Evaluation of Bile Salts on the Survival and Modulation of Virulence of Aliarcobacter butzleri
Abstract
:1. Introduction
2. Results
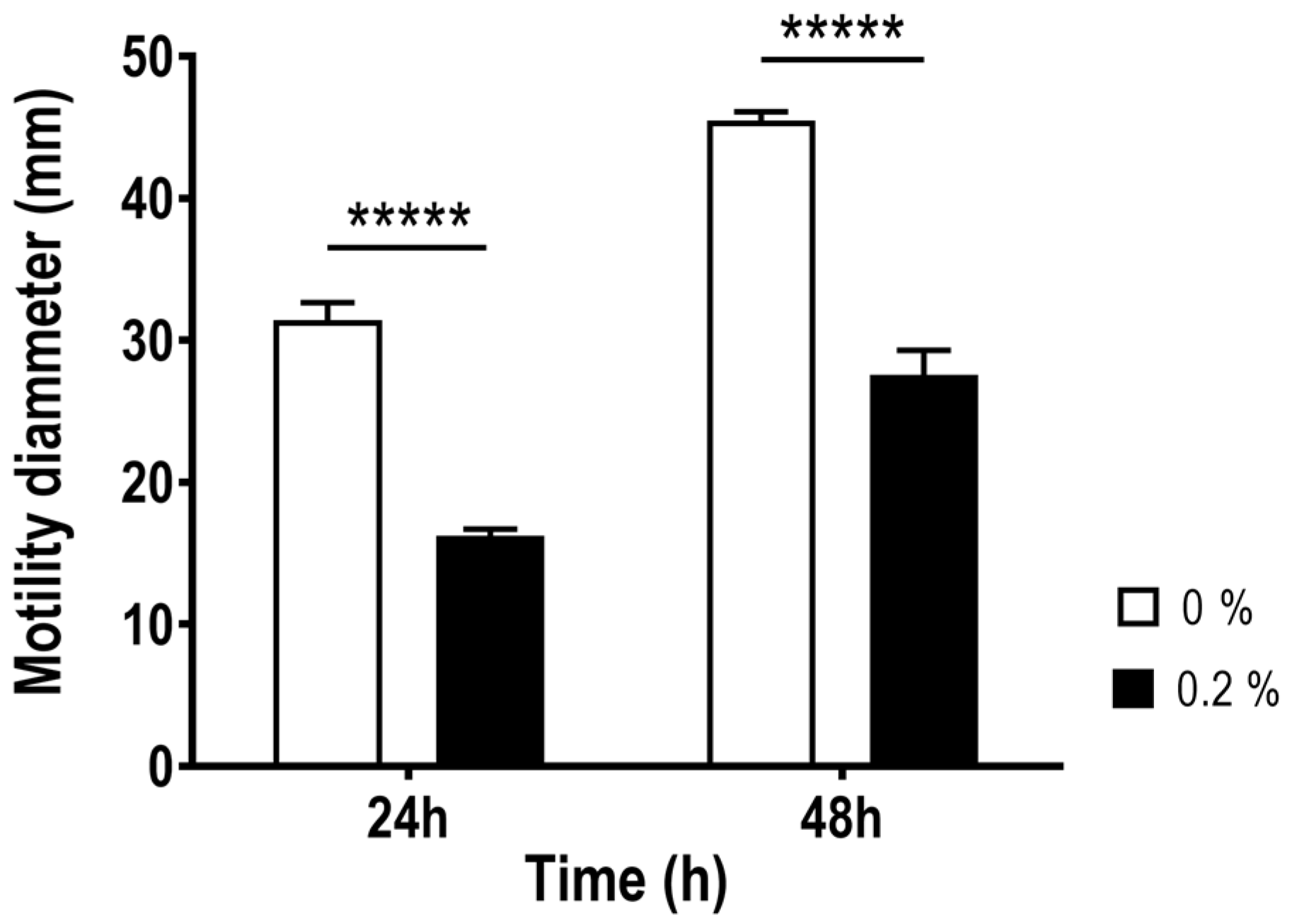
2.1. Effect of Bile Salts on the Growth, Aggregation and Motility of Aliarcobacter butzleri
2.2. Impact of Bile Salts on Biofilm Formation of Aliarcobacter butzleri
2.3. Effect of Bile Salts in the Expression of Five Virulence Genes of Aliarcobacter butzleri
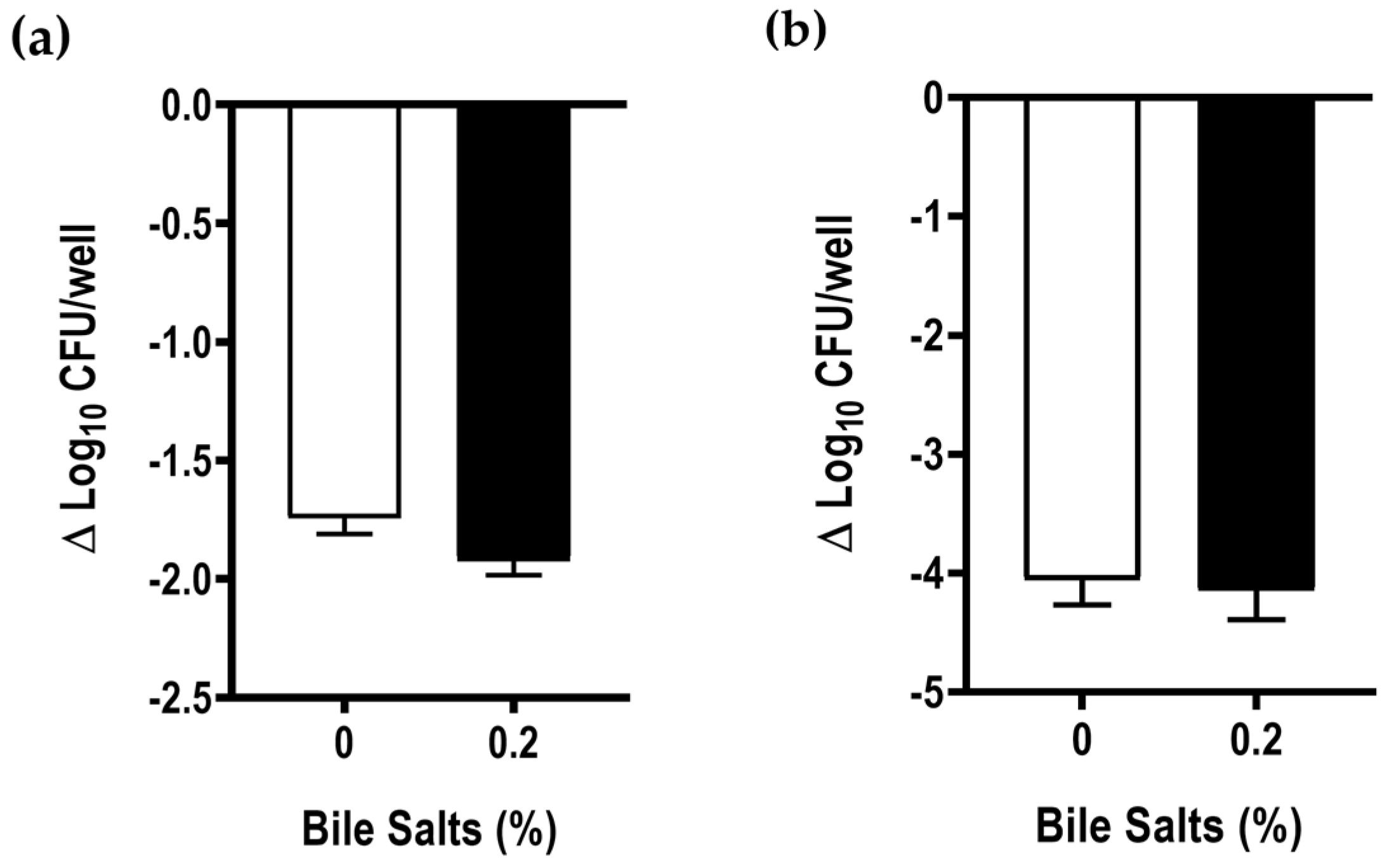
2.4. Effect of Bile Salts in the Adhesion and Invasion Abilities of Aliarcobacter butzleri to Caco-2 Cells
3. Discussion
4. Materials and Methods
4.1. Effect of Bile Salts on the Growth, Aggregation and Motility of Aliarcobacter butzleri
4.2. Growth Curves
4.3. Hydrophobicity Test
4.4. Motility Assay
4.5. Biofilm Formation and Quantification Assay
4.6. Confocal Laser-Scanning Microscopy Imaging of Biofilms
4.7. Relative Expression of Several Virulence Genes
4.8. Adhesion and Invasion Assays to Caco-2 Intestinal Epithelial Cells
4.9. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pérez-Cataluña, A.; Salas-Massó, N.; Diéguez, A.L.; Balboa, S.; Lema, A.; Romalde, J.L.; Figueras, M.J. Revisiting the taxonomy of the Genus Arcobacter: Getting order from the chaos. Front. Microbiol. 2018, 9, 2077. [Google Scholar] [CrossRef]
- Oren, A.; Garrity, G.M. List of new names and new combinations previously effectively, but not validly, published. Int. J. Syst. Evol. Microbiol. 2020, 70, 2960–2966. [Google Scholar] [CrossRef] [PubMed]
- Oren, A.; Garrity, G.M. List of new names and new combinations previously effectively, but not validly, published. Int. J. Syst. Evol. Microbiol. 2020, 70, 1–5. [Google Scholar] [CrossRef]
- Collado, L.; Figueras, M.J. Taxonomy, epidemiology, and clinical relevance of the Genus Arcobacter. Clin. Microbiol. Rev. 2011, 24, 174–192. [Google Scholar] [CrossRef]
- Figueras, M.J.; Levican, A.; Pujol, I.; Ballester, F.; Quilez, M.J.R.; Gomez-Bertomeu, F. A severe case of persistent diarrhoea associated with Arcobacter cryaerophilus but attributed to Campylobacter ap. and a review of the clinical incidence of Arcobacter spp. New Microbes New Infect. 2014, 2, 31–37. [Google Scholar] [CrossRef]
- Van den Abeele, A.M.; Vogelaers, D.; Van Hende, J.; Houf, K.; Van Den Abeele, A.; Vogelaers, D.; Van Hende, J.; Houf, K. Prevalence of Arcobacter species among humans, Belgium, 2008–2013. Emerg. Infect. Dis. 2014, 20, 1746–1749. [Google Scholar] [CrossRef]
