Atwa, M.H.; El-kholy, A.W.; Hetta, H.F.; Hozzein, W.N. Emerging MDR-Pseudomonas Aeruginosa in Fish Commonly Harbor OprL and ToxA Virulence Genes and BlaTEM, BlaCTX-M, and TetA Antibiotic-Resistance Genes. Sci. Rep. 2020, 10, 15961. [Google Scholar] [CrossRef] [PubMed]
- Wolfgang, M.C.; Kulasekara, B.R.; Liang, X.; Boyd, D.; Wu, K.; Yang, Q.; Miyada, C.G.; Lory, S. Conservation of Genome Content and Virulence Determinants among Clinical and Environmental Isolates of Pseudomonas Aeruginosa. Proc. Natl. Acad. Sci. USA 2003, 100, 8484–8489. [Google Scholar] [CrossRef] [PubMed]
- Poulsen, B.E.; Yang, R.; Clatworthy, A.E.; White, T.; Osmulski, S.J.; Li, L.; Penaranda, C.; Lander, E.S.; Shoresh, N.; Hung, D.T. Defining the Core Essential Genome of Pseudomonas Aeruginosa. Proc. Natl. Acad. Sci. USA 2019, 116, 10072–10080. [Google Scholar] [CrossRef] [PubMed]
- Freschi, L.; Vincent, A.T.; Jeukens, J.; Emond-Rheault, J.G.; Kukavica-Ibrulj, I.; Dupont, M.J.; Charette, S.J.; Boyle, B.; Levesque, R.C. The Pseudomonas Aeruginosa Pan-Genome Provides New Insights on Its Population Structure, Horizontal Gene Transfer, and Pathogenicity. Genome Biol. Evol. 2019, 11, 109–120. [Google Scholar] [CrossRef]
- Wei, L.; Wu, Q.; Zhang, J.; Guo, W.; Gu, Q.; Wu, H.; Wang, J.; Lei, T.; Xue, L.; Zhang, Y.; et al. Prevalence, Virulence, Antimicrobial Resistance, and Molecular Characterization of Pseudomonas Aeruginosa Isolates From Drinking Water in China. Front. Microbiol. 2020, 11, 544653. [Google Scholar] [CrossRef]
- Castañeda-Montes, F.J.; Avitia, M.; Sepúlveda-Robles, O.; Cruz-Sánchez, V.; Kameyama, L.; Guarneros, G.; Escalante, A.E. Population Structure of Pseudomonas Aeruginosa through a MLST Approach and Antibiotic Resistance Profiling of a Mexican Clinical Collection. Infect. Genet. Evol. 2018, 65, 43–54. [Google Scholar] [CrossRef]
- Ali, N.G.; Ali, T.E.S.; Aboyadak, I.M.; Elbakry, M.A. Controlling Pseudomonas Aeruginosa Infection in Oreochromis Niloticus Spawners by Cefotaxime Sodium. Aquaculture 2021, 544, 737107. [Google Scholar] [CrossRef]
- Jose, D.; Mohandas, A.; Singh, I.S.B. A Non-Pathogenic Environmental Isolate of Pseudomonas Aeruginosa MCCB 123 with Biotechnological Potential. Int.J.Curr.Microbiol.App.Sci. 2018, 7, 3060–3071. [Google Scholar] [CrossRef]
- Radhapriya, P.; Ramachandran, A.; Anandham, R.; Mahalingam, S. Pseudomonas Aeruginosa RRALC3 Enhances the Biomass, Nutrient and Carbon Contents of Pongamia Pinnata Seedlings in Degraded Forest Soil. PLoS ONE 2015, 10, e0139881. [Google Scholar] [CrossRef]
- Berger, C.; Rückert, C.; Blom, J.; Rabaey, K.; Kalinowski, J.; Rosenbaum, M.A. Estimation of Pathogenic Potential of an Environmental Pseudomonas Aeruginosa Isolate Using Comparative Genomics. Sci. Rep. 2021, 11, 1370. [Google Scholar] [CrossRef]
- Ebadi, A.; Khoshkholgh Sima, N.A.; Olamaee, M.; Hashemi, M.; Ghorbani Nasrabadi, R. Effective Bioremediation of a Petroleum-Polluted Saline Soil by a Surfactant-Producing Pseudomonas Aeruginosa Consortium. J. Adv. Res. 2017, 8, 627–633. [Google Scholar] [CrossRef] [PubMed]
- Yasmin, S.; Hafeez, F.Y.; Mirza, M.S.; Rasul, M.; Arshad, H.M.I.; Zubair, M.; Iqbal, M. Biocontrol of Bacterial Leaf Blight of Rice and Profiling of Secondary Metabolites Produced by Rhizospheric Pseudomonas Aeruginosa BRp3. Front. Microbiol. 2017, 8, 1895. [Google Scholar] [CrossRef] [PubMed]
- Llamas, M.A.; van der Sar, A.M. Assessing Pseudomonas Virulence with Nonmammalian Host: Zebrafish. Methods Mol. Biol. 2014, 1149, 709–721. [Google Scholar] [CrossRef]
- Lorenz, A.; Pawar, V.; Häussler, S.; Weiss, S. Insights into Host–Pathogen Interactions from State-of-the-Art Animal Models of Respiratory Pseudomonas Aeruginosa Infections. FEBS Lett. 2016, 590, 3941–3959. [Google Scholar] [CrossRef] [PubMed]
- Pont, S.; Blanc-Potard, A.B. Zebrafish Embryo Infection Model to Investigate Pseudomonas Aeruginosa Interaction With Innate Immunity and Validate New Therapeutics. Front. Cell Infect. Microbiol. 2021, 11, 745851. [Google Scholar] [CrossRef] [PubMed]
- Meijer, A.H.; Spaink, H.P. Host-Pathogen Interactions Made Transparent with the Zebrafish Model. Curr. Drug Targets 2011, 12, 1000–1017. [Google Scholar] [CrossRef] [PubMed]
- Gomes, M.C.; Mostowy, S. The Case for Modeling Human Infection in Zebrafish. Trends Microbiol. 2020, 28, 10–18. [Google Scholar] [CrossRef]
- Benard, E.L.; van der Sar, A.M.; Ellett, F.; Lieschke, G.J.; Spaink, H.P.; Meijer, A.H. Infection of Zebrafish Embryos with Intracellular Bacterial Pathogens. J. Vis. Exp. 2012, 61, 3781. [Google Scholar] [CrossRef]
- Carvalho, R.; de Sonneville, J.; Stockhammer, O.W.; Savage, N.D.L.; Veneman, W.J.; Ottenhoff, T.H.M.; Dirks, R.P.; Meijer, A.H.; Spaink, H.P. A High-Throughput Screen for Tuberculosis Progression. PLoS ONE 2011, 6, e16779. [Google Scholar] [CrossRef]
- Csenki, Z.; Garai, E.; Risa, A.; Cserháti, M.; Bakos, K.; Márton, D.; Bokor, Z.; Kriszt, B.; Urbányi, B. Biological Evaluation of Microbial Toxin Degradation by Microinjected Zebrafish (Danio Rerio) Embryos. Chemosphere 2019, 227, 151–161. [Google Scholar] [CrossRef]
- Nogaret, P.; El Garah, F.; Blanc-Potard, A.B. A Novel Infection Protocol in Zebrafish Embryo to Assess Pseudomonas Aeruginosa Virulence and Validate Efficacy of a Quorum Sensing Inhibitor in Vivo. Pathogens 2021, 10, 401. [Google Scholar] [CrossRef] [PubMed]
- Rowe, H.M.; Withey, J.H.; Neely, M.N. Zebrafish as a Model for Zoonotic Aquatic Pathogens. Dev. Comp. Immunol. 2014, 46, 96. [Google Scholar] [CrossRef] [PubMed]
- Díaz-Pascual, F.; Ortíz-Severín, J.; Varas, M.A.; Allende, M.L.; Chávez, F.P. In Vivo Host-Pathogen Interaction as Revealed by Global Proteomic Profiling of Zebrafish Larvae. Front. Cell Infect. Microbiol. 2017, 7, 334. [Google Scholar] [CrossRef] [PubMed]
- Poplimont, H.; Georgantzoglou, A.; Boulch, M.; Walker, H.A.; Coombs, C.; Papaleonidopoulou, F.; Sarris, M. Neutrophil Swarming in Damaged Tissue Is Orchestrated by Connexins and Cooperative Calcium Alarm Signals. Curr. Biol. 2020, 30, 2761–2776.e7. [Google Scholar] [CrossRef]
- Kaszab, E.; Radó, J.; Kriszt, B.; Pászti, J.; Lesinszki, V.; Szabó, Á.; Tóth, G.; Khaledi, A.; Szoboszlay, S. Groundwater, Soil and Compost, as Possible Sources of Virulent and Antibiotic-Resistant Pseudomonas Aeruginosa. Int. J. Environ. Health Res. 2019, 31, 848–860. [Google Scholar] [CrossRef]
- Lizewski, S.E.; Lundberg, D.S.; Schurr, M.J. The Transcriptional Regulator AlgR Is Essential for Pseudomonas Aeruginosa Pathogenesis. Infect. Immun. 2002, 70, 6083. [Google Scholar] [CrossRef]