- Ferreira, S.; Queiroz, J.A.; Oleastro, M.; Domingues, F.C. Insights in the pathogenesis and resistance of Arcobacter: A review. Crit. Rev. Microbiol. 2016, 42, 364–383. [Google Scholar] [CrossRef]
- Kerkhof, P.J.; Van den Abeele, A.M.; Strubbe, B.; Vogelaers, D.; Vandamme, P.; Houf, K. Diagnostic approach for detection and identification of emerging enteric pathogens revisited: The (Ali)Arcobacter lanthieri case. New Microbes New Infect. 2021, 39, 100829. [Google Scholar] [CrossRef]
- Chieffi, D.; Fanelli, F.; Fusco, V. Arcobacter Butzleri: Up-to-date taxonomy, ecology, and pathogenicity of an emerging pathogen. Compr. Rev. Food Sci. Food Saf. 2020, 19, 2071–2109. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, S.; Oleastro, M.; Domingues, F. Current insights on Arcobacter butzleri in food chain. Curr. Opin. Food Sci. 2019, 26, 9–17. [Google Scholar] [CrossRef]
- Collado, L.; Gutiérrez, M.; González, M.; Fernández, H. Assessment of the prevalence and diversity of emergent Campylobacteria in human stool samples using a combination of traditional and molecular methods. Diagn. Microbiol. Infect. Dis. 2013, 75, 434–436. [Google Scholar] [CrossRef]
- Ferreira, S.; Júlio, C.; Queiroz, J.A.; Domingues, F.C.; Oleastro, M. Molecular diagnosis of Arcobacter and Campylobacter in diarrhoeal samples among portuguese patients. Diagn. Microbiol. Infect. Dis. 2014, 78, 220–225. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, S.; Queiroz, J.A.; Oleastro, M.; Domingues, F.C. Genotypic and phenotypic features of Arcobacter butzleri pathogenicity. Microb. Pathog. 2014, 76, 19–25. [Google Scholar] [CrossRef]
- Bücker, R.; Troeger, H.; Kleer, J.; Fromm, M.; Schulzke, J.D. Arcobacter butzleri induces barrier dysfunction in intestinal HT-29/B6 cells. J. Infect. Dis. 2009, 200, 756–764. [Google Scholar] [CrossRef]
- Ducarmon, Q.R.; Zwittink, R.D.; Hornung, B.V.H.; van Schaik, W.; Young, V.B.; Kuijper, E.J. Gut microbiota and colonization resistance against bacterial enteric infection. Microbiol. Mol. Biol. Rev. 2019, 5, e00007-19. [Google Scholar] [CrossRef] [PubMed]
- Begley, M.; Gahan, C.G.M.; Hill, C. The interaction between bacteria and bile. FEMS Microbiol. Rev. 2005, 29, 625–651. [Google Scholar] [CrossRef]
- Flint, A.; Butcher, J.; Stintzi, A. Stress responses, adaptation, and virulence of bacterial pathogens during host gastrointestinal colonization. Microbiol. Spectr. 2016, 4, VMBF-0007-2015. [Google Scholar] [CrossRef] [PubMed]