- Roser, D.J.; Van Den Akker, B.; Boase, S.; Haas, C.N.; Ashbolt, N.J.; Rice, S.A. Dose–Response Algorithms for Water-Borne Pseudomonas Aeruginosa Folliculitis. Epidemiol. Infect. 2015, 143, 1524. [Google Scholar] [CrossRef]
- Deredjian, A.; Colinon, C.; Hien, E.; Brothier, E.; Youenou, B.; Cournoyer, B.; Dequiedt, S.; Hartmann, A.; Jolivet, C.; Houot, S.; et al. Low Occurrence of Pseudomonas Aeruginosa in Agricultural Soils with and without Organic Amendment. Front. Cell Infect. Microbiol. 2014, 4, 53. [Google Scholar] [CrossRef]
- Kaszab, E.; Szoboszlay, S.; Dobolyi, C.; Háhn, J.; Pék, N.; Kriszt, B. Antibiotic Resistance Profiles and Virulence Markers of Pseudomonas Aeruginosa Strains Isolated from Composts. Bioresour. Technol. 2011, 102, 1543–1548. [Google Scholar] [CrossRef]
- Mena, K.D.; Gerba, C.P. Risk Assessment of Pseudomonas Aeruginosa in Water. Rev. Env. Contam. Toxicol. 2009, 201, 71–115. [Google Scholar]
- Kaszab, E.; Kriszt, B.; Atzél, B.; Szabó, G.; Szabó, I.; Harkai, P.; Szoboszlay, S. The Occurrence of Multidrug-Resistant Pseudomonas Aeruginosa on Hydrocarbon-Contaminated Sites. Microb. Ecol. 2010, 59, 37–45. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.Y.; Li, N.J.; Cheng, F.Y.; Hsueh, J.F.; Huang, C.C.; Lu, F.I.; Fu, T.F.; Yan, S.J.; Lee, Y.H.; Wang, Y.J. The Effect of the Chorion on Size-Dependent Acute Toxicity and Underlying Mechanisms of Amine-Modified Silver Nanoparticles in Zebrafish Embryos. Int. J. Mol. Sci. 2020, 21, 2864. [Google Scholar] [CrossRef]
- Diggle, S.P.; Whiteley, M. Microbe Profile: Pseudomonas Aeruginosa: Opportunistic Pathogen and Lab Rat. Microbiology 2020, 166, 30–33. [Google Scholar] [CrossRef] [PubMed]
- Rosen, J.N.; Sweeney, M.F.; Mably, J.D. Microinjection of Zebrafish Embryos to Analyze Gene Function. J. Vis. Exp. 2009, 9, 1115. [Google Scholar] [CrossRef]
- Walker, M.K.; Hufnagle, L.C.; Clayton, M.K.; Peterson, R.E. An Egg Injection Method for Assessing Early Life Stage Mortality of Polychlorinated Dibenzo-p-Dioxins, Dibenzofurans, and Biphenyls in Rainbow Trout, (Oncorhynchus Mykiss). Aquat. Toxicol. 1992, 22, 15–37. [Google Scholar] [CrossRef]
- Schubert, S.; Keddig, N.; Hanel, R.; Kammann, U. Microinjection into Zebrafish Embryos (Danio Rerio)—A Useful Tool in Aquatic Toxicity Testing? Environ. Sci. Eur. 2014, 26, 32. [Google Scholar] [CrossRef]
- Xie, J.; He, J.B.; Shi, J.W.; Xiao, Q.; Li, L.; Woo, P.C.Y. An Adult Zebrafish Model for Laribacter Hongkongensis Infection: Koch’s Postulates Fulfilled. Emerg. Microbes Infect. 2014, 3, e73. [Google Scholar] [CrossRef]
- Bose, S.; Ghosh, A.K. Diagnosis of Biofilm-Associated Infections in Medical Devices. Biomater. Med. Device-Assoc. Infect. 2015, 71–82. [Google Scholar] [CrossRef]
- Velikova, N.; Kavanagh, K.; Wells, J.M. Evaluation of Galleria Mellonella Larvae for Studying the Virulence of Streptococcus Suis. BMC Microbiol. 2016, 16, 291. [Google Scholar] [CrossRef] [PubMed]
- Clatworthy, A.E.; Lee, J.S.W.; Leibman, M.; Kostun, Z.; Davidson, A.J.; Hung, D.T. Pseudomonas Aeruginosa Infection of Zebrafish Involves Both Host and Pathogen Determinants. Infect. Immun. 2009, 77, 1293–1303. [Google Scholar] [CrossRef]