- Joyce, S.A.; Gahan, C.G.M. Bile Acid modifications at the microbe-host interface: Potential for nutraceutical and pharmaceutical interventions in host health. Annu. Rev. Food Sci. Technol. 2016, 7, 313–333. [Google Scholar] [CrossRef] [PubMed]
- Long, S.L.; Gahan, C.G.M.; Joyce, S.A. Interactions between gut bacteria and bile in health and disease. Mol. Asp. Med. 2017, 56, 54–65. [Google Scholar] [CrossRef]
- Merritt, M.E.; Donaldson, J.R. Effect of bile salts on the DNA and membrane integrity of enteric bacteria. J. Med. Microbiol. 2009, 58, 1533–1541. [Google Scholar] [CrossRef]
- Reshetnyak, V.I. Physiological and molecular biochemical mechanisms of bile formation. World J. Gastroenterol. 2013, 19, 7341–7360. [Google Scholar] [CrossRef]
- Chiang, J.Y.L. Bile Acid metabolism and signaling. Compr. Physiol. 2013, 3, 1191–1212. [Google Scholar] [CrossRef] [PubMed]
- Sistrunk, J.R.; Nickerson, K.P.; Chanin, R.B.; Rasko, D.A.; Faherty, C.S. Survival of the fittest: How bacterial pathogens utilize bile to enhance infection. Clin. Microbiol. Rev. 2016, 29, 819–836. [Google Scholar] [CrossRef] [PubMed]
- Urdaneta, V.; Casadesús, J. Interactions between bacteria and bile salts in the gastrointestinal and hepatobiliary tracts. Front. Med. 2017, 4, 163. [Google Scholar] [CrossRef] [PubMed]
- Malik-Kale, P.; Parker, C.T.; Konkel, M.E. Culture of Campylobacter jejuni with sodium deoxycholate induces virulence gene expression. J. Bacteriol. 2008, 190, 2286–2297. [Google Scholar] [CrossRef]
- Fitzgerald, C.; Nachamkin, I. Campylobacter and Arcobacter. In Manual of Clinical Microbiology, 10th ed.; Versalovic, J., Carroll, K.C., Funke, G., Jorgensen, J.H., Landry, M.L., Warnock, D.W., Eds.; ASM Press: Washington, DC, USA, 2011; Volume 1, pp. 885–899. [Google Scholar] [CrossRef]
- Nickerson, K.P.; Chanin, R.B.; Sistrunk, J.R.; Rasko, D.A.; Fink, P.J.; Barry, E.M.; Nataro, J.P.; Faherty, C.S. Analysis of Shigella flexneri resistance, biofilm formation, and transcriptional profile in response to bile salts. Infect. Immun. 2017, 85, e01067-16. [Google Scholar] [CrossRef] [PubMed]
- Joffre, E.; Nicklasson, M.; Álvarez-Carretero, S.; Xiao, X.; Sun, L.; Nookaew, I.; Zhu, B.; Sjöling, Å. The Bile Salt Glycocholate induces global changes in gene and protein expression and activates virulence in enterotoxigenic Escherichia coli. Sci. Rep. 2019, 9, 108. [Google Scholar] [CrossRef]
- Rivera-Amill, V.; Kim, B.J.; Seshu, J.; Konkel, M.E. Secretion of the virulence-associated Campylobacter invasion antigens from Campylobacter jejuni requires a stimulatory signal. J. Infect. Dis. 2001, 183, 1607–1616. [Google Scholar] [CrossRef] [PubMed]