- Llamas, M.A.; Van Der Sar, A.; Chu, B.C.H.; Sparrius, M.; Vogel, H.J.; Bitter, W. A Novel Extracytoplasmic Function (ECF) Sigma Factor Regulates Virulence in Pseudomonas Aeruginosa. PLoS Pathog. 2009, 5, e1000572. [Google Scholar] [CrossRef]
- Wood, S.J.; Kuzel, T.M.; Shafikhani, S.H. Pseudomonas Aeruginosa: Infections, Animal Modeling, and Therapeutics. Cells 2023, 12, 199. [Google Scholar] [CrossRef]
- Kukavica-Ibrulj, I.; Bragonzi, A.; Paroni, M.; Winstanley, C.; Sanschagrin, F.; O’Toole, G.A.; Levesque, R.C. In Vivo Growth of Pseudomonas Aeruginosa Strains PAO1 and PA14 and the Hypervirulent Strain LESB58 in a Rat Model of Chronic Lung Infection. J. Bacteriol. 2008, 190, 2804–2813. [Google Scholar] [CrossRef]
- Van Heeckeren, A.M.; Schluchter, M.D. Murine Models of Chronic Pseudomonas Aeruginosa Lung Infection. Lab. Anim. 2002, 36, 291–312. [Google Scholar] [CrossRef]
- Kaszab, E.; Szoboszlay, S.; Dura, G.; Radó, J.; Kovács, B.; Kriszt, B. Pathogenic and Phylogenetic Features of 2 Multiresistant Pseudomonas Aeruginosa Strains Originated from Remediated Sites. Int. J. Occup. Med. Environ. Health 2016, 29, 503–516. [Google Scholar] [CrossRef]
- Fauvarque, M.O.; Bergeret, E.; Chabert, J.; Dacheux, D.; Satre, M.; Attree, I. Role and Activation of Type III Secretion System Genes in Pseudomonas Aeruginosa-Induced Drosophila Killing. Microb. Pathog. 2002, 32, 287–295. [Google Scholar] [CrossRef]
- Tan, M.W.; Rahme, L.G.; Sternberg, J.A.; Tompkins, R.G.; Ausubel, F.M. Pseudomonas Aeruginosa Killing of Caenorhabditis Elegans Used to Identify P. Aeruginosa Virulence Factors. Proc. Natl. Acad. Sci. USA 1999, 96, 2408–2413. [Google Scholar] [CrossRef]
- Tan, M.W.; Mahajan-Miklos, S.; Ausubel, F.M. Killing of Caenorhabditis Elegans by Pseudomonas Aeruginosa Used to Model Mammalian Bacterial Pathogenesis. Proc. Natl. Acad. Sci. USA 1999, 96, 715–720. [Google Scholar] [CrossRef]
- Miyata, S.; Casey, M.; Frank, D.W.; Ausubel, F.M.; Drenkard, E. Use of the Galleria Mellonella Caterpillar as a Model Host to Study the Role of the Type III Secretion System in Pseudomonas Aeruginosa Pathogenesis. Infect. Immun. 2003, 71, 2404–2413. [Google Scholar] [CrossRef]
- Kumar, S.S.; Tandberg, J.I.; Penesyan, A.; Elbourne, L.D.H.; Suarez-Bosche, N.; Don, E.; Skadberg, E.; Fenaroli, F.; Cole, N.; Winther-Larsen, H.C.; et al. Dual Transcriptomics of Host-Pathogen Interaction of Cystic Fibrosis Isolate Pseudomonas Aeruginosa PASS1 With Zebrafish. Front. Cell Infect. Microbiol. 2018, 8, 406. [Google Scholar] [CrossRef]
- Rocker, A.J.; Weiss, A.R.E.; Lam, J.S.; Van Raay, T.J.; Khursigara, C.M. Visualizing and Quantifying Pseudomonas Aeruginosa Infection in the Hindbrain Ventricle of Zebrafish Using Confocal Laser Scanning Microscopy. J. Microbiol. Methods 2015, 117, 85–94. [Google Scholar] [CrossRef]
- Kiani, A.K.; Pheby, D.; Henehan, G.; Brown, R.; Sieving, P.; Sykora, P.; Marks, R.; Falsini, B.; Capodicasa, N.; Miertus, S.; et al. Ethical Considerations Regarding Animal Experimentation. J. Prev. Med. Hyg. 2022, 63, E255. [Google Scholar] [CrossRef]
- Directive 2010/63/EU of the European Parliament and of the Council of 22 September 2010 on the Protection of Animals Used for Scientific Purposes (Text with EEA Relevance). Available online: https://eur-lex.europa.eu/LexUriServ/LexUriServ.do?uri=OJ:L:2010:276:0033:0079:en:PDF (accessed on 10 December 2023).