- Pumbwe, L.; Skilbeck, C.A.; Nakano, V.; Avila-Campos, M.J.; Piazza, R.M.F.; Wexler, H.M. Bile Salts enhance bacterial co-aggregation, bacterial-intestinal epithelial cell adhesion, biofilm formation and antimicrobial resistance of Bacteroides fragilis. Microb. Pathog. 2007, 43, 78–87. [Google Scholar] [CrossRef]
- Pace, J.L.; Chai, T.J.; Rossi, H.A.; Jiang, X. Effect of bile on Vibrio parahaemolyticus. Appl. Environ. Microbiol. 1997, 63, 2372–2377. [Google Scholar] [CrossRef] [PubMed]
- Prouty, A.M.; Gunn, J.S. Salmonella enterica serovar Typhimurium invasion is repressed in the presence of bile. Infect. Immun. 2000, 68, 6763–6769. [Google Scholar] [CrossRef] [PubMed]
- De Jesus, M.C.; Urban, A.A.; Marasigan, M.E.; Foster, D.E.B. Acid and Bile-Salt stress of enteropathogenic Escherichia coli enhances adhesion to epithelial cells and alters glycolipid receptor binding specificity. J. Infect. Dis. 2005, 192, 1430–1440. [Google Scholar] [CrossRef]
- Vandamme, P.; Vancanneyt, M.; Pot, B.; Mels, L.; Hoste, B.; Dewettinck, D.; Vlaes, L.; Van den Borre, C.; Higgins, R.; Hommez, J.; et al. Polyphasic taxonomic study of the emended Genus Arcobacter with Arcobacter butzleri comb. nov. and Arcobacter skirrowii sp. nov., an aerotolerant bacterium isolated from veterinary specimens. Int. J. Syst. Bacteriol. 1992, 42, 344–356. [Google Scholar] [CrossRef]
- Poole, K. Bacterial stress responses as determinants of antimicrobial resistance. J. Antimicrob. Chemother. 2012, 67, 2069–2089. [Google Scholar] [CrossRef]
- Kim, S.-H.; Chelliah, R.; Ramakrishnan, S.R.; Perumal, A.S.; Bang, W.S.; Rubab, M.; Daliri, E.B.M.; Barathikannan, K.; Elahi, F.; Park, E.; et al. Review on stress tolerance in Campylobacter jejuni. Front. Cell. Infect. Microbiol. 2021, 10, 1596570. [Google Scholar] [CrossRef] [PubMed]
- Fox, E.M.; Raftery, M.; Goodchild, A.; Mendz, G.L. Campylobacter jejuni response to ox-bile stress. FEMS Immunol. Med. Microbiol. 2007, 49, 165–172. [Google Scholar] [CrossRef]
- Lin, J.; Overbye Michel, L.; Zhang, Q. CmeABC functions as a multidrug efflux system in Campylobacter jejuni. Antimicrob. Agents Chemother. 2002, 46, 2124–2131. [Google Scholar] [CrossRef] [PubMed]
- Ma, D.; Cook, D.N.; Alberti, M.; Pon, N.G.; Nikaido, H.; Hearst, J.E. Genes acrA and acrB encode a stress-induced efflux system of Escherichia coli. Mol. Microbiol. 1995, 16, 45–55. [Google Scholar] [CrossRef] [PubMed]
- Buckley, A.M.; Webber, M.A.; Cooles, S.; Randall, L.P.; La Ragione, R.M.; Woodward, M.J.; Piddock, L.J.V. The AcrAB-TolC Efflux system of Salmonella enterica serovar Typhimurium plays a role in pathogenesis. Cell. Microbiol. 2006, 8, 847–856. [Google Scholar] [CrossRef]
- Ferreira, S.; Silva, A.L.; Tomás, J.; Mateus, C.; Domingues, F.; Oleastro, M. Characterization of AreABC, a RND-type efflux system involved in antimicrobial resistance of Aliarcobacter butzleri. Antimicrob. Agents Chemother. 2021, 65, e00729-21. [Google Scholar] [CrossRef]
- Mateus, C.; Nunes, A.R.; Oleastro, M.; Domingues, F.; Ferreira, S. RND Efflux systems contribute to resistance and virulence of Aliarcobacter butzleri. Antibiotics 2021, 10, 823. [Google Scholar] [CrossRef] [PubMed]
- Guttenplan, S.B.; Kearns, D.B. Regulation of flagellar motility during biofilm formation. Bone 2006, 23, 849–871. [Google Scholar] [CrossRef] [PubMed]
- Kristoffersen, S.M.; Ravnum, S.; Tourasse, N.J.; Økstad, O.A.; Kolstø, A.B.; Davies, W. Low concentrations of bile salts induce stress responses and reduce motility in Bacillus cereus ATCC 14570. J. Bacteriol. 2007, 189, 5302–5313. [Google Scholar] [CrossRef]
- Prouty, A.M.; Brodsky, I.E.; Manos, J.; Belas, R.; Falkow, S.; Gunn, J.S. Transcriptional regulation of Salmonella enterica serovar Typhimurium genes by bile. FEMS Immunol. Med. Microbiol. 2004, 41, 177–185. [Google Scholar] [CrossRef] [PubMed]
- Allen, K.J.; Griffiths, M.W. Effect of environmental and chemotactic stimuli on the activity of the Campylobacter jejuni FlaA Σ28 promoter. FEMS Microbiol. Lett. 2001, 205, 43–48. [Google Scholar] [CrossRef] [PubMed]
- Svensson, S.L.; Pryjma, M.; Gaynor, E.C. Flagella-mediated adhesion and extracellular DNA release contribute to biofilm formation and stress tolerance of Campylobacter jejuni. PLoS ONE 2014, 9, e106063. [Google Scholar] [CrossRef]
- Hung, D.T.; Zhu, J.; Sturtevant, D.; Mekalanos, J.J. Bile Acids stimulate biofilm formation in Vibrio cholerae. Mol. Microbiol. 2006, 59, 193–201. [Google Scholar] [CrossRef] [PubMed]
- Crawford, R.W.; Gibson, D.L.; Kay, W.W.; Gunn, J.S. Identification of a bile-induced exopolysaccharide required for Salmonella biofilm formation on gallstone surfaces. Infect. Immun. 2008, 76, 5341–5349. [Google Scholar] [CrossRef]
- Branda, S.S.; Vik, Å.; Friedman, L.; Kolter, R. Biofilms: The matrix revisited. Trends Microbiol. 2005, 13, 20–26. [Google Scholar] [CrossRef] [PubMed]