- Commission Implementing Decision (EU) 2020/569 of 16 April 2020 Establishing a Common Format and Information Content for the Submission of the Information to Be Reported by Member States Pursuant to Directive 2010/63/EU of the European Parliament and of the Council on the Protection of Animals Used for Scientific Purposes and Repealing Commission Implementing Decision 2012/707/EU (Notified under Document C(2020) 2179) (Text with EEA Relevance), C/2020/2179. 2020. Available online: https://eur-lex.europa.eu/legal-content/EN/TXT/?uri=CELEX%3A32020D0569 (accessed on 10 December 2023).
- Bondue, T.; Berlingerio, S.P.; van den Heuvel, L.; Levtchenko, E. The Zebrafish Embryo as a Model Organism for Testing MRNA-Based Therapeutics. Int. J. Mol. Sci. 2023, 24, 11224. [Google Scholar] [CrossRef]
- Zhang, X.; Zhao, Y.; Wu, Q.; Lin, J.; Fang, R.; Bi, W.; Dong, G.; Li, J.; Zhang, Y.; Cao, J.; et al. Zebrafish and Galleria Mellonella: Models to Identify the Subsequent Infection and Evaluate the Immunological Differences in Different Klebsiella Pneumoniae Intestinal Colonization Strains. Front. Microbiol. 2019, 10, 470231. [Google Scholar] [CrossRef]
- Liew, S.M.; Rajasekaram, G.; Puthucheary, S.D.A.; Chua, K.H. Antimicrobial Susceptibility and Virulence Genes of Clinical and Environmental Isolates of Pseudomonas Aeruginosa. PeerJ 2019, 7, e6217. [Google Scholar] [CrossRef]
- Woods, D.E.; Lam, J.S.; Paranchych, W.; Speert, D.P.; Campbell, M.; Godfrey, A.J. Correlation of Pseudomonas Aeruginosa Virulence Factors from Clinical and Environmental Isolates with Pathogenicity in the Neutropenic Mouse. Can. J. Microbiol. 2011, 43, 541–551. [Google Scholar] [CrossRef]
- Vives-Flórez, M.; Garnica, D. Comparison of Virulence between Clinical and Environmental Pseudomonas Aeruginosa Isolates. Int. Microbiol. 2006, 9, 247–252. [Google Scholar]
- Wang, Y.; Li, C.; Gao, C.; Ma, C.; Xua, P. Genome Sequence of the Nonpathogenic Pseudomonas Aeruginosa Strain ATCC 15442. Genome Announc. 2014, 2, 421–435. [Google Scholar] [CrossRef]
- Atzél, B.; Szoboszlay, S.; Mikuska, Z.; Kriszt, B. Comparison of Phenotypic and Genotypic Methods for the Detection of Environmental Isolates of Pseudomonas Aeruginosa. Int. J. Hyg. Environ. Health 2008, 211, 143–155. [Google Scholar] [CrossRef]
- PubMLST—Public Databases for Molecular Typing and Microbial Genome Diversity. Available online: https://pubmlst.org/ (accessed on 21 July 2023).
- Radó, J.; Kaszab, E.; Petrovics, T.; Pászti, J.; Kriszt, B.; Szoboszlay, S. Characterization of Environmental Pseudomonas Aeruginosa Using Multilocus Sequence Typing Scheme. J. Med. Microbiol. 2017, 66, 1457–1466. [Google Scholar] [CrossRef]
- EUCAST: Clinical Breakpoints and Dosing of Antibiotics. Available online: https://www.eucast.org/clinical_breakpoints/ (accessed on 9 May 2022).