- Reuter, M.; Ultee, E.; Toseafa, Y.; Tan, A.; Arnoud, H.; Vliet, M. Van Inactivation of the core CheVAWY chemotaxis genes disrupts chemotactic motility and organised biofilm formation in Campylobacter jejuni. FEMS Microbiol. Lett. 2020, 367, fnaa198. [Google Scholar] [CrossRef] [PubMed]
- Plummer, P.J. LuxS and Quorum-Sensing in Campylobacter. Front. Cell. Infect. Microbiol. 2012, 2, 22. [Google Scholar] [CrossRef] [PubMed]
- Tsai, M.H.; Liang, Y.H.; Chen, C.L.; Chiu, C.H. Characterization of Salmonella resistance to bile during biofilm formation. J. Microbiol. Immunol. Infect. 2020, 53, 518–524. [Google Scholar] [CrossRef] [PubMed]
- Sela, S.; Frank, S.; Belausov, E.; Pinto, R. A mutation in the luxS gene influences Listeria monocytogenes biofilm formation. Appl. Environ. Microbiol. 2006, 72, 5653–5658. [Google Scholar] [CrossRef]
- Reeser, R.J.; Medler, R.T.; Billington, S.J.; Jost, B.H.; Joens, L.A. Characterization of Campylobacter jejuni biofilms under defined growth conditions. Appl. Environ. Microbiol. 2007, 73, 1908–1913. [Google Scholar] [CrossRef]
- Gunn, J.S. Mechanisms of bacterial resistance and response to bile. Microbes Infect. 2000, 2, 907–913. [Google Scholar] [CrossRef] [PubMed]
- Doig, P.; Yao, R.; Burr, D.H.; Guerry, P.; Trust, T.J. An environmentally regulated pilus-like appendage involved in Campylobacter pathogenesis. Mol. Microbiol. 1996, 20, 885–894. [Google Scholar] [CrossRef]
- Gupta, S.; Chowdhury, R. Bile affects production of virulence factors and motility of Vibrio cholerae. Infect. Immun. 1997, 65, 1131–1134. [Google Scholar] [CrossRef] [PubMed]
- Tan, B.M.; Tu, Q.V.; Kovach, Z.; Raftery, M.; Mendz, G.L. Wolinella succinogenes response to ox-bile stress. Antonie Van Leeuwenhoek 2007, 92, 319–330. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, S.; Fraqueza, M.J.; Queiroz, J.A.; Domingues, F.C.; Oleastro, M. Genetic diversity, antibiotic resistance and biofilm-forming ability of Arcobacter butzleri isolated from poultry and environment from a Portuguese slaughterhouse. Int. J. Food Microbiol. 2013, 162, 82–88. [Google Scholar] [CrossRef]
- Misawa, N.; Blaser, M.J. Detection and characterization of autoagglutination activity by Campylobacter jejuni. Infect. Immun. 2000, 68, 6168–6175. [Google Scholar] [CrossRef]
- Martins, R.; Mateus, C.; Domingues, F.; Bücker, R.; Oleastro, M.; Ferreira, S. Effect of atmospheric conditions on pathogenic phenotypes of Arcobacter butzleri. Microorganisms 2022, 10, 2409. [Google Scholar] [CrossRef]