- Garai, E.; Risa, A.; Varga, E.; Cserháti, M.; Kriszt, B.; Urbányi, B.; Csenki, Z. Evaluation of the Multimycotoxin-Degrading Efficiency of Rhodococcus Erythropolis NI1 Strain with the Three-Step Zebrafish Microinjection Method. Int. J. Mol. Sci. 2021, 22, 724. [Google Scholar] [CrossRef]
- Garai, E.; Risa, A.; Varga, E.; Cserháti, M.; Kriszt, B.; Urbányi, B.; Csenki, Z. Qualifying the T-2 Toxin-Degrading Properties of Seven Microbes with Zebrafish Embryo Microinjection Method. Toxins 2020, 12, 460. [Google Scholar] [CrossRef] [PubMed]
- Csenki, Z.; Bartók, T.; Bock, I.; Horváth, L.; Lemli, B.; Zsidó, B.Z.; Angeli, C.; Hetényi, C.; Szabó, I.; Urbányi, B.; et al. Interaction of Fumonisin B1, N-Palmitoyl-Fumonisin B1, 5-O-Palmitoyl-Fumonisin B1, and Fumonisin B4 Mycotoxins with Human Serum Albumin and Their Toxic Impacts on Zebrafish Embryos. Biomolecules 2023, 13, 755. [Google Scholar] [CrossRef] [PubMed]
- R: The R Project for Statistical Computing. Available online: https://www.r-project.org/ (accessed on 6 September 2022).
- Royston, J.P. An Extension of Shapiro and Wilk’s W Test for Normality to Large Samples. Appl. Stat. 1982, 31, 115. [Google Scholar] [CrossRef]
- Properties of Sufficiency and Statistical Tests. Proc. R. Soc. Lond. A Math. Phys. Sci. 1937, 160, 268–282. [CrossRef]



| Levels of Dilution and Bacterial Density (CFU) of the Final Injection Volume (SD = ±3.33%) | |||||
|---|---|---|---|---|---|
| Exposition | Drop Size | 10−1 | 10−2 | 10−3 | 10−4 |
| Perivitelline space (PV) | 100 µM (0.52 nL) | 2.4 × 101 | 2.4 × 100 | 2.4 × 10−1 | |
| 150 µM (1.77 nL) | 8.4 × 101 | 8.4 × 100 * | 8.4 × 10−1 | ||
| 200 µM (4.17 nL) | 2.0 × 102 | 2.0 × 101 | 2.0 × 100 | ||
| Yolk (Y) | 100 µM (0.52 nL) | 2.4 × 100 | 2.4 × 10−1 | 2.4 × 10−2 | |
| 150 µM (1.77 nL) | 8.4 × 100 * | 8.4 × 10−1 | 8.4 × 10−2 | ||
| 200 µM (4.17 nL) | 2.0 × 101 | 2.0 × 100 | 2.0 × 10−1 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kaszab, E.; Jiang, D.; Szabó, I.; Kriszt, B.; Urbányi, B.; Szoboszlay, S.; Sebők, R.; Bock, I.; Csenki-Bakos, Z. Evaluating the In Vivo Virulence of Environmental Pseudomonas aeruginosa Using Microinjection Model of Zebrafish (Danio rerio). Antibiotics 2023, 12, 1740. https://doi.org/10.3390/antibiotics12121740
Kaszab E, Jiang D, Szabó I, Kriszt B, Urbányi B, Szoboszlay S, Sebők R, Bock I, Csenki-Bakos Z. Evaluating the In Vivo Virulence of Environmental Pseudomonas aeruginosa Using Microinjection Model of Zebrafish (Danio rerio). Antibiotics. 2023; 12(12):1740. https://doi.org/10.3390/antibiotics12121740
Chicago/Turabian StyleKaszab, Edit, Dongze Jiang, István Szabó, Balázs Kriszt, Béla Urbányi, Sándor Szoboszlay, Rózsa Sebők, Illés Bock, and Zsolt Csenki-Bakos. 2023. "Evaluating the In Vivo Virulence of Environmental Pseudomonas aeruginosa Using Microinjection Model of Zebrafish (Danio rerio)" Antibiotics 12, no. 12: 1740. https://doi.org/10.3390/antibiotics12121740