- Wang, F.; Deng, L.; Huang, F.; Wang, Z.; Lu, Q.; Xu, C. Flagellar motility is critical for Salmonella enterica serovar Typhimurium biofilm development. Front. Microbiol. 2020, 11, 1695. [Google Scholar] [CrossRef]
- Heydorn, A.; Nielsen, A.T.; Hentzer, M.; Sternberg, C.; Givskov, M.; Ersboll, B.K.; Molin, S. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 2000, 146, 2395–2407. [Google Scholar] [CrossRef]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, 16–21. [Google Scholar] [CrossRef] [PubMed]
- Medina, G.; Neves, P.; Flores-Martin, S.; Manosalva, C.; Andaur, M.; Otth, C.; Lincopan, N.; Fernández, H. Transcriptional analysis of flagellar and putative virulence genes of Arcobacter butzleri as an endocytobiont of Acanthamoeba castellanii. Arch. Microbiol. 2019, 201, 1075–1083. [Google Scholar] [CrossRef] [PubMed]
- Muyzer, G.; De Waal, E.C.; Uitterlinden, A.G. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 1993, 59, 695–700. [Google Scholar] [CrossRef] [PubMed]




| Primers | Target Gene | Sequence | Reference |
|---|---|---|---|
| cadF_F | cadF | 5′-CTCCAGTTGCTGCACCAAAA-3′ | This study |
| cadF_R | 5′-CCAATATTGTCAACTTTTGCACC-3′ | ||
| ciaB_F | ciaB | 5′-TTGGCAAACTTCATGGACTGC-3′ | This study |
| ciaB_R | 5′-AGCAGTAATTCCTCCATGTCCT-3′ | ||
| cheV_F | cheV | 5′-TGAAGAGGTGGCGATAAATG-3′ | This study |
| cheV_R | 5′-GCTTTTAATCCAAGCCAAGC-3′ | ||
| flaA_F | flaA | 5′-AGTTGCACCAGCTGACATTT-3′ | [66] |
| flaA_R | 5′-AGTTGGTGAAGGAAGTTCCGA-3′ | ||
| luxS_F | luxS | 5′-GAGCACCTTTTTGCTGGATT-3′ | This study |
| luxS_R | 5′-TTCCAAGCAACTGCAACTTC-3′ | ||
| P338_F | 16S rRNA | 5′-ACTCCTACGGGAGGCAGCAG-3′ | [67] |
| P518_R | 5′-ATTACCGCGGCTGCTGG-3′ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mateus, C.; Maia, C.J.; Domingues, F.; Bücker, R.; Oleastro, M.; Ferreira, S. Evaluation of Bile Salts on the Survival and Modulation of Virulence of Aliarcobacter butzleri. Antibiotics 2023, 12, 1387. https://doi.org/10.3390/antibiotics12091387
Mateus C, Maia CJ, Domingues F, Bücker R, Oleastro M, Ferreira S. Evaluation of Bile Salts on the Survival and Modulation of Virulence of Aliarcobacter butzleri. Antibiotics. 2023; 12(9):1387. https://doi.org/10.3390/antibiotics12091387
Chicago/Turabian StyleMateus, Cristiana, Cláudio J. Maia, Fernanda Domingues, Roland Bücker, Mónica Oleastro, and Susana Ferreira. 2023. "Evaluation of Bile Salts on the Survival and Modulation of Virulence of Aliarcobacter butzleri" Antibiotics 12, no. 9: 1387. https://doi.org/10.3390/antibiotics12091387







